NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Bradley LA, Palomaki G, Gutman S, et al. PCA3 Testing for the Diagnosis and Management of Prostate Cancer [Internet]. Rockville (MD): Agency for Healthcare Research and Quality (US); 2013 Apr. (Comparative Effectiveness Reviews, No. 98.)
This publication is provided for historical reference only and the information may be out of date.
Background
Cancer of the prostate is the second most common cancer and the second leading cause of cancer deaths in men in the United States.1,2 Most patients have slow-growing tumors, and may live for years with no or minimal effects, ultimately dying of other causes.3,4 The lifetime risk of being diagnosed with prostate cancer is 16 percent, but lifetime risk of dying from the disease is only 3 percent.1,3,5 However, some patients have aggressive tumors that spread beyond the prostate, resulting in significant morbidity and death. A challenge in managing clinically localized disease is distinguishing between men who have aggressive disease and need immediate therapy, and those who have less aggressive disease that can be safely managed by active surveillance.
Production of serum total prostate specific antigen (tPSA) was found to be increased in men with prostate cancer as many as 5 to 10 years prior to symptoms of clinical disease.6 The rationale for initiating prostate cancer screening using tPSA was to reduce the prevalence of advanced prostate cancer and prostate cancer-related mortality through early detection, and improve quality of life.3,7,8 Prostate cancer mortality has decreased,1,2 but at what cost in overdiagnosis and potential harms related to treatment?11 Also, issues such as who to test, when to test and retest, and the most effective clinical tPSA threshold continue to be debated. A recent U.S. Preventive Services Task Force recommendation concluded that the potential benefits do not outweigh the harms.9 However, the balance of benefits and harms of tPSA screening remains controversial.9,10
In 1999, researchers reported that the prostate cancer antigen 3 gene (PCA3; also known as DD3), was highly overexpressed in prostate cancer relative to normal prostate or benign prostatic hyperplasia tissue.12 Subsequently, PCA3 tests on messenger RNA from urine were developed.13,14 Two proposed intended uses of PCA3 and comparator tests were to inform decisionmaking about initial or repeat biopsy of men with elevated tPSA and/or other risk factors. The third was to inform decisions about management and treatment (e.g., active surveillance, prostatectomy, radiotherapy) by classifying disease in men with positive biopsies as insignificant or aggressive.
The U.S. Food and Drug Administration (FDA) recently approved a PCA3 assay for use in men 50 years of age or older who have had one or more previous negative biopsies, but did not have a finding of atypical small acinar proliferation in the most recent biopsy. The intended use of the test is to inform decisionmaking about repeat biopsy.
Scope and Key Questions
Biomarker comparators for detection of prostate cancer at biopsy considered in this review are tPSA, specific isoforms of tPSA, and validated risk-assessment calculators or nomograms.
- Serum tPSA is widely available as a screening and monitoring test using set (e.g., 2.5 or 4 ng/mL) or age-specific cutoffs.15
- One tPSA isoform is free PSA, reported as a ratio of free to total PSA or percent free PSA (%fPSA). Low levels (less than 25 percent) are associated with cancer and high levels with benign disease. Percent fPSA may be useful in decisionmaking about biopsy, particularly for men whose tPSA levels are in the “grey zone” (2.5 to 10 ng/mL).
- A second isoform is PSA bound to serum antiproteases, or complexed PSA (cPSA). Data are limited but performance may be similar to %fPSA.
- PSA density is the ratio of tPSA concentration to prostate volume. Addition to tPSA may improve the prediction of positive biopsy or insignificant cancer, but this has not been confirmed.
- Externally validated nomograms are risk assessment tools that combine multiple clinical and laboratory risk factors to inform clinical decisionmaking about biopsy, risk classification, and/or treatment options. Despite variability and lack of validation in some cases, such tools may provide better information than use of individual markers.
For risk classification, PCA3 comparators in a prognostic workup include Gleason score, prostate volume, risk factors, biochemical markers, and clinical/pathological staging.
The Key Questions (KQs) relate to the three proposed scenarios described above:
KQ1: In patients with elevated PSA and/or an abnormal digital rectal examination who are candidates for initial prostate biopsy, what is the comparative effectiveness of PCA3 testing as a replacement for, or supplement to, standard tests, including diagnostic accuracy (clinical validity) for prostate cancer, intermediate outcomes (e.g., improved decision making about biopsy), and long-term health outcomes (clinical utility), including mortality/morbidity, quality of life, and potential harms?
