NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Bradley LA, Palomaki G, Gutman S, et al. PCA3 Testing for the Diagnosis and Management of Prostate Cancer [Internet]. Rockville (MD): Agency for Healthcare Research and Quality (US); 2013 Apr. (Comparative Effectiveness Reviews, No. 98.)
This publication is provided for historical reference only and the information may be out of date.
Literature Search
Of the 1,556 citations identified through the literature searches, 1,514 were excluded at various stages of review. One additional study was identified through grey literature searches. No additional studies were identified from one identified systematic review on PCA380; this review was excluded from analyses as having no primary data. The 43 included articles reported the results of observational cohort studies with matched comparisons of PCA3 and other selected biomarkers. The PRISMA flow diagram (Figure 3) illustrates the review process for published studies, exclusions at each step and the selection results.
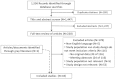
Figure 3
PRISMA diagram for selection of published studies.
For Key Question (KQ) 1 and KQ 2, no randomized or comparative intervention trials were identified that included the use of PCA3 testing and reported long-term outcomes, or intermediate outcomes other than diagnostic accuracy. Of the 43 articles included, six studies addressing KQ 1 and KQ 2 were found to have duplicate data and were excluded from analyses34,47,81-84. Two other studies85,86 were excluded because reported data were not in a format usable for these analyses. Of the remaining 34 studies, 24 addressed KQ 1 and/or KQ 2 (Table 1, Table 2a).
Table 1
General descriptions of the included studies in matched populations.
Table 2
Reported criteria for specific inclusion/exclusion of subjects for studies.
For KQ 3, no randomized or comparative intervention trials were identified that included the use of PCA3 testing and reported intermediate or long-term outcomes. Twelve observational studies were identified that addressed KQ 3 (Table 1, Table 2b), but one was excluded due to duplicate data.87 Two studies reported on short-term health outcomes (i.e., biochemical recurrence and time to progression to treatment from active surveillance).88,89Table 1 provides general descriptive information on all studies. Table 2 describes study inclusion/exclusion criteria, and Table 3 describes the populations studied. Table 4 provides key information on PCA3 testing. In later sections, Table 17 details the characteristics of matched biopsy and prostatectomy studies addressing KQ 3; Table 18 provides information on comparators investigated along with PCA3 scores in studies addressing KQ 3.
Table 3
Characteristics of populations studied.
Table 4
PCA3/DD3 assay characteristics.
Table 17
Characteristics of matched studies addressing KQ3 with biopsy and prostatectomy results.
Table 18
Comparators investigated with PCA3 scores in matched studies addressing KQ 3.
Grey Literature Search
The process for evaluation of grey literature search results is summarized in Figure 4 and Appendix C. Two clinical trial registry citations were potentially relevant to the review:
- Prostate Cancer Antigen 3 (PCA-3) Gene Project (NCT01177436) – The status of this trial is unclear (last update August, 2010). Of interest was the use of three housekeeping genes for PCA3 testing in addition to KLK3 (PSA): ACTB (beta-actin), TUA (Ka 1 tubulin), and GAPDH (glyceraldehyde-3-phosphate). Results may resolve remaining concerns about potential bias related to the use of KLK3 as the housekeeping gene.
- Clinical Evaluation of the Progensa® PCA3 Assay in Men With a Previous Negative Biopsy Result (NCT01024959) – This trial, conducted by GenProbe and completed in April, 2011, provided data for the premarket (PMA) submission to FDA that was approved. However, the published article reporting the results of this clinical trial was not available for review.
Overall, the search of grey literature yielded one study, reported in the FDA Summary of Safety and Effectiveness Data for Gen-Probe's PROGENSA® PCA3 Assay (PMA P100033).
Potential Biases in Included Studies
The populations in the included studies were largely drawn from academic medical centers where patients with elevated tPSA results and/or other risk factors (e.g., positive DRE, family history, African American race) were seeking referral or specialty care. Observational studies of such opportunistic cohorts are subject to specific biases.
Verification Bias
Men will be offered prostate biopsy based on the extent of tPSA elevations, suspicious findings on a digital rectal exam (DRE), a combination of the two or, less commonly, other risk factors such as family history or race. In order to obtain an unbiased estimate of diagnostic accuracy for tPSA at specific cutoffs, it is necessary that the identification of prostate cancer not be related to tPSA levels. This is a potential problem as studies have shown that higher tPSA levels are indicative of a higher likelihood for the presence of prostate cancer. Men are more likely to undergo prostate biopsy, if the tPSA is high (e.g., 10-20 ng/mL), rather than close to lower cutoffs used to define a positive tPSA screening test (e.g., 3-4 ng/mL). If a study reports results in which biopsy is tPSA-related, the sensitivity and specificity at select tPSA cutoffs will not be accurate. If those not accepting biopsy are considered missing, this is considered “partial verification” bias. All studies included in the evidence review are opportunistic cohorts of men agreeing to biopsy, and will be subject to this bias. However, no study addressed this potential bias. We addressed this bias through modeling the effect of verification bias on the tPSA measurements. A detailed discussion of this bias and the modeling performed can be found in Appendix J.
Spectrum Bias
Spectrum effects should also be considered when evaluating diagnostic tests generated from convenience samples collected at referral centers. For example, such studies are likely to represent men at higher risk of prostate cancer than in the total cohort of screened men, and might be at higher risk of more aggressive cancers as well (e.g., those with rapid rise in PSA). The positive biopsy rate in such referral populations will depend on multiple factors, including the tPSA cutoff, the number of men with elevated tPSA who opt out of biopsy (e.g., men with lower tPSA levels and lower risk), and/or the proportions of men with other important risk factors. In 17 included studies, biopsy positive rates ranged from 16.9 to 72.9 percent, with a median of 36 percent. Although this may not influence the clinical sensitivity and specificity estimates it certainly will influence the positive and negative predictive values (as the disease prevalence varies).
A second spectrum effect of more concern relates to the range of severity of disease between those identified with an elevated PCA3 score compared with those with a positive comparator test. For example, suppose two tests have the same sensitivity and specificity estimated in a cohort of biopsied men. In order to show the true clinical validity, it would be necessary to examine the men with positive biopsies having discordant test results (i.e., positive by one test but negative by the other). If the men positive by one test have similar tumor characteristics and/or severity of disease to those positive by the other test, then the sensitivity/specificity estimates could be both statistically and clinically equivalent. However, if one test identifies a difference in tumor characteristics and/or severity of disease that the other test does not, the estimates could be statistically equivalent but clinically different.
Sampling Bias
Analysis of tPSA (and related comparators) was also subject to a sampling bias. A subset of studies restricted enrollment to tPSA results in the “grey zone” (e.g., 2.5 ng/mL to less than 10 ng/mL). The impact of this restriction would be to reduce the prevalence of disease in the study group, as there is a positive correlation between tPSA and prevalence of prostate cancer. In addition this would also reduce the tPSA test performance (sensitivity/specificity) as those men with higher tPSA levels are not enrolled in the study. It is at the higher tPSA levels that the test is most predictive. This would reduce the apparent performance of tPSA measurements, and increase the difference between the PCA3 and tPSA performance estimates. Overall, this bias reduces the external validity (generalizability) of studies in a general population. This bias cannot be avoided by statistical analysis. These studies could be removed from consideration. Instead, we have chosen to address this bias by stratifying analyses by selection criteria. That is, studies of the “grey zone” were summarized separately from the studies that include all levels of elevated tPSA.
