Related Resources
NCBI Genome Assembly Model
Why we need a model
Fig. 1: An ideogram representation of the human genome.
The genome of an organism consists of a set of chromosomes. While bacteria can have a single chromosome, often accompanied by extra-chromosomal plasmids, eukaryotes often have multiple chromosomes, with each chromosome being represented greater than 1 time. While many eukaryotes tend to exist in a diploid state (2 copies of each autosome and 2 sex chromosomes) other organisms, such as plants, can have many copies (e.g. tetraploid or hexaploid). However, most representations of an organism's genome tend to only represent the haploid state, as shown in figure 1.
However, current sequencing technology does not allow for the complete sequencing of a single chromosome. To accomplish genome sequencing, the genome is fragmented and small pieces are sequenced many times. These sequences are then assembled to try to recreate the chromosome sequences (see Assembly Basics for more information). Because of the complexities found in many organisms, it is currently not possible to obtain a complete chromosome sequence. In fact, the output of most assembly algorithms is a set of contigs and scaffolds that are then ordered and oriented using external data (such as mapping information). Typically, not all sequences can be can be ordered and oriented. Developing a robust assembly model allows us to tie together the output of assembly algorithms with the biological model that has developed over years of genomic research.
The assembly model
When genome sequencing initially started it was thought that the genome assembly could be represented by a single 'Golden Path'. That is, a single set of overlapping sequences could be selected to produce a non-redundant chromosome sequence (with gaps) that would fully represent the sequence at all loci. It was thought that the predominant form of variation was single nucleotide polymorphism (SNP) and these polymorphisms would be represented as annotation on the chromosome sequence. Subsequent genome analysis has shown that this model will not work for some parts of the genome. Large-scale structural variations, often in the form of Copy Number Variation (CNVs) are more prevalent than originally thought (see dbVar for more information). If a genome contains regions with complex allelic diversity, it may be necessary to produce more than 1 sequence path to fully represent that region. For example, the current public human reference assembly (GRCh38) has 8 different paths through the MHC region, a region known to have a high degree of allelic complexity. To accommodate this increased complexity, we have developed a more robust data model. Terms used in this model are defined below and a graphical representation is shown in figure 2.
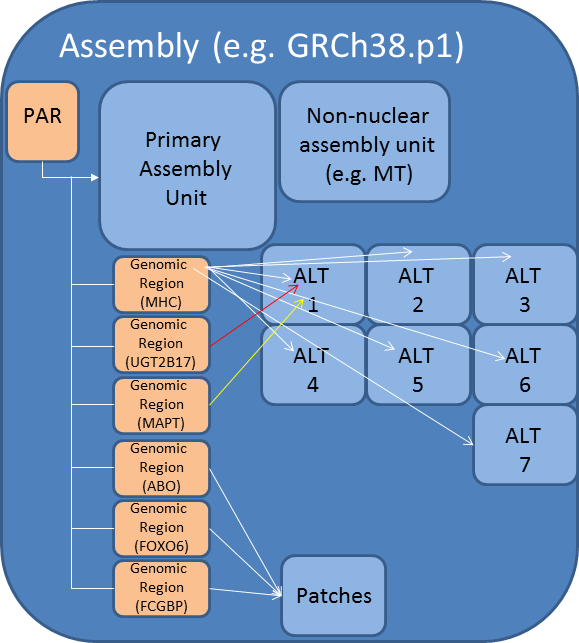
Fig 2. A graphical representation of the NCBI assembly model. The assembly model has been extended to formally deal with sequences that don't fit neatly into chromosome space but that are still critical pieces of data. Genomic regions are defined on the Primary Assembly. These regions point to scaffolds contained within the alternate locus units. Note: not all alternate locus scaffolds will be associated with a region.
Definitions
Assembly Types
- Assembly:
- The set of chromosomes, unlocalized and unplaced (sometimes called 'random') and alternate sequences used to represent an organism's genome. Assemblies are constructed from 1 or more assembly units.