KQ 2: In patients with elevated PSA and/or an abnormal digital rectal examination who are candidates for repeat prostate biopsy, what is the comparative effectiveness of PCA3 testing as a replacement for, or supplement to, standard tests, including diagnostic accuracy (clinical validity) for prostate cancer, intermediate outcomes (e.g., improved decisionmaking about biopsy), and long-term health outcomes (clinical utility), including mortality/morbidity, quality of life, and potential harms?
KQ 3: In patients with a positive biopsy for prostate cancer who are being evaluated to distinguish between insignificant/indolent and aggressive disease, what is the effectiveness of using PCA3 testing alone, or in combination with the standard prognostic workup (e.g., tumor volume, Gleason score, clinical staging) or monitoring tests (e.g., PSA, PSA velocity), with regard to diagnostic accuracy (clinical validity) for aggressive (high-risk) prostate cancer, intermediate outcomes (e.g., improved decisionmaking about prognosis and triage for active surveillance and/or aggressive treatment), and long-term health outcomes (clinical utility), including mortality/morbidity, quality of life, and potential harms?
Corresponding analytic frameworks are presented in the full report.
Methods
Literature Search Strategy
We searched PubMed®, Embase®, and the Cochrane Central Register of Controlled Trials for the timeframe January 1, 1990 to August 15, 2011; updated searches were performed for the timeframe ending May 15, 2012. The grey literature searches included regulatory information, clinical trial registries, conference papers, and selected Web sites. We included studies that were in English, reported primary data, addressed KQs, and fulfilled the criteria for: (1) study design (matched studies in the same clinical setting in which PCA3 and comparators were assessed in all men in a study population); (2) study subjects/populations (at-risk men or men with a positive biopsy); (3) study interventions (biomarker testing, biopsy, risk classification); (4) study comparators; and (5) intermediate (diagnostic accuracy, impact on decisionmaking, harms) and long-term (e.g., mortality, morbidity, function, quality of life, harms) outcomes.
For title/abstract and full-article review, one reviewer read and determined eligibility and a second reviewer audited a subset of abstracts (and all marked uncertain) and all full articles; discrepancies were resolved by discussion or a third reviewer when needed. Data were extracted by a single reviewer, and then fully audited by a second senior reviewer. Disagreements were resolved through review team discussion.
Quality Assessment of Individual Studies and Strength of Evidence
In adherence with the Methods Guide,19 grading the methodological quality of individual comparative studies was performed based on study design-specific criteria. The quality of diagnostic accuracy studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) tool.20 The QUADAS ratings were summarized into general quality classes of good, fair, and poor.19 The strength of evidence for outcomes was evaluated using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) system.21 GRADE addresses four domains of evidence (risk of bias, consistency, directness, and precision), and rates each body of evidence as High, Moderate, Low, or Insufficient.19,21 In all cases, two independent reviewers assessed the quality of individual studies and the strength of evidence. Discordant decisions were resolved through discussion or third-party adjudication.
Data Synthesis
For KQ 1 and KQ 2, PCA3 scores were evaluated against all comparators for which published data were available. Analyses included clinical sensitivity, clinical specificity (or the false positive rate equal to 1-specificity), and positive and negative predictive values. When data were available, the following analyses were performed for PCA3 and one or more comparators:
- Differences in area under the receiver operating characteristic (ROC) curve (AUC)a, including direction and magnitude of differences.
- Reported medians and standard deviations in positive and negative biopsy populations (reported as z scores), including direction and strength of effect.
- Performance at a PCA3 cutoff score of 35 (sensitivity and specificity).
- Receiver operator characteristic (ROC) curves (sensitivity and specificity), to evaluate fixed specificities and compare corresponding sensitivities.
- Regression analysis using regression coefficients and associated relative odds ratios.
Based on the limited number of studies identified that address KQ 3, we anticipated focusing on a qualitative analysis (e.g., descriptive narrative, summary tables, identification of themes in content).
Applicability
Applicability of the results presented in this review was assessed in a systematic manner using the PICOTS framework (Population, Intervention, Comparison, Outcome, Timing, Setting).
Results
Detailed description of analyses with tables and figures are included in the full report.
Results of Literature Searches
Our inclusion criteria restricted the analyses to matched studies that provide data on PCA3 and at least one other comparator in the same patient population. Population matching was preserved by computing differences between PCA3 test results and comparator test results within each study, and comparing these differences across studies. Searches identified 1,556 citations, of which 220 underwent full text review and 42 were included. Grey literature search identified 1 additional study for a total of 43.