Analyses Relating to KQs 1 and 2
KQ 1. Testing PCA3 and Comparators To Identify Prostate Cancer in Men Having an Initial Biopsy
Among the 17 studies addressing KQ 1 (Table 1), only two reported results in populations where all men were having initial biopsies (Table 5).95,98 Both studies reported data on tPSA and %fPSA; one95 also reported on PSA density. The actual data will be presented in later analyses (KQ 1/KQ 2 combined), but there were too few data for reliable interpretations. The five matched analyses performed (see Table 5 footnote a) are outlined in Methods, and discussed in detail as part of the analyses.
Table 5
Summary of matched analyses (of five) that can be performed for each PCA3 comparator, with analysis restricted to studies only on initial biopsies.
Strength of Evidence
When data were restricted to the two studies reporting only on populations of men having an initial prostate biopsy, only one comparison could be made, the D analysis for %fPSA. Both studies were poor quality. It is not possible to evaluate consistency (between-study results). In addition, estimates of effect size will be imprecise. This results in assigning grades of “insufficient” for the reported comparisons of PCA3 with tPSA, %fPSA, and PSA density, and other comparators (PSA velocity, complexed PSA and externally validated nomograms) with no matched studies.
KQ 2. Testing PCA3 and Comparators in Men Having Repeat Biopsy
Among the 21 studies addressing KQ 2 (Table 1, Table 2b), seven65,91,92,97,105,106,111 reported results in populations where all of the men were having a repeat biopsy (Table 6). Studies are ranked by number of patients enrolled. Five studies reported on tPSA,65,91,92,106,111 four on %fPSA91,92,105,106 and two on externally validated nomograms.65,97 All studies were poor quality. The actual data were included in later analyses, but there were too few data for any one analysis to provide reliable interpretations.
Table 6
Summary of matched analyses (of five) that can be performed for each PCA3 comparator, with analysis restricted to studies reporting results only in repeat biopsy.
Strength of Evidence
When data were restricted to these seven studies, the number of comparisons possible for each matched analysis remained small due to three “grey zone” studies.91,105,106 For example, the “D” analysis for tPSA has three sets of data, but two are for all levels of tPSA and one is restricted to the “grey zone.” All studies were poor quality. Due to these differences in inclusion criteria, it is difficult to evaluate consistency. Estimates of effect size will, necessarily, also be imprecise. Strength of evidence was deemed insufficient for all comparisons of PCA3 with tPSA, %fPSA, PSA velocity, PSA density, complexed PSA and externally validated nomograms in this population of men.
Potential To Combine KQs 1 and 2: Testing PCA3 and Comparators in Men Having Initial or Repeat Biopsy
The sections above addressed the nine studies that exclusively studied men having an initial (KQ 1) or repeat biopsy (KQ 2). However, 15 additional studies included matched results of PCA3 and the comparators (Table 7). Eleven reported the proportion of men having initial and repeat biopsies,31,90,93,96,99,102,103,107-110 but four did not report biopsy history.94,100,101,104 The results from these studies were most often not stratified by biopsy history.
Table 7
Summary of matched analyses (of five) that can be performed for each PCA3 comparator, with analysis restricted to studies on initial and repeat biopsies.
At this point, one could have ignored the data in these 15 additional studies, as they did not directly apply to either KQ 1 or KQ 2. Instead, based on the inadequate strength of evidence found for the prior individual analyses that focused on only those men with initial or with repeat biopsies, we chose to examine whether data from the studies that could be stratified by biopsy history might be suitable for a combined analysis (Table 7). Prior to performing this combined analysis, however, it was necessary to determine whether biopsy status was an important covariate that could bias the findings. An examination of Table 7 found that the most common comparator was tPSA, and the most common analysis, by far, was the area under the curve (AUC), indicated by an “A.”
Fifteen of the 19 studies that reported AUC results for both PCA3 and tPSA also provided the proportion of study subjects with no previous prostate biopsies. A regression analysis of AUC difference (PCA3 – tPSA) versus the proportion of men with an initial biopsy would provide evidence regarding suitability of the combined analysis. The raw data for this figure can be found in Table 10.
Table 10
Comparing PCA3 scores and tPSA elevations in matched studies via AUC analysis.
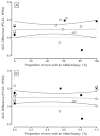
Figure 5A shows the analysis. Based on linear regression, the slope (-0.00227) was not significant (p=0.97), indicating that there was no significant relationship between the biopsy status and AUC difference for PCA3 versus tPSA elevations. In addition, a subset of three of the 15 studies reported AUCs stratified by initial and repeat biopsy status (Table 10). Figure 5B shows the analysis with the replacement of the three “composite” AUCs with initial and repeat biopsy subgroup AUCs. The slope (-0.01307) was also not significant (p=0.81).
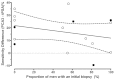
Examining Table 7 also indicated that 14 studies (16 datasets) reported the ROC curves for both PCA3 and tPSA. A regression analysis of (PCA3 – tPSA) sensitivities at a constant specificity of 50 percent versus the proportion of men with an initial biopsy would also provide evidence regarding the suitability of the combined analysis. The raw data for this figure can be found in Table 13. Figure 6 shows the described analysis. Based on linear regression, the slope (0.02956) is not significant (p=0.79). Again, there appears to be little or no association between the biopsy history and the relative performance of PCA3 and tPSA.
Table 13
Differences in PCA3 and tPSA sensitivities (difference) at PCA3 false positive (1-specificity) rates from 20% to 80%.
Together, these two analyses shown in Figures 5 and 6 provided evidence that combining results from studies of initial biopsies, repeat biopsies, and mixtures of initial and repeat biopsies did not appear to impact the comparison of PCA3 with tPSA elevations.
One other matched analysis provided additional support for this finding. Two studies93,99 provided matched PCA3/tPSA ROC curves separately for men who had an initial biopsy, and those receiving a repeat biopsy (also Table 13). Therefore, these were matched within-study comparisons of the relative effectiveness of the two markers in these biopsy-specific subgroups. The first study93 found that tPSA performed much better in initial compared with repeat biopsies, while PCA3 performed much worse in initial compared with repeat biopsied men. This is consistent with some who have argued that tPSA would be expected to perform poorly in the repeat biopsy setting, as those tumors associated with high tPSA were identified in the initial round of testing and would not be present in a population having repeat biopsies. However, the second study99 found much different patterns. The tPSA performed almost equally as well in initial and repeat biopsy settings, and PCA3 performed much better in initial compared with repeat biopsies. These two studies reported almost opposite findings.
Such analyses cannot be performed for any of the other comparators. However, given the lack of data for those comparisons, we chose to comprehensively list all potentially relevant results, regardless of the biopsy status of the enrolled men. The following sections provide the results of the combined analysis of KQ 1 and KQ 2.
Description of Included Studies for KQ 1/KQ 2 “Combined”
As noted in Methods, the inclusion criteria restricted study inclusion to matched studies. These were defined as studies that provided estimates of diagnostic test performance for PCA3 and at least one other comparator (e.g., tPSA elevations or %fPSA) using the same patient population. Thus, a study of PCA3 alone, or a comparator alone, would not be included. In examining the included studies, it was clear that, although the same population was used, the reports rarely applied a true matched analysis. However, the results were still considered as being “matched,” due to the application of the test(s) to the same underlying population. We preserved this population matching by computing differences between PCA3 test results and comparator test results within each study. These matched differences could then be compared across studies.
