| |
| |
CDTree is...
- a powerful tool to aid in the classification of protein sequences and investigate their evolutionary relationships
- a web-based helper application used by the CDD on-line search service to permit user interaction with pre-defined protein domain hierarchies
- an integrated software environment organized to help users assimilate large amounts of biological data from various resources by access to a suite of analysis methods
- an alignment editor and 3D structure visualization program through its integration with Cn3D 4.3
 Download CDTree 3.1 for the PC or Mac
Download CDTree 3.1 for the PC or Mac
|
|
 |
| |
|
|
 |
| |
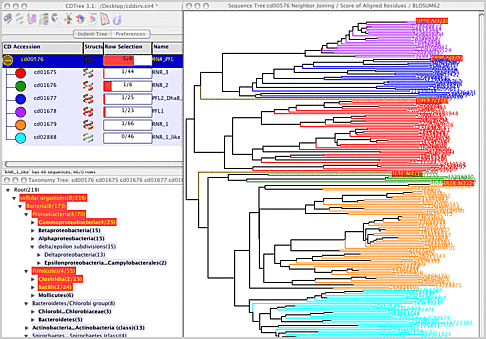
The screenshot above results from launching CDTree with the 'Interactive Display' buttons on the CDD page for the RNR_PFL domain hierarchy.
Ribonucleotide reductase (RNR) and pyruvate formate lyase (PFL) are structurally similar and are thought to have diverged from a common ancestral domain.
CDTree has been designed to discover and explore such relationships in detail, and is a central tool used to craft all of the curated domain hierarchies in the CDD.
|
|
|
|
|
| |
| |
CDTree features include ...
- PSI-BLAST interface for recruiting new sequences into an existing model
- Use BLAST to seed a domain model from a single sequence
- Phylogenetic tree analysis
- Taxonomy interface
- Domain architecture interface
- Cross-hit analysis between multiple protein domain models
- Cn3D 4.3 for multiple alignment editing and 3D structure visualization. See the Cn3D home page for details about the program's features and functions as well as a tutorial
- Eliminate redundancy in a domain model, for example, based on sequence similarity and/or taxonomic representation
- Build a hierarchy of related protein domains whose multiple alignments are 'self-consistent' (i.e., reflect the common evolutionary relationship modeled by the domain hierarchy)
- Selection mechanism allows for identification of sequences across multiple viewers
- In-Program Help documentation
Features added in CDTree 3.1
- Mac OSX supported CDTree and Cn3D have been ported to Macintosh computers running OSX 10.4 and higher
- Annotation matrix Viewer conveniently record and review annotations and literature references for all domains in a hierarchy
- Multi-CD operations new select and edit operations that can act across multiple domain models
- Sequence tree coloring greater flexibilty in visualizing evolutionary relationships between domains
- Speed improvements optimized computations to improve response time in large domains and hierarchies
|
|
|
|
|
|
| |
|
| |
 |
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015 Jan 28;43(Database issue):D222-2. doi: 10.1093/nar/gku1221. Epub 2014 Nov 20. [PubMed PMID: 25414356] [Full Text] |
| |
|
| |
|
| |
 |
Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007 Jan;35(Database issue):D237-40. Epub 2006 Nov 29. [PubMed PMID: 17135202] [Full Text] |
| |
|
| |
|
(See all publications about NCBI's Conserved Domains Resources, including those listed above plus articles by the NCBI Structure group describing the results of computational biology research, such as comparison of sequence and 3D structure alignments for protein domains, annotation of functional sites on proteins, and more.) |
| |
|
|
|
|
|
|