Potential Biases in Included Studies
Subjects in the included studies were drawn from academic medical centers where patients with elevated tPSA results and/or other risk factors were seeking referral or specialty care. Observational studies of such opportunistic cohorts are subject to specific biases.
- Verification bias: Men are most often offered prostate biopsy based on the extent of tPSA elevation. Higher tPSA levels indicate higher likelihood of prostate cancer, and men are more likely to undergo prostate biopsy if tPSA is high (e.g., 10-20 ng/mL) rather than closer to cutoffs (e.g., 3-4 ng/mL). Estimates of sensitivity and specificity at select tPSA cutoffs will be impacted in studies in which biopsy decisions are tPSA-related. If those not accepting biopsy are considered missing, this is considered “partial verification” bias.
- Spectrum bias: Convenience samples can also predispose to spectrum effects, as they may represent men at higher risk of prostate cancer than the total cohort of screened men. Of more concern is that the range of severity of disease predicted by PCA3 and comparators could be different, for example, if men positive on one test were found to have different characteristics or severity of disease than those positive on another test.
- Sampling bias: A subset of studies restricted enrollment to tPSA results in the “grey zone” (e.g., 2.5 ng/mL to < 10 ng/mL). The effect was to reduce both the prevalence of disease in the study group and tPSA test performance, as those men with higher tPSA levels (where tPSA is most predictive) are not enrolled in the study. This bias cannot be avoided by statistical analysis, but was addressed by stratifying analyses and summarizing “grey zone” studies separately.
KQ 1. Initial Biopsy
Two matched studies reported results in populations where all men were having initial biopsies.22,23 Both reported comparisons of PCA3 with tPSA and %fPSA; one22 also reported on PSA density. All studies were graded as poor, and strength of evidence was rated insufficient because there were too few data for reliable interpretation. No studies addressed other comparators or outcomes; all other comparisons were graded insufficient.
KQ 2. Repeat Biopsy
Seven matched studies addressed diagnostic accuracy for KQ 2, reporting results in populations where all men were having a repeat biopsy.24-30 Five studies24-28 reported on PCA3 and tPSA, four25-27,29 on %fPSA, one on PSA velocity,26 and two on externally validated nomograms.24,30 However, the numbers of comparisons possible for each of these matched analyses remained small. For example, one of three tPSA studies providing AUC data restricted recruitment to men with tPSA levels in the “grey zone.”27 No studies addressed other comparators or outcomes. Strength of evidence was insufficient for all comparisons.
KQ 1 and 2. Initial and Repeat Biopsies
In addition to the 9 studies described above, another 15 studies provided matched PCA3 and tPSA data and reported the proportion of men having initial and repeat biopsies.31-45 Given inadequate strength of evidence for analyses focused on men with initial or repeat biopsy only, we examined all studies to determine suitability for a combined analysis (Table A). Using the most commonly reported comparator and analysis (tPSA and AUC), we performed a regression analysis of AUC difference (PCA3 – tPSA) versus the proportion of study subjects on whom prostate biopsy. Based on linear regression, the slope was not significant (p=0.97), indicating no significant relationship between biopsy status and AUC difference for PCA3 versus tPSA elevations. Three of the 15 studies also reported AUCs stratified by initial and repeat biopsy status that could replace the composite AUCs; analysis with these data showed that the slope was again not significant (p=0.81).
Table A
PCA3 versus comparators—analyses and strength of evidence for the intermediate outcome of diagnostic accuracy.
Fourteen studies also provided ROC curves for both PCA3 and tPSA. Regression analysis of (PCA3 – tPSA) sensitivities at a specificity of 50 percent versus biopsy status again showed little or no association between biopsy history and relative performance of PCA3 and tPSA (p=0.79). No similar analyses can be made for any other comparator for diagnostic accuracy. This was considered sufficient to proceed with a combined analysis for KQ 1/KQ 2, without the biopsy history restriction. The same regression analysis conducted in different datasets (e.g., including/ excluding “grey zone” studies, stratified by assay type) consistently found no significant slope. In addition, very similar median AUC differences (PCA3 – tPSA) were found for studies enrolling all men having initial biopsy and studies enrolling all men having repeat biopsy.
Total PSA (tPSA) Elevations, PCA3 Score, and Diagnostic Accuracy for Combined KQ 1/KQ 2 Analysis
Subsets of 20 studies provided sufficient data to compare the diagnostic accuracy of PCA3 with tPSA elevations, using the five described analyses (Table A). We identified two important biases: verification bias and sampling bias. Verification bias occurred for the comparator tPSA (and related measures), as the extent of those elevations was often the basis for deciding on biopsy. Modeling was used to account for the potential impact of verification bias. A sampling bias occurred for the comparator tPSA (and related measures) when some studies only enrolled men with tPSA elevations in the “grey zone” (e.g., upper limit of 10 ng/mL). This results in diminished test performance for tPSA, as it is most predictive of a positive biopsy when very elevated. This bias was accounted for by stratification.