For example, one study of biopsied patients might report an AUC for PCA3 and then separately report an AUC for tPSA in the same population. The difference in the two would then be computed and compared with the difference in AUCs from other similarly matched studies. Although this restriction limited the number of included publications, it was aimed at improving the consistency of results. For example, an analysis of unmatched studies might have provided sufficient information to stratify PCA3 performance by number of previous biopsies. Similar data could be obtained from the literature for tPSA. Comparing the results between these unmatched studies might have shown differences related to variations in study populations or design rather than the variable of interest.
All studies were judged to be of poor quality, based on reasons including: use of convenience data (i.e., opportunistic cohorts of men having prostate biopsy); potential biases (e.g., verification, selection, spectrum); incomplete or unclear study protocol (e.g., inclusion criteria, missing key variables); limited analyses and no matched analyses (or raw data from which to conduct matched analysis); and/or lack of blinding or reporting on blinding. In addition, four of the 24 studies addressing KQ 1 and KQ 2 were funded by GenProbe and a third of the 24 (N=8) indicated conflicts of interest for investigators (Table 1). All of these studies focused on determining the diagnostic accuracy of PCA3 testing using biopsy results as the reference or gold standard. No studies were identified that reported on intermediate outcomes other than diagnostic accuracy, or long-term clinical outcomes.
Evaluation of PCA3 and Other Comparators To Identify KQs 1 and 2 Intermediate and Long-Term Outcomes
Comparator: Total Serum PSA
Study design was a crucial criterion for this comparison, because tPSA measurements were integral to decisionmaking regarding uptake of prostate biopsy after the finding of an initial tPSA elevation through prostate cancer screening. Men were likely offered biopsy based on the extent of tPSA elevations, suspicious findings on a digital rectal exam (DRE), a combination of the two or, less commonly, other risk factors such as family history or race. This led to only a subset of initially identified men having the “gold standard” test (biopsy) that defines one of the outcomes of interest – diagnostic accuracy. This association of test result with uptake of the diagnostic test has been labeled verification bias.
As noted, verification bias would have occurred in this setting because men with higher tPSA elevation were more likely to undergo biopsy compared with men with lower levels. Thus, test sensitivity would have been overestimated (as a higher proportion of cancers with lesser elevations would not have been identified by biopsy). This bias would have underestimated specificity (or overestimated the false positive rate), because the larger number of men without cancer and negative biopsy results were not identified via biopsy. See Appendix J for a more complete description of verification bias, an example relevant to prostate cancer and tPSA elevations, a review of directly relevant literature, and an evaluation of what will, or will not be compromised in this comparison. Appendix J also contains a more complete description of the modeling used to overcome the major impact of verification bias and a more extensive comparison of PCA3 and tPSA test performance characteristics.
These analyses indicated that the relative performance of tPSA elevations (sensitivity at a given specificity) was, at most, modestly influenced by verification bias, but the tPSA cutoff level at which this performance occurred cannot be directly observed. We employed a simple model to determine approximate tPSA cutoff levels in the presence of verification bias. This bias would be less likely to have been an issue for the other comparators (e.g., %fPSA, PSA density), but the extent of this bias is likely related to the correlation between that comparator and tPSA measurements. In addition, this correlation may be low because these comparators were not routinely used in all men with a tPSA/DRE positive result and may, therefore, not be strongly associated with biopsy uptake. A second known bias, sampling bias, was related to a subset of studies that limited their reporting to men in the “grey zone” of tPSA measurements and was variably defined in these studies as between 2.5 and 10 ng/mL,91,95,106 2.0 and 20 ng/mL98 or 4 and 10 ng/mL.105 These studies would have underestimated the performance of tPSA compared with studies that included all men with elevated results. We accounted for this bias by stratifying results, when possible.
Total PSA and the Intermediate Outcome of Diagnostic Accuracy
Key Points
The extent of tPSA elevations was compared with PCA3 scores to determine their diagnostic accuracy to predict prostate biopsy results (cancer/no cancer). Measures included in the analyses were the sensitivity, specificity (or the false-positive rate equal to 1-specificity), and positive and negative predictive values. As a reminder, only studies in which the performance estimates for both comparators were made in the same population were included in the five analyses listed below.
- Area under the curve (AUC). Twenty studies (Table 10) reported AUC estimates for tPSA and PCA3 in the same population and the difference of the two [AUC(PCA3) – AUC(tPSA)] was computed. Overall, 18 of the 20 studies found a positive difference. The two90,102 studies finding tPSA elevations to have a greater AUC were among the smaller studies. Removing the four studies31,91,95,106 that restricted recruitment to the tPSA “grey zone” resulted in an AUC difference of 0.0865 in the remaining 16 studies (Table 10).
- Reported median, interquartile range, range and estimated logarithmic means/standard deviations (SD). Eight studies (Table 11) provided sufficient data for analysis, and none of these directly reported a logarithmic SD (most, if not all studies examining the distribution found both PCA3 and tPSA to be highly right skewed). The logs SDs were estimated from the ranges or inter-quartile ranges. The differences, reported as z-scores, indicated that one study102 (the smallest) found tPSA to be slightly better than PCA3 at separating populations of positive and negative prostate biopsies, while the remaining seven others found a larger difference in favor of PCA3.
- Performance at a PCA3 cutoff score of 35. Nine studies (Table 12) reported the sensitivity and specificity of PCA3 at this cutoff. We computed the difference in sensitivity (PCA3 – tPSA) when tPSA was held at the PCA3-related specificity. Eight of the nine studies reported a positive difference (median 16.3 percent, range -9.5 to 35 percent) favoring PCA3 (Table 12).
- ROC curves - sensitivity/specificity. Fourteen studies and 16 datasets (Table 13) provided a ROC curve, or data representing a ROC curve, for both markers. At a specificity of 50 percent, the difference in corresponding specificities (PCA3 – tPSA) was zero or positive for all included studies except the one performed in a majority black population.90 Removing the four studies that restricted recruitment to the tPSA “grey zone”31,91,95,106 and the one study performed in a majority black population90, the difference in sensitivity favored PCA3 by 20 percent (range 0 to 39 percent).
- Regression analysis. Only one study provided sufficient data to apply the respective regression coefficients to create a relative odds ratio (OR) between the 25th and 75th centiles of the two distributions.31 A second study95 reported all but the inter-quartile range, and that was estimated from the first study so that both datasets could be evaluated. In both studies, the ratio of the ORs (PCA3 / tPSA) was greater than 1 (1.38 and 1.97). These two studies31,95 both restricted recruitment to the tPSA “grey zone,” so the results were likely to overestimate the relative superiority of PCA3 by underestimating tPSA performance.
Table 11
Comparison of PCA3 and tPSA differences in central estimates in men with positive and negative prostate biopsy results, after accounting for study-specific variability in measurements.
Table 12
Differences in PCA3 and tPSA sensitivities at the fixed specificity associated with the commonly used PCA3 score cutoff of 35.
Interpretation
The results of analyzing the literature regarding the matched analyses of PCA3 score versus extent of tPSA elevations was summarized in Table 8. A more complete description of how these data were computed has been provided in Appendix J. Table 8 compares the diagnostic accuracy of PCA3 scores and tPSA elevations to independently identify men who would have a positive biopsy (prostate cancer). In Table 8A, the false-positive rate (1-specificity) was held constant, while in Table 8B, the sensitivity (detection rate) was held constant. This display was chosen because an undetected cancer was not considered equivalent to a falsely positive prostate biopsy and, therefore, comparing a loss in sensitivity with a gain in specificity was difficult. The last column shows the difference between the two estimates (PCA3 – tPSA). When comparing the sensitivities (Table 8A), this column contains the improvement in prostate cancer detection. When comparing the false-positive rates, it contains the reduction in biopsies performed on men without prostate cancer.