- Haploid Assembly:
- The collection of chromosome assemblies, unlocalized and unplaced sequences that represent an organism's genome. Any locus may be represented 0 or 1 time, and entire chromosomes are only represented 0 or 1 times.
- Haploid-with-alt-loci:
- The collection of chromosome assemblies, unlocalized and unplaced sequences and alternate loci that represent an organism's genome. Any locus may be represented 0, 1 or >1 time, but entire chromosomes are only represented 0 or 1 times.
- Diploid Assembly:
- A genome assembly for which a chromosome assembly is available for both sets of an individual's chromosomes. It is anticipated that a diploid genome assembly represents the genome of an individual, therefore, it is not expected that alternate loci will be defined for this assembly, although it is possible that unlocalized or unplaced sequences could be part of the assembly.
- Unresolved Diploid:
- A genome assembly from a diploid in which many of the haplotypic sequences have been resolved but the two haplotypes have not been separated. Consequently, the assembly will be much larger than the expected haploid genome size and many genes will be present in two copies.
- Linked Pseudohaplotype Assemblies:
- A genome assembly from a diploid in which many of the haplotypic sequences have been resolved, phased and the two haplotypes have been separated. The current state of the technology is that most assemblers produce blocks that are phased, separated by blocks where the haplotype cannot be distinguished. The typical result is that the "Principal Pseudohaplotype" assembly is a mosaic of haplotype blocks linked by unresolved segments and the "Alternate Pseudohaplotype" assembly is the other haplotype wherever the haplotypes can be distinguished. A pair of pseudohaplotype assemblies derived from the same diploid individual can be linked with a cross-reference.
Assembly Parts
- Assembly Unit:
- The collection of sequences used to define discrete parts of an assembly.
- Alternate Locus:
- A sequence that provides an alternate representation of a locus found in the haploid assembly ("Primary Assembly").
- Alternate Locus Group:
- A set of alternate loci that have been grouped together for annotation purposes. This may be because they are from the same haplotype or strain, or it may be for convenience in annotation.
- Genome Patches:
- Sequence updates that are released outside of the major assembly cycle. These are instantiated as independent scaffolds that are aligned to the primary assembly to provide chromosome context. There are two types of patches: Fix patch: This patch is made in a region where the Tiling Path File (TPF) will change in the next major assembly update. These scaffolds will be withdrawn at the next major assembly update, the accessions will be made secondary to the chromosome and the sequence will be incorporated into the Primary Assembly TPF. Novel patch: These represent new alternate loci. At the next major assembly update, these sequences will be moved to the appropriate assembly-unit and the accession will remain stable.
- Regions:
- Regions are locations on the primary assembly (typically on the chromosome sequences) for which alternate representations or genome patches exist.
- PAR:
- Pseudo-autosomal region. A region found on the X and Y chromosomes of mammals that allows recombination between the sex chromosomes. In human, the regions are defined on the X chromosome and the sequence from the X chromosome is copied onto the Y, but this is not a requirement for representing the PAR.
All assemblies must contain one assembly unit that represents the "Primary Assembly". The primary assembly unit contains non-redundant sequence (chromosome and/or scaffolds) that represents an organism's haploid genome and contains the sequences most people think of when they think of an assembly. For eukaryotic assemblies, we typically separate the non-nuclear sequences (e.g. MT) into a separate assembly unit as many annotation pipelines wish to treat these sequences differently.
Sequence Types
- Placed sequence:
- Sequence that has been ordered and oriented on the chromosome. The locations of these sequences can usually be expressed in chromosome coordinates.
- Unlocalized sequence:
- A sequence found in an assembly that is associated with a specific chromosome but cannot be ordered or oriented on that chromosome. The location of these sequences cannot be expressed in chromosome coordinates.
- Unplaced sequences:
- A sequence found in an assembly that is not associated with any chromosome. These sequences cannot be expressed in chromosome coordinates.