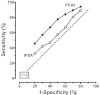
Figure A shows the consensus observed ROC curves for PCA3 and tPSA using data from the 13 studies with suitable analyses. Based on other internal analyses and modeling, it was possible to generate smooth overlapping logarithmic Gaussian curves that fitted these observed data well. Table B shows select data from this modeling that compares the ability of PCA3 and tPSA to identify prostate cancer among men at increased risk. The first row of Table B shows that at a set false-positive rate of 80 percent (specificity of 20 percent), the corresponding sensitivity for PCA3 scores is 95.8 percent. This is 5.1 percentage points higher than the 90.7 percent sensitivity for tPSA measurements. The table then compares sensitivities of the two markers at lower false positive rates. The bottom of Table B displays differences in the false positive rates for selected sensitivities. This is just another way to view the data from the fitted ROC curves.
Table B
Comparison of PCA3 and tPSA measurements to identify men with prostate cancer, holding constant either the false-positive rate (1-specificity) or sensitivity.
Both Figure A and Table B indicate that PCA3 is associated with higher sensitivity at any given specificity than tPSA elevations, and higher specificity at any given sensitivity. This combined approach made it possible to reliably compare PCA3 and tPSA measurements for diagnostic accuracy. Quality of all individual studies was poor and strength of evidence was low (Table A). For all other comparators (Table A), and all other intermediate (impact on decisionmaking, harms of biopsy) and long-term outcomes (morbidity/mortality, quality of life, potential harms), analyses were not possible or were constrained by variability of study populations and limited numbers of studies. All individual studies were graded poor and strengths of evidence for all other outcomes were insufficient.
KQ 3. Testing PCA3 and Comparators To Identify Men With Insignificant Cancer Who May Benefit From Active Surveillance
Thirteen studies were identified that addressed KQ 3 and reported on PCA3 and other preoperative/pretreatment markers for stratification of prostate cancer by risk.22,42,43,46-55 Two studies based analyses on biopsy markers without prostatectomy22,42 and eight reported prostatectomy results as an endpoint. Two studies46,47 were conducted on subjects in an active surveillance program and included short-term followup. One46 predicted outcome based on identification of micrometastases through measurement of tPSA and PCA3 in lymph node extracts, and reported decreased 4- to 6-year biochemical recurrence-free survival in patients with identified micrometastases. Another47 reported 2-year followup of progression from active surveillance to treatment in men with prostate cancer, based on results of yearly biopsy.
Quality of all studies was poor. Strength of evidence was insufficient for diagnostic accuracy due to the inability to compare or combine the two studies on different sample types, using different parameters (e.g., sensitivity/specificity, mean/median biomarker levels) and addressing different outcomes. For this outcome: risk of bias was high; consistency was unknown with two studies; directness was indirect; and precision could not be assessed (imprecise). Strength of evidence was insufficient for any other outcomes or comparators, as no studies were identified.
Key Findings and Strength of Evidence
Strength of evidence was insufficient for KQ 3 and for all comparators and outcomes for KQ 1 and KQ 2 except the comparison of PCA3 and tPSA for the outcome of diagnostic accuracy (Figure A, Table A, Table B). Among men at risk, PCA3 was more discriminatory for detecting prostate cancer at biopsy than tPSA elevations. The finding that the relative performance of PCA3 versus tPSA elevations does not appear to be dependent on biopsy history is a new observation that could impact future studies. The quality of all studies was poor. The strength of evidence was considered low.
Discussion
An important consideration in this conclusion was the potential for spectrum bias, and the associated indirectness of evidence for identifying positive biopsy status. We made the underlying assumption that not all positive biopsies are equal. For example, identifying a positive biopsy associated with a high Gleason score or specific pathological findings may be considered to be clinically more valuable than one with only a low Gleason score. None of the included studies provided a two-way cross tabulation of PCA3 and tPSA positive and negative test results among biopsy positive patients. Of most interest would be the clinical finding for the cases in the off-diagonal (when one test is positive and the other negative).