Table 8
Modeled comparison of PCA3 and tPSA elevations to identify men with prostate cancer, with either the false positive rate (1 – specificity) or the sensitivity held constant.
For example, assume that one would like to set test sensitivities to 85 percent. Using the row with 85 percent sensitivity (shaded row, Table 8B), only 59 percent of men without cancer would be subject to biopsy with PCA3 testing (cutoff score of about 17). Using tPSA elevations, 79 percent of those men without cancer would be biopsied (cutoff of 1.9 ng/mL). This means that using PCA3 instead of tPSA elevations, the same proportion of cancers might be detectable while performing 20 percentage points fewer biopsies. Additional tables at fixed PCA3 and tPSA cutoff levels, individual risks and positive and negative predictive values at several difference prostate cancer rates can be found in Appendix J (Tables J1 through J4).
Characteristics of Studies Reporting Data Used in Five Analyses for KQs 1 and 2 Combined
Twenty two studies addressing KQ 1 and KQ 2 reported PCA3 and tPSA comparisons that could be used in one or more of the five analyses of matched studies (Table 9). Of interest are the five studies that used an upper cutoff for tPSA elevations to define a “grey zone.”31,91,95,98,106 When the tPSA range was truncated, it would reduce the effectiveness of the marker to predict biopsy outcome. In general, this was confirmed in our analyses, and, for that reason, the results may be stratified by this characteristic, the entries shaded in tables, and the observations noted in Figures. All study quality ratings were poor.
Table 9
Summary results for five analytic comparisons of PCA3 versus tPSA in matched populations of men having prostate biopsies.
PCA3 and tPSA: Area Under the Curve
Twenty studies and 22 datasets reported the diagnostic performance of PCA3 and extent of tPSA elevation among men with initially screen positive test results (elevated tPSA with or without positive DRE) to discriminate between positive and negative biopsy test results. These studies and related information are shown in Table 10. Five studies65,91,92,106,111 in which all individuals already had one or more negative biopsies were among the included studies, in order to strengthen the analysis of PCA3 and tPSA elevations. The studies are ordered by effect size, the difference between the matched AUC estimates of PCA3 minus tPSA (positive numbers indicate PCA3 performed better, negative numbers indicate tPSA performed better).
All but two studies90,102 found the matched AUC point estimate for PCA3 higher than that for tPSA. Those two studies were among the five smallest reported, with 62 and 105 enrollees, respectively. The largest effect size was reported by the smallest study of all109, reporting matched results for only 32 men. The median AUC difference for all studies was 0.1055 (range -0.1389 to 0.2150). Only eight31,65,95,99,101-103,108 of the 20 studies reported the matched p-values comparing the two AUCs. Using these as a guide, at least the 10 studies from row 11 (Mearini101) to the end of the table are likely to have been statistically significant. No study reported a statistically significant lower performance for PCA3. The study reporting a difference of -0.139 did not report a p-value, but did provide the respective 95%confidence intervals (CI) on the PCA3 and tPSA AUC estimates. These overlapped, indicating the differences were not likely to be significant (0.705, 95% CI: 0.599 to 0.812; and 0.844, 95% CI: 0.765 to 0.910, respectively).
None of the studies reported a confidence interval or standard deviation for the matched difference of the two AUCs. Although the AUCs for PCA3 and tPSA ranged widely (indicating relatively high heterogeneity), the variability of the differences seemed more consistent. This may be due to the requirement that only paired estimates of the AUCs be included in this analysis.
Four studies31,91,95,106 enrolled only men with tPSA levels less than 10 ng/mL, essentially limiting their population to the so-called “grey zone.” In general, only one to two percent of biopsy negative men had tPSA levels over 10, while about 20 percent of biopsy positive men were in this range.120,121 Removing this subset from the overall population of men with positive tPSA/DRE was likely to have the effect of reducing the ability of tPSA to predict positive prostate biopsies. Thus, one would have expected these studies to show greater differences in favor of PCA3. The four studies focusing on the “grey zone” are highlighted in grey in Table 10. All but one91 was near the bottom of the table, indicating that they did, in fact, find greater differences. The median difference in AUC in the “grey zone” studies was 0.1595. If these four studies were removed, the AUC difference was reduced to 0.0865. Although not formally computed, the heterogeneity would also be expected to be reduced. One could argue that these four “grey zone” studies should have been excluded, as they did not, technically, satisfy fully the inclusion criteria. However, they were included for two reasons. First, the performance in this subset had clinical implications. For example, some may argue that clinicians could intervene based solely on a very elevated tPSA, but use additional markers to evaluate the remaining “grey zone” patients. This assumes that very elevated tPSA results are, by themselves, sufficiently informative for decisionmaking, and performance would not benefit from adding a second useful and independent marker like PCA3. Should PCA3 come into routine practice, it is not clear that use only in the “grey zone” would be an effective approach. Second, the “grey zone” stratification identified a source of heterogeneity and helped demonstrate the validity of these analyses.
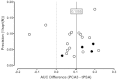
An estimate of the potential for publication bias for this analysis could be generated under the assumption that the standard error of the AUC difference was proportional to the reciprocal of the square root of the number of enrolled men for each study. Figure 7 shows a plot of the computed AUC difference (x-axis) versus its estimated precision (i.e., the reciprocal of the square root of the sample size [y-axis]). The solid vertical line shows the median difference of 0.1055 while the dashed vertical line at 0.000 shows where the AUC would be equivalent. As predicted, the data were found to fit a symmetric “inverted funnel,” suggesting that at least some of the variability was due to the small samples sizes for several of the studies. The results seemed far more consistent for the 11 largest studies31,65,91,93,95,96,99,100,106-108 that provided matched results for 200 or more men.
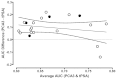
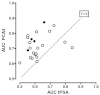
Figure 8a explores the relationship of the AUC difference, this time comparing the results against the average AUC (average of PCA3 and tPSA AUCs). The actual AUC for the markers may be indicative of extraneous factors (e.g., tPSA cutoff level, age of enrollees) that may vary among the 20 studies included in this analysis. Of interest, the two studies90,102 reporting the negative AUC differences were two of the three highest average AUCs. However, regression analysis showed no significant relationship between average AUC and the AUC difference (slope not significant; p=0.72). The median average AUC was 0.6605 (range 0.5525 to 0.7875). Figure 8b displays the tPSA AUC on the x-axis versus the PCA3 AUC on the y-axis for the same studies shown in Figure 8a. The dashed “line of identity” indicates all values where the tPSA and PCA3 AUCs would be equal. On average, the observations fall above the line, showing that the PCA3 AUC is higher than the tPSA AUC within a given study. As expected, the four studies in the “grey zone” of tPSA (filled circles) tend to have higher differences that fall farther from the line of identity.
Eighteen studies identified the methodology used for PCA3 testing (Table 4). Two used Aptima reagents (GenProbe),100,103 10 specified using the Progensa® kit,31,90,92,93,95,96,99,102,106,108 and two reported only using a “GenProbe” test.91,109 Among the remaining six studies, four used a quantitative RT-PCR method,94,101,104,107 The remaining two studies64,106 did not specify the method, but disclosures suggested that both were likely using the current GenProbe assay (Progensa).65,111 AUC differences (PCA3-tPSA) for fourteen studies using GenProbe reagents were compared with the proportion of men having an initial biopsy (figure not shown). This regression analysis showed no significant relationship between the AUC difference and initial biopsy status (slope -0.0472; p=0.52). The slope indicates that over the range of AUCs shown in Figure 8a (0.6 to 0.8), the difference in AUC would be -0.0094 or about a 1 percent lower for all initial biopsied patients compared with all repeat biopsied patients. The same analysis for six studies using other assays also showed no significant relationship (slope 0.0854; p=0.27), but the slope indicated a 1.7 percent higher AUC for the same comparison. Therefore, the analysis does not provide statistically significant evidence that assay methodology is an important consideration.
