NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Bradley LA, Palomaki G, Gutman S, et al. PCA3 Testing for the Diagnosis and Management of Prostate Cancer [Internet]. Rockville (MD): Agency for Healthcare Research and Quality (US); 2013 Apr. (Comparative Effectiveness Reviews, No. 98.)
This publication is provided for historical reference only and the information may be out of date.
Introduction: Partial Verification Bias for Total PSA (tPSA)
Study design is an important inclusion criterion for this comparative review, because tPSA measurements, the main comparator to PCA3, are integral to men's decision-making regarding prostate biopsy. Men will be offered prostate biopsy based on the extent of tPSA elevations, suspicious findings on a digital rectal exam (DRE), a combination of the two or, less commonly, other risk factors such as family history or race. In order to obtain an unbiased estimate of diagnostic accuracy for tPSA at specific cut-offs, it is necessary that the identification of prostate cancer not be related to tPSA levels. This is a potential problem as most clinicians believe that higher tPSA levels are indicative of a higher likelihood for the presence of prostate cancer. Men are more likely to undergo prostate biopsy, if the tPSA is high (e.g., 10-20 ng/mL), rather than close to lower cut-offs used to define a positive tPSA screening test (e.g., 3-4 ng/mL). If a study reports results in which biopsy is tPSA-related, the sensitivity and specificity at select tPSA cut-offs will not be accurate. If those not accepting biopsy are considered missing, this is considered ‘partial verification’ bias. All studies included in the evidence review are opportunistic cohorts of men agreeing to biopsy and will likely be subject to this bias.
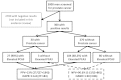
An example of how partial verification bias occurs is demonstrated in Figure J1. These numbers are intended to be representative of clinical practice, but are mainly designed to demonstrate partial verification bias in diagnostic studies of tPSA and prostate cancer. Among a cohort of as many as 3,000 men over 50 years of age, a subset of 300 men have been identified as having tPSA values of 3 ng/mL or higher. The remaining 2,700 men had low tPSA values and are not shown. For simplicity, consider all men with tPSA over three to be categorized into only two groups defined by the extent of tPSA elevation (3-6 ng/mL and ≥7 ng/mL). The prevalence of prostate cancer in the group with modestly elevated tPSA will be somewhat lower (10%) than the prevalence (16%) in the group with high tPSA elevations. At this point, the ‘true’ sensitivity and associated false positive rate (1-specificity) using an arbitrary tPSA cut-off of 7 ng/mL can be determined to be 44% (16/(16+20)) and 32% (84/(84+180)), respectively (shaded box with solid outline). The associated group likelihood ratio is 1.38 (44%/32%). These ‘correct’ results would have been obtained if all 300 men were to have had biopsies, assuming that biopsy is a perfect test for identify prostate cancer. A less biased result could also be obtained if, in addition to the subset having biopsies, all remaining men were followed longitudinally for several years to identify future cancer diagnoses.
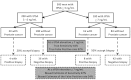
Figure J1
A schematic showing the impact of partial verification bias on the sensitivity and false positive rate for total PSA (tPSA) and prostate biopsy. A complete description is provided in the text.
In clinical practice, however, the uptake rate for prostate biopsy will depend on the group to which the men belong. In the group with lower tPSA levels, the uptake rate may be as low as 20%. In the group with higher tPSA levels, the uptake rate may be as high as 50%. These uptake rates, and results in men choosing biopsy are shown by the dashed arrows and boxes. Among the group of men choosing biopsy based loosely on the tPSA level, the sensitivity and false positive rate are now 67% (8/(8+4)) and 54% (42/(42+36)), respectively. The corresponding cumulative likelihood ratio is 1.24 (67/57).
Figure J1 shows that partial verification bias in this setting using a cut-off of 7 ng/mL tends to overestimate the sensitivity (67% versus 44%), as the men with cancer having with low tPSA are less likely to be identified (false negatives). The bias will also overestimate the false positive rate (54% versus 32%) as the men without cancer having low tPSA are also less likely to be identified (true negatives).