For KQ 3, the literature review revealed few relevant matched studies and a lack of clinical followup after patients were placed into risk categories defined by the results of PCA3 and other biomarker and pathological tests. In 11 of 13 studies, a reference clinical endpoint (or validated surrogate) was lacking. The quality of all individual studies was poor and strength of evidence was insufficient. It is likely that more time will be needed for studies to assess the diagnostic accuracy of predicting long-term outcomes for patients based on categorization as having low-risk or high-risk disease.
Applicability
The populations studied in the included articles were largely drawn from academic medical centers where patients with elevated tPSA results and/or other risk factors (e.g., positive digital rectal examination, family history, race) often seek, or are referred for, specialty care. Performance of PCA3 and comparators in a broader range of health care settings may differ from that described in this review. It is not yet clear how PCA3, alone or in combination with other biomarkers/risk factors would be integrated into diagnostic or management pathways. The level of acceptance by physicians (and consumers) may well be impacted by Food and Drug Administration approval of a test kit to inform decisions about repeat biopsy in men with a specific clinical history that includes previous negative biopsies. While there was evidence that PCA3 performed better than tPSA with regard to diagnostic accuracy as a secondary test for men with increased risk, it is important to note that neither PCA3 nor tPSA have high performance. A combination of biomarkers and other risk information may be needed to improve overall performance in predicting prostate cancer at biopsy, or informing treatment based on risk classification. The intermediate outcome of diagnostic accuracy is key, as improvement could directly impact the number of biopsies performed in men without prostate cancer and the number of men with prostate cancer who are missed. It is also important to understand other potential harms, as well as the impact of the information on decisionmaking. The effect of even a great test is limited if uptake is low. Longer-term outcomes are challenging, due to the difficulty of following patients and collecting the necessary information.
Research Gaps
With the exception of analyses that include PCA3 and tPSA for the intermediate outcome of diagnostic accuracy, evidence was insufficient to answer the KQs. These questions, therefore, articulate remaining gaps in evidence. Other gaps in knowledge include:
- How much improvement in diagnostic accuracy is needed for any new test to impact biopsy decisionmaking?
- What is the potential of adding PCA3 alone or with other biomarkers to change decisionmaking in practice?
- How does PCA3 compare with the two more frequently used add-on tests (free PSA, PSA velocity) that have appeared in guidance documents?
- Matched studies not derived from “convenience” populations (e.g., biopsy referral centers), and more data on how key demographic factors (family history, race) impact on the performance of PCA3 and comparators.
- Outcome studies to determine how well PCA3 and other comparators used to categorize risk as insignificant/indolent or aggressive to predict the behavior of tumors over time.
- A range of methodological and statistical questions relating to modeling, assessing impact of verification bias, identifying most effective cutoffs for tests based on ROC analysis, and designs for future studies.
Conclusions
For diagnostic accuracy, there was a low strength of evidence that PCA3 had better diagnostic accuracy than tPSA elevations, but insufficient evidence that this led to improved intermediate or long-term health outcomes. In men at risk for prostate cancer based on elevated serum tPSA levels and/or suspicious digital rectal exam or other risk factor (e.g., family history), PCA3 was found to be more discriminatory for predicting prostate cancer at biopsy than tPSA elevations (i.e., at any sensitivity, the specificity is higher, or at any specificity, the sensitivity is higher). The finding that the relative performance of PCA3 versus tPSA elevations is not dependent on biopsy history (i.e., initial biopsy or repeat biopsy after one or more negative biopsies) is a new observation that allowed more studies on KQ 1 and KQ 2 to be combined for analyses. Strength of evidence was insufficient for all other comparators and all other outcomes of interest in KQ 1 and KQ 2.
Eleven of 13 studies addressing KQ 3 lacked a defined reference clinical endpoint (or validated surrogate), and the other two addressed different outcomes. Strength of evidence was Insufficient. There was insufficient evidence for all other comparators and for all other outcomes of interest in KQ 3. With one exception, these three questions continue to identify important gaps in knowledge, with other gaps identified in the review. Current uncertainty about the utility of tPSA screening for prostate cancer30-34 makes understanding followup tests (e.g., PCA3, other biomarkers, and algorithms) for assessing risk prior to biopsy and/or treatment particularly important.35,36
References
- 1.
- Brawley OW. Prostate cancer epidemiology in the United States. World J Urol. 2012 Apr;30(2):195–200. [PubMed: 22476558]
- 2.
- Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010 Sep-Oct;60(5):277–300. [PubMed: 20610543]
- 3.
- National Comprehensive Cancer Network Guidelines—Prostate Cancer Early Detection, Version I.2011. www
.NCCN.org. - 4.