Among the six of 20 studies that reported the racial/ethnic distribution in the study population (Table 3),65,90,94,96,103,104 one was in an Asian (Japanese) population103 and this group's AUC difference of 0.126 was slightly higher than the summary estimate of 0.1055 Another Asian study94 (China) had an AUC difference estimate of 0.173. Only one study performed in South Africa reported on a population composed of a majority of black men (68.6 percent, Table 3),90 and the group's AUC difference of -0.139 was the lowest observed in all studies (Table 10). The four North American studies reporting a small black population (5.396,110 and 2 percent65,104) had AUC differences near the consensus estimate.
Each reviewed study was assigned a QUADAS quality score of good, fair, or poor. Among the 20 included studies in the AUC differences computation (Table 10), all were rated poor. Only one of the studies was blinded in both directions (i.e., laboratory blinded to outcome, and clinicians blinded to laboratory results), and only two were blinded in a single direction (one in each direction).
PCA3 and tPSA: Reported Medians and Standard Deviations
Eight studies (Table 11) reported some information concerning the distributions of PCA3 and tPSA levels among men with screen positive test results (elevated tPSA with or without positive DRE) who subsequently had positive or negative biopsy results. Results from these studies and related information are also shown in Table 11. The distributions of both markers are highly right skewed and have been shown to be reasonably Gaussian after a logarithmic transformation. For this reason, we chose to include for analysis only those studies in which the median or logarithmic mean could be determined along with the logarithmic standard deviation. In some instances the standard deviation was estimated using reported centiles (e.g., inter-quartile range). If a study only reported the range, the standard deviation was computed assuming the range represented 6 standard deviations.122 For each study, the difference in marker levels in those with positive or negative biopsies was expressed as a z-score, using a study-specific pooled standard deviation.
It was possible to obtain a median and pooled log standard deviation for both markers using data from eight studies (Table 11). Studies were sorted by the difference in z-scores. The two studies that truncated results above 10 ng/mL31 or 20 ng/mL98 are shown in grey. One study65 incorrectly reported the median PCA3 score in men with a negative biopsy; the corrected value of 19.4 is shown. Two additional studies91,95 had partial data, and were also summarized at the bottom of Table 11 to allow for comparison of median levels only. This analysis would have been more robust had the authors actually reported the median and logarithmic standard deviations for their populations, provided raw data (in the form of a scatterplot) or fitted the data to some other distribution.
Figure 9 shows the overlapping distributions from the eight studies shown in Table 11. The overlapping curves were drawn based on the log Gaussian parameters described there. The individual figures show each set of paired distributions. Given the fact that only eight studies were analyzed, it was not possible to stratify results by race, region or test methodology. Note that the very tight distributions for tPSA can be seen for the two “grey zone” studies.31,98
PCA3 and tPSA: Performance at a PCA3 Cutoff Score of 35
Nine studies (Table 12) reported the sensitivity and specificity of PCA3 score at a cutoff of 35 among men with positive initial screening results (elevated tPSA with or without positive DRE) who subsequently had positive or negative biopsy results. Table 12 shows the sensitivity and false positive rates (1 – specificity) for PCA3 with the corresponding sensitivity of tPSA at the same specificity found for the PCA3 cutoff level. The table was sorted by effect size. The difference in the two sensitivities (with the specificity held constant) provided a comparison of the ability to distinguish prostate cancer between the two markers. In some instances, the tPSA results were estimated from a published ROC curve. In one study, the specificity/1-specificity was incorrectly reported, as evidenced by the additive inverse found on the accompanying ROC curve. In another study102, the reported sensitivity/specificity did not match the corresponding ROC curve, and the reason for the discrepancy could not be identified. Those data were excluded from analysis. The most appropriate analysis that compared two tests on the same population was to use a matched analysis of the 2×2 table. However, all nine studies reported only independent evaluations of each marker.
Among the nine studies (Table 12), the PCA3 score cutoff level of 35 was associated with false-positive rates (1-specificity) ranging between 20 and 50 percent, with corresponding sensitivities (detection rates) between 38 and 77 percent. For each study, the corresponding sensitivity for tPSA (at the same false positive rate) was subtracted from the PCA3 sensitivity. For one study90, the difference was negative, while the eight remaining studies showed PCA3 having higher sensitivities with increases ranging from 3 to 35 percent. The median increase in sensitivity was 16.3 percent.
Given that only nine studies were analyzed, it was not possible to stratify results by race, region or test methodology. Of interest, however, is that the one study90 that found PCA3 to be least useful was performed in a largely black population and was quite small (45 positive biopsies), leading to a wide confidence interval on the sensitivity estimate.
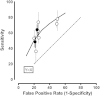
As a way of estimating whether the nine studies were reasonably consistent in their estimates of sensitivity and specificity, a summary analysis was performed for PCA3 (Figure 10). There was high and significant heterogeneity (I2=100 percent. p<0.001). This can be seen in the figure and the table with the two studies90,93 having much higher false positive rates, but only modestly higher sensitivities. Thus, a better summary of the data is the fitted ROC curve shown in Figure 10 [Spearman correlation between the logit (sensitivity) and logit (1-specificity) = 0.76, p=0.01]. This presentation is not subject to the usual strong bias introduced by the tPSA upper cutoff of 10 ng/mL used in two of the studies,91,95 as PCA3 is essentially independent of tPSA measurements (Table 15), and Figure 10 focuses only on the PCA3 results.
Table 15
Measures of independence of PCA3 and tPSA in identifying men with a positive biopsy.
PCA3 and tPSA: ROC Curves-Sensitivity/Specificity
Fourteen studies (Table 13) provided ROC curves for both PCA3 scores and tPSA elevations among men with positive initial screening test results (elevated tPSA with or without positive DRE) who subsequently had positive or negative biopsy results. Two of these studies93,99 reported ROC curves separately for initial and repeat biopsies and, therefore, there are 16 rows/datasets. The performance of PCA3 and tPSA testing are presented in Table 13, sorted from smallest to largest number of enrolled men. For each study, the sensitivities of each marker at preselected false positive (1-specificity) rates were estimated from published ROC curves. These values were recorded to the nearest percent (e.g., sensitivity of 55 percent). The table entries showing test performance are the PCA3 sensitivity, followed, in parentheses, by the incremental increase, or decrease, of tPSA sensitivity. For example, at a false-positive rate (1-specificity) of 20 percent, the first study found a PCA3 sensitivity of 57 percent, which was 19 percent higher than tPSA sensitivity, (i.e., 57 - 19 = 38 percent). Negative numbers indicated that tPSA is performing better; positive numbers indicated PCA3 is performing better.
The last three lines in Table 13 are the median results ignoring matching. That is, the median PCA3 sensitivity is provided along with the median difference computed separately. The first of the three lines summarizes all 13 studies. The next summarizes the four tPSA “grey zone” studies, while the last summarizes the nine remaining studies (non-shaded rows) after the one study90 performed in a mainly black population was removed.