- Chromosome context:
- A term used to describe alternate loci and patches that have been aligned to the chromosome sequences defined in the Primary Assembly. While these sequences cannot strictly be expressed as chromosome coordinates, they can be related to the chromosome sequence via their alignment to the chromosome.
Tracking assembly data
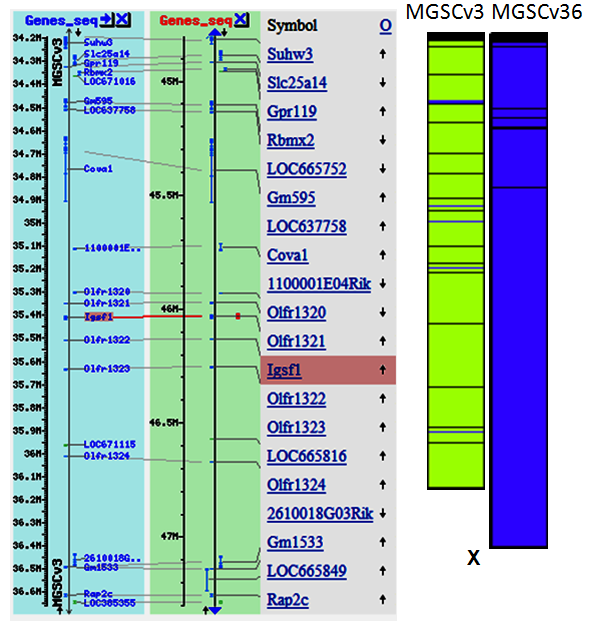
Fig. 3: The left panel shows the gene map for mouse chromosome X from 35M - 36M in MGSCv3 compared to the same region in MGSCv36. The ruluer on the left of each map shows that the coordinates for this region in MGSCv36 have moved approximately 10Mb distal on the chromosome. The graphic to the right shows a complete overview in the sequence lengths of the representations of the X chromosome in MGSCv3 (on the left) and MGSCv36 (on the right). During the improvement of the assembly, approximately 17 Mb of sequence have been added to the chrX assembly. Chromosome coloration: green = Whole Genome Shotgun sequence, blue = Finished sequence.
Typically, people refer to specific regions in an assembly by providing chromosome coordinates, for example chrX:35,000,000-36,000,000. However, this is not a very precise way to refer to a genomic region (as illustrated in figure 3). Assemblies can and do change and a more robust way of providing precise locations is needed. When assemblies are submitted to an INSDC database , all sequences associated with the assembly are assigned a unique accession number (e.g. CM000663). The first instance of this accession is assigned version 1. If this sequence is updated in anyway, the accession will remain the same, but the version will increment. This allows specific versions of any sequence to be uniquely identified. Genomic locations should always be expressed using an accession.version, followed by the chromosome start and stop.
While the INSDC databases are great at tracking revisions of individual sequences, they are not well suited to tracking revisions of collections of sequences, which is what an assembly is. To fulfill this need, we have created an Assembly Database. This database provides a unique and trackable identifier (in the form of an accession.version) for both assemblies and assembly units. In this way, users can unambiguously track changes to an assembly.
The assembly accession starts with a three letter prefix, GCA for GenBank assemblies and GCF for RefSeq assemblies. This is followed by an underscore and 9 digits. A version is then added to the accession. For example, the assembly accession for the GenBank version of the public human reference assembly (GRCh38.p11) is GCA_000001405.26.
There are two different types of assembly updates.
- Major Release: Any update that changes the sequence and/or changes the chromosome coordinate system defined in the primary assembly.
- Minor Release: Updates that don't change the coordinate system, but may add or modify information. Such events include addition of genome patches, assignment of unplaced sequences to a chromosome or new placements for alternate loci.
Accessing data
Genome sequences and annotation can be accessed by following the links to the FTP site from the page for the assembly of interest in the NCBI Assembly resource or by navigating the NCBI genomes FTP site. See the Genomes Download FAQ for more details.