Literature Regarding Partial Verification Bias for tPSA elevations
An important paper on partial verification bias and diagnostic accuracy of tPSA addressed this population-based screening test.1 That study focused on a data set of men having tPSA testing in which men with both normal and elevated tPSA levels were offered biopsies. This differs from the setting of the current review which focuses on using the extent of tPSA elevations among men already identified as being ‘high risk’ due to at least modest elevations of tPSA (e.g., >2.5 ng/mL, ≥3 ng/mL) and/or other factors already described. Punglia and colleagues reported that partial verification bias has two separate and important effects: 1) the performance of tPSA measurements is actually better than the reported performance subject to partial verification bias, and 2) the cut-off level at which select sensitivities and associated false positive rates occur are quite different in biased versus unbiased studies.1 For example, they reported the area under the ROC curve for men under age 60 years to be 0.69 with partial verification bias, and 0.86 after accounting for the bias (p<0.001). In that same group, a tPSA cut-off of 4.1 ng/mL was associated with a sensitivity and false positive rate of about 43% and 22% with bias; 18% and 2% after adjustment. One limitation to that study was the use of a categorical uptake rate to model the presence of cancer in men without biopsies. This may have caused at least some of the difference in performance.
A later study2 also adjusted for verification and found that the ROC curve was essentially unchanged (AUC 0.682 among verified, and 0.678 after accounting for partial verification). However, a relatively high proportion (64%) of the population was biopsied. We verified by in-house modeling that in the setting of the current review, it is likely that the ROC will not be substantially impacted (modeled uptake proportional to tPSA, ranging from 5% to 50%, with 30% overall verification). Several additional studies have assumed that a single cut-off will be used, and focus only on correcting the estimates of sensitivity and specificity at that cut-off, and do not consider the issue of whether the biased estimates of sensitivity and specificity falls on the true ROC curve or not.3-7
There are two important factors that appear to influence the extent of partial verification bias:
- the differential rates of cancer (positive prostate biopsy) in the group below the tPSA cut-off compared to the group above the cut-off. If the rates of cancer are the same in the two groups, no partial verification bias will occur. The greater the relative difference (rate ratio) between groups, the larger effect the partial verification bias will have.
- the differential biopsy uptake rates in the two groups. If the biopsy uptake rates are the same, there will be no partial verification bias, even if the rates of cancer are different. Analogously, if there is no difference in the cancer rates, there will be no partial verification bias even if the uptake rates differ.
The setting of the current evidence review differs from that explored by Puglia and colleagues1 in two important ways. First, the differential rate of cancer in studies relevant to this review (all having at least modest elevations) is likely to be considerably less. This suggests that the partial verification bias might have less impact in the current setting. Second, the biopsy uptake across the more limited range of tPSA in this review/setting is likely to be more similar. Lastly, the overall uptake rate will also be higher, as all of the men in our setting would all have an elevated tPSA (and/or positive DRE or other factor). All of these differences would tend to reduce the effect of partial verification bias. Of most importance is our finding that the ROC curves (and, by extension, AUC and sensitivity/specificity estimates) are likely to not be impacted by partial verification bias for tPSA measurements, confirming a previous finding2.
Review of Methodology Specific to the PCA3/tPSA Comparison
The aims of the following interpretative analyses are to estimate the unbiased performance of tPSA and PCA3 to identify ‘at risk’ men who will have a negative, or positive, biopsy. The analysis will be anchored by two important findings. First, the ROC curves for tPSA (and for PCA3) are not influenced by the partial verification bias. Thus, any set of parameters (distribution descriptors such as means and standard deviations for PCA3 and comparators in both biopsy negative and positive men) we might generate, would have to fit that data. Second, the literature contains sufficient information to estimate the needed parameters, but those estimates will be subject to the partial verification bias for tPSA and related markers, but not for PCA3. If the reported tPSA parameters can be ‘unbiased’ and fitted to the relevant ROC curve, then a direct comparison between tPSA and PCA3 at selected cut-off levels could be made. Based on published data, the distributions of these markers are likely to be Gaussian, after a logarithmic transformation. To simplify the analysis, we have also chosen to force the logarithmic standard deviations within a study to be the same for each marker. This was accomplished by an un-weighted pooling of the estimated variances. According to the literature, this seems to be a reasonable assumption. The secondary aims of these analyses are to provide improved templates for more reliably exploring the comparison of prostate cancer markers and assist in providing methods to more fully inform decision-making by men and their health care providers.