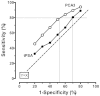
Figure 11 displays the summary ROC curves computed using the PCA3 median sensitivities and median differences provided in the last row of Table 13. This can be taken as a simple summary of performance for the two tests, under similar circumstances in a general population of men with elevated tPSA.
PCA3 and tPSA: Regression Analysis
Two studies31,95 reported sufficient results of regression analysis separately for PCA3 and for tPSA elevations in the same population of men to be included in these analyses. Both studies included in this analysis restricted tPSA levels to less than 10 ng/mL (“grey zone”). Each of the studies reported the odds ratio (OR) for each marker when that marker was assumed to be a continuous variable. That is, the antilog of the OR will be the regression coefficient per unit increase of the marker (e.g., increase of PCA3 score from 30 to 31). This makes comparison of PCA3 and tPSA difficult, as the range of results for the two markers differs. To account for this, the coefficients will be used to compute the ratio of the ORs at the 25th and 75th centiles for each marker. This is a measure of the change in odds over the inter-quartile range. This ratio of ORs for PCA3 will then be divided by the corresponding ratio for tPSA. Values greater than 1 indicates that PCA3 provided more discrimination than tPSA. This normalization also allows for comparisons between studies, where the coefficient is dependent on the range of tPSA values studied.
Only one of the included studies31 provided the inter-quartile ranges for both markers. It was necessary to estimate those ranges for the second study.95 For PCA3, this was done by extrapolating the log mean and SD from two centiles provided as part of the sensitivity/ specificity results. For tPSA, this was done by using the inter-quartile range from the first study31 and adjusting for a minor difference in the mean values reported.
Two additional studies provide some further insight. One91 showed similar coefficients for PCA3 and tPSA, but it was not possible to compute the ratio of the ORs for tPSA because no data were provided to estimate the 25th and 75th centiles. However, given that the inter-quartile range of PCA3 scores were generally larger than the corresponding range of tPSA results, these coefficients were likely to have shown an overall finding of PCA3 being more discriminatory. Another study94 provided only the continuous OR estimates. The PCA3 OR was the highest reported among the four studies in Table 14, and the corresponding OR for tPSA was slightly under 1.0. This would have to be associated with PCA3 being more discriminatory, but estimates of effect size could not be provided because information about the distributions were not provided.
Table 14
Comparison of modeled univariate continuous odds ratios (OR) for PCA3 and tPSA in matched studies.
Five studies31,91,94,95,103 provided some information on the independence of PCA3 and tPSA as markers for prostate biopsy status (Table 15). Two specific measures were sought. Thought to be most useful were the bivariate correlations (parametric or non-parametric) between the two markers for those with positive, and for those with negative, prostate biopsies. Alternatively, logistic regression coefficients (or the corresponding ORs) reported with, and without, adjustment for tPSA were evaluated. In many of the studies reporting logistic regression models, additional factors such as history and prostate volume were also included. If both PCA3 and tPSA coefficients remained essentially constant after adjusting for the other marker (and possibly additional markers), this was taken as evidence that the two markers together were more predictive than either alone (independent).
Three studies31,94,95 reported information on correlation coefficients (Table 15). One reported the two correlation coefficients (non-parametric estimates),94 two reported a single merged correlation (parametric)95, and the third just reported that the correlations were “low” for both groups.31 One potential problem with these estimates is that reliable correlation estimates for both PCA3 and tPSA would require a logarithm transformation prior to computing a parameter estimate such as the Pearson's correlation coefficient. None reported that the data were transformed. Overall, the two markers were not highly correlated in either of the groups of interest.
Five studies31,91,94,95,103 provided information on coefficients for PCA3 and/or tPSA from univariate and multivariate logistic regression modeling (Table 15). Four of the five31,91,95,103 found the PCA3 coefficients unchanged after accounting for tPSA (and often other variables as well). The remaining study94 found a reduction in the coefficient, but it was still the most significant predictor. In addition, this study did not include tPSA in the multivariate model, as it was not statistically significant in the univariate logistic regression (p=0.08).
The results were less consistent for tPSA. Three studies31,91,95 found tPSA essentially unchanged after accounting for PCA3. Interestingly, these three studies all restricted tPSA levels to under 10 ng/mL. This may reduce the correlation between the two markers, if PCA3 and tPSA are concordant when tPSA elevations are relatively high. A fourth study103 did not report the coefficients but did report that the p-value was reduced from being highly significant (p<0.001) to no significance (p=0.52). The fifth94 did not report results for tPSA after adjustment, as it only included variables found to be significant in univariate modeling.
PCA3 and tPSA Elevations: Diagnostic Accuracy
PCA3 and tPSA GRADE Strength of Evidence: LOW
The rationale for “low” follows the GRADE assumption that the high risk of bias in observational studies correlates with a starting strength of evidence of low. The results were deemed to be Consistent, but Indirect. Precision was supported by the ability to observe the expected selection bias of “grey zone” studies and the differences in PCA3 and tPSA performance, but could not be directly measured (e.g., confidence intervals). Strength of association was weak. The results for these domains do not warrant either downgrading to Insufficient or upgrading to moderate.
- Risk of Bias: HIGHThe quality of individual studies was poor. Three biases were identified that could potentially impact this analysis: partial verification bias, spectrum bias and a sampling bias. Partial verification bias was clearly present for tPSA elevation, but our analyses and a review of the literature indicated that in this setting, the ROC curve was unlikely to be biased (Appendix J). Thus, the focus was towards the ROC and related measurements. Monte Carlo modeling was used to account for the verification bias related to the specific cutoff level at which a certain performance was obtained (Appendix J). Sampling bias was accounted for by stratifying the analyses, when possible. Although there was a relatively high potential for bias to affect select measurements and their interpretation, the measures taken as part of the analyses result in a low risk of those biases influencing the final interpretation. Spectrum bias needs to be considered in addition to the performance estimates (sensitivity, specificity). For example, even though PCA3 has a higher sensitivity at any given specificity, the included studies provided no evidence that those identified as positive with either test had similar or different distribution of disease severity. Although PCA3 appears to be statistically better, it does not necessarily follow that it is clinically superior. Publication bias was informally evaluated and not considered to be an important source of potential bias. However, given the poor quality of all the individual included studies, there is potential for unidentified biases to have occurred.
- Consistency: CONSISTENTOverall, analysis showed that PCA3 measurements had higher sensitivity at any specificity compared with tPSA, and higher specificity at any sensitivity. However, it was not possible to formally test for heterogeneity, as original data were not available. No study reported a matched analysis.
- Directness: INDIRECTThe intermediate outcome of diagnostic accuracy (PCA3 and tPSA) shows both types of indirectness: (1) one body of evidence links the test to the intermediate outcome of diagnostic accuracy and another body of evidence is needed to link the test-related intervention(s) to health outcomes; and (2) based on the lack of matched analyses, it is not possible to determine the extent to which PCA3 and tPSA (or other comparators) are identifying cancer with the same or different characteristics (e.g., aggressiveness) within the spectrum of the disease, and yet another body of evidence is needed to resolve this question.
- Precision: PRECISEA formal analysis of precision (e.g., confidence intervals) was not able to be computed due to the matched nature of our analyses and the lack of original data. In one analysis that included 20 studies (AUC difference), it was possible to see the reduction in performance for tPSA in a subset of four “grey zone” studies where the AUC difference expanded to a median of 16 percent, compared with the 8.7 percent found in the 16 studies with no sampling bias.
- Strength of Association: WEAKAlthough there is evidence that PCA3 will be slightly better at identifying high risk individuals with a prostate cancer, both PCA3 and tPSA are relatively weak predictors with low sensitivity and low specificity.