- Freedland SJ. Screening, risk assessment, and the approach to therapy in patients with prostate cancer. Cancer. 2011 Mar 15;117(6):1123–35. [PubMed: 20960523]
- 5.
- Yin M, Bastacky S, Chandran U, et al. Prevalence of incidental prostate cancer in the general population: a study of healthy organ donors. J Urol. 2008 Mar;179(3):892–5. discussion 95. [PubMed: 18207193]
- 6.
- Draisma G, Boer R, Otto SJ, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95(12):868–78. [PubMed: 12813170]
- 7.
- Basch E, Oliver TK, Vickers A, et al. Screening for prostate cancer with prostate-specific antigen testing: American Society of Clinical Oncology Provisional Clinical Opinion. J Clin Oncol. 2012 Jul 16 [PMC free article: PMC3776923] [PubMed: 22802323]
- 8.
- Catalona WJ, Smith DS, Ratliff TL, et al. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med. 1991 Apr 25;324(17):1156–61. [PubMed: 1707140]
- 9.
- Moyer VA., on behalf of the USPSTF. Screening for Prostate Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 2012 Jul 17;157(2):120–34. [PubMed: 22801674]
- 10.
- Schroder FH, Hugosson J, Roobol MJ, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012 Mar 15;366(11):981–90. [PMC free article: PMC6027585] [PubMed: 22417251]
- 11.
- Chou R, Croswell JM, Dana T, et al. Screening for prostate cancer: a review of the evidence for the U.S. Preventive Services Task Force. Ann Intern Med. 2011 Dec 6;155(11):762–71. [PubMed: 21984740]
- 12.
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999 Dec 1;59(23):5975–9. [PubMed: 10606244]
- 13.
- Durand X, Moutereau S, Xylinas E, et al. Progensa PCA3 test for prostate cancer. Expert Rev Mol Diagn. 2011 Mar;11(2):137–44. [PubMed: 21405964]
- 14.
- Makarov DV, Loeb S, Getzenberg RH, et al. Biomarkers for prostate cancer. Annual review of medicine. 2009 Feb;60:139–51. [PubMed: 18947298]
- 15.
- Oesterling JE, Jacobsen SJ, Chute CG, et al. Serum prostate-specific antigen in a community-based population of healthy men. Establishment of age-specific reference ranges. JAMA. 1993 Aug 18;270(7):860–4. [PubMed: 7688054]
- 16.
- Lee R, Localio AR, Armstrong K, et al. A meta-analysis of the performance characteristics of the free prostate-specific antigen test. Urology. 2006 Apr;67(4):762–8. [PubMed: 16600352]
- 17.
- Vickers AJ, Savage C, O'Brien MF, et al. Systematic review of pretreatment prostate-specific antigen velocity and doubling time as predictors for prostate cancer. J Clin Oncol. 2009 Jan 20;27(3):398–403. [PMC free article: PMC2645854] [PubMed: 19064972]
- 18.
- Vickers AJ, Brewster SF. PSA velocity and doubling time in diagnosis and prognosis of prostate cancer. Br J Med Surg Urol. 2012 Jul 1;5(4):162–68. [PMC free article: PMC3375697] [PubMed: 22712027]
- 19.
- Methods Guide for Effectiveness and Comparative Effectiveness Reviews. Rockville, MD: Agency for Healthcare Research and Quality; Aug, 2011. AHRQ Publication No. 10(11)-EHC063-EF. www
.effectivehealthcare.ahrq.gov. - 20.
- Whiting PF, Weswood ME, Rutjes AW, et al. Evaluation of QUADAS, a tool for the quality assessment of diagnostic accuracy studies. BMC Med Res Methodol. 2006;6(Mar 6):529–36. [PMC free article: PMC1421422] [PubMed: 16519814]
- 21.
- Owens DK, Lohr KN, Atkins D, et al. AHRQ series paper 5: grading the strength of a body of evidence when comparing medical interventions--Agency for Healthcare Research and Quality and the Effective Health-care Program. J Clin Epidemiol. 2010 May;63(5):513–23. [PubMed: 19595577]
- 22.
- de la Taille A, Irani J, Graefen M, et al. Clinical evaluation of the PCA3 assay in guiding initial biopsy decisions. J Urol. 2011 Jun;185(6):2119–25. [PubMed: 21496856]
- 23.
- Ferro M, Bruzzese D, Perdona S, et al. Predicting prostate biopsy outcome: prostate health index (phi) and prostate cancer antigen 3 (PCA3) are useful biomarkers. Clin Chim Acta. 2012 Aug 16;413(15-16):1274–8. [PubMed: 22542564]
- 24.