Interpretative Analysis for PCA3
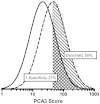
Figure J2 shows how overlapping distributions of PCA3 can be used to generate sensitivity and false positive rates (1-specificity). The baseline data used to estimate the median and logarithmic SDs are provided in the body of the evidence review (Table 11). Eight studies8-15 reported sufficient data for this analysis, after excluding the two studies14, 15 that focused on the grey zone of tPSA. Although the reported median values are somewhat variable, the PCA3 scores are about 2.2 (median ratio) times as high in those with a positive versus a negative biopsy. These results are not subject to partial verification bias because tPSA and PCA3 levels are essentially independent markers (Evidence Review, Table 15). There is more consensus on the pooled logarithmic SDs being at about 0.420. The median PCA3 score among those with a negative biopsy was 21. Using the 2.2 multiplier, a reasonable expected median PCA3 score among those with a positive biopsy would be around 46 (2.2 * 21). These population parameters fit the consensus ROC data well, as shown in the next paragraph and Figure J3.
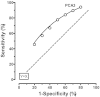
Figure J3 shows seven points on the ROC curve for PCA3 derived from the literature review (Evidence review, Table 13). These points correspond to false positive rates of 20% through 80%, in 10% intervals. The dashed line indicates a ‘useless’ test as a reference (sensitivity equal to the false positive rate). The solid line has been derived from the overlapping distributions shown in Figure J2, and fit well. It is not possible to formally test the fit, as most of the original data were obtained from the published figures, rather than from original data. This model allows the derivation of expected sensitivity and false positive rates across the range of specific cut-off values, and provides an estimate of what PCA3 score is associated with that test performance. According to the relevant studies, PCA3 scores in men with negative (and positive) biopsies vary between reports, so the modeled PCA3 cut-offs may not be appropriate in all settings. For this reason, the results of this analysis should be used only as a guide. This gap in knowledge could be addressed in future research by examining normalizing functions such as multiple of the median PCA3 score, where the median level is determined in a healthy male population without prostate cancer.
Interpretative Analysis for tPSA
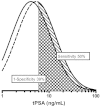
Similar data were available to create a tPSA model, but with the caveat that partial verification bias must be accounted for in determining the parameters. The tPSA literature was more consistent than PCA3 in the reported median and logarithmic SDs for tPSA (Evidence Review, Table 9). For the same six studies8-13, the median tPSA in those with a negative biopsy was 6.3 ng/mL. The median ratio between the tPSA in positive versus negative biopsies was 1.30. Thus, the expectation is that the tPSA level would be about 8.2 ng/mL (6.3 × 1.3) in men with positive biopsies. The consensus logarithmic SD was about 0.31. Unfortunately, partial verification bias will inflate the median value, as lower tPSA measurements in men both with and without cancer will be under-represented. The bias will also tend to shrink the logarithmic standard deviation. In order to estimate how much lower and broader to make the distributions, we generated log Gaussian distributions and subjected them to partial verification bias to study the effect. In one of these models, the uptake rate was estimated to be the tPSA value divided by 20, with an upper limit of 0.5 (50% uptake). For example, men with a tPSA of 8 ng/mL would have an uptake rate of 40% (8/20) while those with a tPSA of 4 ng/mL would have a 20% uptake rate. We found the variance increased by about 40%, while the median value was reduced by about 35%. When we applied these correction factors to the consensus reported medians and logarithmic SD described above, slight adjustments in the reduction in median values were still needed to more closely match the reported tPSA ROC curve. The final parameters are shown in Figure J5. These data must be viewed as approximate. However, knowing that the necessary data to produce an observed and unbiased estimate is likely to be unattainable, it may be preferable to reporting that the analysis cannot proceed due to the existence of partial verification bias.
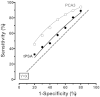
Figure J5 shows seven points on the ROC curve for tPSA derived from the literature review. Only data from the six studies8-13 that did not restrict tPSA measurements to the ‘grey zone’ were included (Evidence Review, Table 11). These points will be essentially unchanged whether or not partial verification bias is accounted for, and correspond to false positive rates of 20% through 80%. The dashed line indicates a ‘useless’ test as a reference (sensitivity equal to the false positive rate). The solid line has been derived from the overlapping distributions show in Figure J4, and fits well.