PCA3 and tPSA Elevations—Other Intermediate and Long-Term Outcomes
No studies were identified that reported PCA3 and tPSA levels along with specific information on intermediate (impact on decisionmaking about initial or repeat biopsy, biopsy-related harms) or long-term (morbidity/mortality, quality of life, harms) outcomes.
Strength of Evidence: Insufficient
Summary of the Remaining KQs 1 and 2 “Combined” Analyses
Table 16 provides a summary of the numbers of available studies available for each comparator and outcome, as well as the domains (see footnotes) and strength of evidence for each. More detailed descriptions of the data and limited findings for all outcomes and all PCA3 comparators can be found in Appendix K.
Table 16
KQs: Summary of available studies and strength of evidence for four outcomes and all PCA3 comparators.
KQ 3. Testing for PCA3 and Comparators To Identify Patients with Insignificant Cancer Who May Be Candidates for Active Surveillance
KQ 3 presented a complex clinical scenario. Based on the implementation of tPSA screening and followup, many more prostate cancers are being diagnosed early in the natural history of the disease. The result is the diagnosis of a proportion of cancers that would otherwise not have been diagnosed clinically during the men's lifetimes. Effective risk stratification could inform decisions about whether/when treatment is warranted for such cancers. Alternatively, if risk stratification provides sufficient certainty that the tumor poses little risk to life and health, the patient might benefit from active surveillance and delayed treatment if the disease progresses. The importance of effective schemes for risk stratification was reemphasized by the recent Prostate Cancer Intervention versus Observation Trial (PIVOT)123 report on 12 year followup of men with histologically confirmed localized prostate cancer (mean age 67 years, stage T1-T2, any grade, tPSA <50 ng/mL). They found no difference in all-cause or prostate cancer-specific mortality between men assigned to observation (watchful waiting) versus those randomly assigned to radical prostatectomy treatment.
The identified studies for KQ 3 investigated the performance of PCA3 and comparators in placing men with biopsy confirmed prostate cancer into categories of clinical risk or significance. Reviewing these studies was complicated by variability in terminology and definitions. Low risk tumors were variably referred to as “low risk,”94 “indolent,”89,95,100,117 “insignificant,”100,112 or “low volume/low grade.”115,118 High risk tumors were referred to as “intermediate or high risk,”29,94 “significant,”95,100,115,118 “unfavorable,”89 and “aggressive.”118
Ploussard provides a conceptual definition of insignificant disease as “…a low-grade, small-volume, and organ-confined PCa that is unlikely to progress to clinical and biologic significance without treatment,” and that is diagnosed in clinical practice “...in the absence of cancer-related symptoms that would not have caused disease-specific mortality during the patient's life if untreated.” 67 Indolent cancers have been characterized as those identified early in the natural history of prostate cancer, possibly prospectively detected by pathologic criteria using tools such as nomograms, and having a good chance of positive outcome with active/aggressive treatment.14,67 However, these terms have been used interchangeably. We have chosen to use the term “insignificant” to denote the cancers for which active surveillance may be considered.
The first challenge is identifying individuals with insignificant disease who are eligible for active surveillance.20 The most commonly used criteria used to define insignificant cancer are the Epstein criteria (and modifications).66 The key prognostic factors are Gleason score ≤6 without Gleason pattern 4 or 5, organ-confined disease (no extraprostatic extension, seminal vesicle invasion or lymph node involvement) and tumor volume less than 0.5 cubic centimeters (cc) (sometimes less than 0.2 cc).14,66,116 Other criteria may include clinical stage T1c, PSA density less than 0.15 ng/mL/gram, fewer than three positive cores, and less than 50 percent cancer per core.124 NCCN and others suggests a similar definition for “very low risk.”16 D'Amico low risk criteria are tPSA ≤10 ng/mL, clinical stage T1-T2a, and Gleason score ≤6.125 This review addresses the potential performance of PCA3 score as a criterion for insignificant disease, but also as a potential marker for an aggressive form of cancer.
The second challenge is determining how to effectively identify progression of disease, to get to a measurable clinical endpoint.20 How effective is the risk classification system in identifying men with insignificant cancer (clinical sensitivity and specificity)? What are the harms related to misclassification? Answering these questions requires for each risk classification (e.g., insignificant/very low risk, low risk, intermediate risk, high risk) specified measures of progression over time, with and without treatment, as well as assessment of harms and all-cause and prostate-cancer-specific mortality rates.
A validation study of Epstein criteria for insignificant disease in European men found that classification by biopsy criteria “may underestimate the true nature of prostate cancer.” At radical prostatectomy, 24 percent of patients with “insignificant” disease had Gleason sum 7-10 scores and 34 percent had non-organ-confined disease.126 A recent systematic review reported on the accuracy of the Epstein criteria in predicting insignificant prostate cancer.127 Five of six studies defined insignificance by biopsy criteria and used concordance with prostatectomy pathology to determine accuracy; one study followed biochemical recurrence-free survival for six years. They found significant heterogeneity among the validation studies that was attributed in part to different criteria, variable application of criteria, and changes in the Gleason scoring system. Lack of clinical followup may also be a factor. They concluded that Epstein criteria have suboptimal accuracy for predicting insignificant prostate cancer and require additional, better quality validation studies127 So, in addition to finding new and most effective markers, better designed validation studies are also needed.
Description of Included Studies
The inclusion criteria for KQ 3 were also set to select only matched studies. These are defined as studies that provide estimates of test performance or other outcomes for PCA3 and at least one other comparator using the same sample set. Studies of PCA3 alone, or of other comparators without PCA3, were, therefore, excluded. Thirteen studies were identified that addressed KQ 3 and reported on PCA3 and other preoperative/pretreatment markers for stratifying tumors by risk (Table 17).88,89,94,95,100,112-119 Two studies based analyses on biopsy markers without prostatectomy94,95 and eight reported prostatectomy results as an endpoint.88,100,112,114-116,118,119 Two studies were conducted on subjects with longitudinal data including short-term followup.88,89Tables 1 through Table 4 include descriptive information about these studies. Table 17 provides information and results. Table 18 provides detailed information about the wide variety of markers investigated in these studies for association with low and high risk disease.
Prostatectomy is not useful as an endpoint for determining diagnostic accuracy because it is not a clinical outcome, but rather an intermediate step. Pathological testing of prostatectomy specimens adds data to further assess the tumor as high or low risk, including tumor volume, prostatectomy Gleason score and stage, possible upgrading from biopsy, and other pathological findings (e.g., extracapsular extension, perineural invasion, positive surgical margins). However, the association of PCA3 with these markers, or the ability of PCA3 to predict them at prostatectomy, relates to the determination of risk category, but does not provide the formal evidentiary link between the risk assigned and specific intermediate and long-term clinical outcomes. Without even short-term specified clinical endpoints or validated surrogates, these data cannot be used to provide estimates of diagnostic accuracy.