- Ankerst DP, Groskopf J, Day JR, et al. Predicting prostate cancer risk through incorporation of prostate cancer gene 3. J Urol. 2008 Oct;180(4):1303–8. discussion 08. [PubMed: 18707724]
- 25.
- Aubin SM, Reid J, Sarno MJ, et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol. 2010 Nov;184(5):1947–52. [PubMed: 20850153]
- 26.
- Auprich M, Augustin H, Budaus L, et al. A comparative performance analysis of total prostate-specific antigen, percentage free prostate-specific antigen, prostate-specific antigen velocity and urinary prostate cancer gene 3 in the first, second and third repeat prostate biopsy. BJU Int. 2012 Jun;109(11):1627–35. [PubMed: 21939492]
- 27.
- Ploussard G, Haese A, Van Poppel H, et al. The prostate cancer gene 3 (PCA3) urine test in men with previous negative biopsies: does free-to-total prostate-specific antigen ratio influence the performance of the PCA3 score in predicting positive biopsies? BJU Int. 2010 Oct;106(8):1143–7. [PubMed: 20230386]
- 28.
- Wu AK, Reese AC, Cooperberg MR, et al. Utility of PCA3 in patients undergoing repeat biopsy for prostate cancer. Prostate Cancer Prostatic Dis. 2012 Mar;15(1):100–5. [PubMed: 22042252]
- 29.
- Pepe P, Aragona F. PCA3 score vs PSA free/total accuracy in prostate cancer diagnosis at repeat saturation biopsy. Anticancer Res. 2011 Dec;31(12):4445–9. [PubMed: 22199313]
- 30.
- U.S. Food and Drug Administration. Summary of Safety and Effectiveness Data: PROGENSA PCA3 Assay. 2012. www
.accessdata.fda.gov /cdrh_docs/pdf10/P100033b.pdf. - 31.
- Adam A, Engelbrecht MJ, Bornman MS, et al. The role of the PCA3 assay in predicting prostate biopsy outcome in a South African setting. BJU Intl Epub. 2011 Apr 20 [PubMed: 21507188]
- 32.
- Bollito E, De Luca S, Cicilano M, et al. Prostate cancer gene 3 urine assay cutoff in diagnosis of prostate cancer: a validation study on an Italian patient population undergoing first and repeat biopsy. Anal Quant Cytol Histol. 2012 Apr;34(2):96–104. [PubMed: 22611765]
- 33.
- Deras IL, Aubin SM, Blase A, et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol. 2008 Apr;179(4):1587–92. [PubMed: 18295257]
- 34.
- Goode RR, Marshall SJ, Duff M, et al. Use of PCA3 in detecting prostate cancer in initial and repeat prostate biopsy patients. Prostate. 2012 May 14 [PubMed: 22585386]
- 35.
- Nyberg M, Ulmert D, Lindgren A, et al. PCA3 as a diagnostic marker for prostate cancer: a validation study on a Swedish patient population. Scand J Urol Nephrol. 2010 Dec;44(6):378–83. [PubMed: 20961267]
- 36.
- Ochiai A, Okihara K, Kamoi K, et al. Prostate cancer gene 3 urine assay for prostate cancer in Japanese men undergoing prostate biopsy. Int J Urol. 2011 Mar;18(3):200–5. [PubMed: 21332814]
- 37.
- Perdona S, Cavadas V, Di Lorenzo G, et al. Prostate cancer detection in the “grey area” of prostate-specific antigen below 10 ng/ml: head-to-head comparison of the updated PCPT calculator and Chun's nomogram, two risk estimators incorporating prostate cancer antigen 3. European Urol. 2011 Jan;59(1):81–7. [PubMed: 20947244]
- 38.
- Rigau M, Morote J, Mir MC, et al. PSGR and PCA3 as biomarkers for the detection of prostate cancer in urine. Prostate. 2010 Dec 1;70(16):1760–7. [PubMed: 20672322]
- 39.
- Roobol MJ, Schroder FH, van Leeuwen P, et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. European urology. 2010 Oct;58(6):475–81. [PubMed: 20637539]
- 40.
- Schilling D, Hennenlotter J, Munz M, et al. Interpretation of the prostate cancer gene 3 in reference to the individual clinical background: implications for daily practice. Urol Int. 2010;85(2):159–65. [PubMed: 20424427]
- 41.
- Wang Y, Sun G, Pan JG, et al. Performance of tPSA and f/tPSA for prostate cancer in Chinese. A systematic review and meta-analysis. Prostate Cancer Prostatic Dis. 2006;9(4):374–8. [PubMed: 16926855]
- 42.