The included articles address a wide range of comparators (Table 18), many different combinations of criteria defining individuals with “low” or “high” risk prostate cancers, and varying presentations and analyses of the data. However, one result was most consistently reported, and that was an association between PCA3 score and tumor volume (Table 17). Most studied PCA3 and comparators as potential predictors of insignificant cancer, while others reported possible use for identifying aggressive cancer.116,117 Three studies reported that PCA3 was an independent predictor of tumor volume, though cutoffs and endpoints (greater than or less than the 0.5cm3 tumor volume cutoff) differed.113,116,119 Five studies113,115,116,118,119 reported higher correlations between PCA3 and tumor volume (r = 0.27 to 0.41; p ≤ 0.04) than for other comparators (e.g., tPSA, %fPSA, PSA density, clinical stage, biopsy Gleason score). Unfortunately, those correlations may be suspect. Most studies did not provide scatterplots of the data. The two that did89,116 clearly show that the data for both PCA3 and tumor volume should be subject to transformation prior to computing the correlation. Figure 12a redisplays data published by Ploussard.116 These data were digitized from a provided figure and should be considered reasonably accurate, but not as reliable as original raw data. This analysis is aimed at demonstrating a more appropriate analytic methodology. The correlation is lower after transformation (but still significant). However, examining the data (Figure 12b), it appears that most of the “prediction” is confined to very high PCA3 scores (>120) associated with very large tumors (>2 cm3).
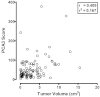
Figure 12a
Scatterplot illustrating the correlation between PCA3 scores and tumor volume in prostatectomy specimens on a linear scale (data from Ploussard et al., 2011). PCA3 = prostate cancer antigen 3 gene Note: For each subject, PCA3 score is shown on the x-axis (more...)
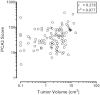
Figure 12b
Scatterplot illustrating the correlation between PCA3 scores and tumor volumes in prostatectomy specimens after logarithmic transformation (reanalysis of data from Ploussard et al., 2011). PCA3 = prostate cancer antigen 3 gene Note: For each subject, (more...)
Only two of the studies reviewed for this report had a longitudinal component and described a clinical outcome other than pathological results of prostatectomy.88,89
- Lymph node involvement in a prostate cancer patient is an indicator of poor clinical outcome. One study88 attempted to identify “micrometastases,” based on identifying tumor cells within the lymph nodes that are producing the prostate cancer markers tPSA and PCA3. The method used to quantify these markers in lymph node extracts was real time reverse transcriptase PCR (RT-PCR) for both PCA3 and PSA mRNA. The study followed 120 patients with localized prostate cancer for 4 to 6 years and used biochemical recurrence (any serum tPSA greater than 0.2 ng/mL) as the surrogate outcome of interest. As expected, they found significantly decreased biochemical recurrence free survival among the 11 subjects with histologically confirmed lymph node metastases, compared with 77 subjects with no lymph node involvement. Among the remaining 32 patients with biochemical recurrence, many were identified as having micrometastases based on either tPSA or PCA3 (or both) testing. tPSA had a sensitivity for biochemical recurrence of 73 percent and a false positive rate of 22 percent (p<0.001). PCA3 had a lower detection (42 percent) and a comparable false positive rate (23 percent), but the effect was not significant (p=0.095). While this appears to indicate that PSA testing is more predictive, the use of PSA mRNA as the test, and a rise in serum tPSA levels as the outcome suggests an important risk of bias. The authors provided no information on validation of quantitative testing for these biomarkers in this sample type or on confirmation of the results using a published method.
- Based on no more than 2-year followup of patients in an active surveillance program, Tosoian et al. reported PCA3 and tPSA results (mean, standard deviation, median) for the 38 of 294 patients progressing to treatment based on yearly biopsy results.88,89 Epstein criteria were used for initial enrollment in the surveillance program. Progression to treatment was recommended for “unfavorable” findings, defined as any Gleason pattern 4 or 5, greater than 2 positive biopsy cores, or more than 50 percent involvement of any core with cancer. No difference in PCA3 and tPSA levels was observed between the 13 percent who progressed and those remaining in active surveillance (p=0.13). However, the authors state that only 140 of the 294 study subjects submitted a urine sample, and did not report how many of these 140 men had an unfavorable result on biopsy. This study did not provide matched results for all subjects (partially matched).
No studies were identified that reported on other intermediate outcomes (e.g., diagnostic accuracy, decisionmaking, harms) or long-term clinical outcomes (e.g., mortality/survival, morbidity, quality of life). All studies were judged to be poor quality, mainly due to lack of clinical followup, but also to lack of information on study subjects. Six studies were funded by GenProbe and six disclosed authors with potential conflicts of interest (Table 1); others did not report on source of funding or conflicts of interest (Table 1). The detailed results of assessment of quality of individual studies addressing KQ 3 are presented in Table F-2 in Appendix F.
PCA3 and Comparators—Intermediate Outcome: Diagnostic Accuracy
- Risk of Bias: HIGHThe quality of individual studies was poor. All studies were observational, raising a high potential for biases to have occurred.
- Consistency: UNKNOWNNo studies were identified that reported on matched data for PCA3 and comparator results, and also reported specific clinical outcomes of patients with tumors characterized as low risk and high risk, who:
- –
opted for active surveillance and never progressed to treatment;
- –
opted for active surveillance and progressed to treatment; or
- –
opted for immediate treatment.
No effect(s) could be measured. - Directness: DIRECTThis should be Direct, as the evidence would ideally determine diagnostic accuracy by linking the risk assignment based on testing/pathological results directly to health outcomes.
- Precision: IMPRECISEThis cannot be assessed, as no comparisons were possible based on the two studies of different populations, using different assays, and reporting different surrogate outcomes.
Strength of Evidence: Insufficient
Strength of evidence could not be evaluated. Only two studies were identified, and they did not perform the studies in the same setting, have the same sample type or have comparable outcome measures.
PCA3 and Comparators—Intermediate Outcome: Impact on Decisionmaking
No studies were identified that reported PCA3 and comparator results and intermediate outcome data (e.g., physician or patient surveys, chart review) on the degree to which PCA3 or comparator test results and categorization of risk as high or low impacted decisions made with regard to selection of active surveillance versus aggressive treatment.
Strength of Evidence: Insufficient
PCA3 and Comparators—Intermediate and Long-Term Outcome: Treatment-Related Harms
Studies have been conducted that document treatment-related clinical harms such as incontinence and impotence. Based on general studies on potential psychosocial harms of diagnostic testing, it is possible to generalize that patients facing treatments such as radical prostatectomy might also experience anxiety or perceive a reduction in quality of life. However, no studies were identified that reported PCA3 and comparator test results and intermediate outcome data (e.g., physician or patient-reported adverse events, biochemical recurrence, progression to treatment) on the degree to which categorization of risk as high or low and choice of active surveillance or treatment related to the occurrence of adverse clinical events.
Strength of Evidence: Insufficient
PCA3 and Comparators—Intermediate and Long-Term Health Outcomes
No studies were identified that reported PCA3 and comparator results and the association of low and high risk categorization with long-term outcomes such as mortality/survival and morbidity (e.g., function, quality of life) of the selected course of management or treatment. However, two poor quality studies reported on relatively short-term health outcomes, biochemical recurrence and progression from surveillance to treatment in an active surveillance program.
Strength of Evidence: Insufficient
- Results - PCA3 Testing for the Diagnosis and Management of Prostate CancerResults - PCA3 Testing for the Diagnosis and Management of Prostate Cancer
- Alpha-1,6-mannosyl-glycoprotein 6-beta-N-acetylglucosaminyltransferase [Caenorha...Alpha-1,6-mannosyl-glycoprotein 6-beta-N-acetylglucosaminyltransferase [Caenorhabditis elegans]gi|392901762|ref|NP_502715.3|Protein
- Caenorhabditis elegans Unclassified non-coding RNA F59G1.10 (F59G1.10), ncRNACaenorhabditis elegans Unclassified non-coding RNA F59G1.10 (F59G1.10), ncRNAgi|392893011|ref|NR_051125.1|Nucleotide
Your browsing activity is empty.
Activity recording is turned off.
See more...