- Cao DL, Ye DW, Zhang HL, et al. A multiplex model of combining gene-based, protein-based, and metabolite-based with positive and negative markers in urine for the early diagnosis of prostate cancer. Prostate. 2011 May 15;71(7):700–10. [PubMed: 20957673]
- 43.
- Hessels D, van Gils MP, van Hooij O, et al. Predictive value of PCA3 in urinary sediments in determining clinico-pathological characteristics of prostate cancer. Prostate. 2010 Jan 1;70(1):10–6. [PubMed: 19708043]
- 44.
- Mearini E, Antognelli C, Del Buono C, et al. The combination of urine DD3(PCA3) mRNA and PSA mRNA as molecular markers of prostate cancer. Biomarkers. 2009 Jun;14(4):235–43. [PubMed: 19489685]
- 45.
- Ouyang B, Bracken B, Burke B, et al. A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J Urol. 2009 Jun;181(6):2508–13. discussion 13-4. [PMC free article: PMC4372725] [PubMed: 19371911]
- 46.
- Kusuda Y, Miyake H, Kurahashi T, et al. Assessment of optimal target genes for detecting micrometastases in pelvic lymph nodes in patients with prostate cancer undergoing radical prostatectomy by real-time reverse transcriptase-polymerase chain reaction. Urol Oncol. 2011 May 18 [PubMed: 21600799]
- 47.
- Tosoian J, Loeb S. PSA and beyond: the past, present, and future of investigative biomarkers for prostate cancer. The Scientific World Journal. 2010;10(1919) [PMC free article: PMC5763794] [PubMed: 20890581]
- 48.
- Auprich M, Chun FK, Ward JF, et al. Critical assessment of preoperative urinary prostate cancer antigen 3 on the accuracy of prostate cancer staging. Euro Urol. 2011 Jan;59(1):96–105. [PubMed: 20980098]
- 49.
- Durand X, Xylinas E, Radulescu C, et al. The value of urinary prostate cancer gene 3 (PCA3) scores in predicting pathological features at radical prostatectomy. BJU Int. 2012 Jul;110(1):43–49. [PubMed: 22221521]
- 50.
- Liss MA, Santos R, Osann K, et al. PCA3 molecular urine assay for prostate cancer: association with pathologic features and impact of collection protocols. World J Urol. 2011 Oct;29(5):683–8. [PMC free article: PMC3189407] [PubMed: 21152924]
- 51.
- Ploussard G, Durand X, Xylinas E, et al. Prostate cancer antigen 3 score accurately predicts tumour volume and might help in selecting prostate cancer patients for active surveillance. European urology. 2011 Mar;59(3):422–9. [PubMed: 21156337]
- 52.
- van Poppel H, Haese A, Graefen M, et al. The relationship between Prostate Cancer gene 3 (PCA3) and prostate cancer significance. BJU Int. 2012 Feb;109(3):360–6. [PubMed: 21883822]
- 53.
- Vlaeminck-Guillem V, Devonec M, Colombel M, et al. Urinary PCA3 score predicts prostate cancer multifocality. J Urol. 2011 Apr;185(4):1234–9. [PubMed: 21334023]
- 54.
- Whitman EJ, Groskopf J, Ali A, et al. PCA3 score before radical prostatectomy predicts extracapsular extension and tumor volume. J Urol. 2008 Nov;180(5):1975–8. discussion 78-9. [PubMed: 18801539]
- 55.
- Nakanishi H, Groskopf J, Fritsche HA, et al. PCA3 molecular urine assay correlates with prostate cancer tumor volume: implication in selecting candidates for active surveillance. J Urol. 2008 May;179(5):1804–9. discussion 09-10. [PubMed: 18353398]
Footnotes
- a
Area under the curve (AUC) is a common metric that measures the accuracy of diagnostic tests, that is the ability of the tests to discriminate those who have (or will develop) the outcome of interest from those who do not have (or will not develop) the outcome. An AUC of 1.0 indicates a perfect test, and an AUC of 0.5 indicates a worthless test.
- Executive Summary - PCA3 Testing for the Diagnosis and Management of Prostate Ca...Executive Summary - PCA3 Testing for the Diagnosis and Management of Prostate Cancer
- Introduction - Impact of Healthcare Algorithms on Racial and Ethnic Disparities ...Introduction - Impact of Healthcare Algorithms on Racial and Ethnic Disparities in Health and Healthcare
Your browsing activity is empty.
Activity recording is turned off.
See more...