Interpretative Analysis Comparing PCA3 and tPSA Performance
These parameter sets for PCA3 scores and tPSA elevations can now be used to compare their performance in identifying men who will have a positive versus a negative biopsy. The following analyses rely only on the parameters presented in the legends to Figures J3 and J5 along with three selected rates of cancer in the population (5%, 10% and 15%). The reported rates from the reviewed literature ranged between 10% and 30%. However, due to partial verification bias, the actual prostate cancer rates are likely to be somewhat lower.
Table J1 presents a series of PCA3 score cut-off levels with accompanying false positive rates (1-specificity) and sensitivity (detection) rates. For example, using the common PCA3 cut-off score of 35 and higher, 29.9% of men without cancer would still undergo biopsy. Alternatively, this can be looked at as avoiding an unneeded biopsy in70.1% of men without cancer. The trade-off to this reduction in biopsy is that the detection rate is only 61.1%, indicating that this would result in 38.9% of prostate cancers in the population being missed. The last two columns include the cumulative likelihood ratio (sensitivity / false positive rate) and the individual likelihood ratio (ratio of the heights of the two curves at the given value). For example, using the same score of 35 as the cut-off, the screen positive men will have 2.0 times the risk of prostate cancer compared to the entire cohort of men identified with elevated tPSA levels and/or positive DRE or other findings. For men with a score of exactly 35, their risk is increased by only 16% (LR=1.16) over the cohort as a whole.
Table J1
Sensitivity, false positive rates and likelihood ratios for PCA3 and tPSA elevations.
The bottom half of Table J1 presents the data in the same way, but for tPSA measurements. For example, 34.7% of the men who would have a negative biopsy would still have a tPSA of six ng/mL or higher (false positive rate). Correspondingly, about 44.9% of the men with cancer would have levels at or above six ng/mL. (It is important to remember, this 44.9% is a conditional proportion of men with cancer among the cohort of men over age 50 identified with an initially elevated tPSA and/or with other markers. It is not the proportion of all men over age 50 with prostate cancer.) The cumulative likelihood indicates an increase in risk of 30% (LR=1.3), while an individual man with a tPSA of 6 ng/mL would have little change in risk (LR=1.07). To allow for direct comparisons, the range of cut-offs shown for the two markers cover approximately the same proportion of the overlapping distributions. The format of Table J1 makes comparisons between the two markers difficult, as neither the sensitivity nor the false positive rate is being held constant.
Table J2 compares the performance of PCA3 scores and tPSA measurement to identify men who would (or would not) have a positive biopsy. The top half of the Table J2 holds the false positive rate constant, while the bottom half holds the sensitivity constant. In the last column is the difference between the two estimates (PCA3 – tPSA). When comparing the sensitivities (top half) this column contains the improvement in prostate cancer detection at a fixed false positive rate. When comparing the false positive rates, it contains the reduction in unnecessary biopsies.
Table J2
A comparison of PCA3 scores and tPSA elevations to identify men with prostate cancer with either the false positive rate, or the sensitivity held constant.
As an example, assume you would like to choose a cut-off so that no more than 10% of the existing cancers would be missed (i.e., false negative on the PCA3 or tPSA tests). Choose the highlighted row with 90% sensitivity (Table J2). It shows the false positive rate would be 68% using PCA3 scores, but 85% using tPSA measurements. This means that the same proportion (90%) of cases might be detectable while performing 17% fewer biopsies were PCA3 to be used instead of tPSA elevations.
These analyses do not account for the prevalence of cancer in the cohort, so cannot determine the positive or negative predictive values. Figure J6 shows how these data can be used with cancer prevalence to determine predictive values. It also shows how the cohort of interest is limited to the population of men who have elevated tPSA/positive DRE. Assume the prevalence of cancer is 5%, and the PCA3 related sensitivity is chosen to be 90%. Based on Table J2, this implies a PCA3 cut-off score of 13.3 and will have a false positive rate of 68.2%. (As an aside, the analysis is more confident in the performance estimates of 90% sensitivity and corresponding false positive rate than that those rates occur at a score of 13.8.) Depending on the situation, the relevant PCA3 score may be higher or lower, depending on factors that have not yet been clarified.)
The flowchart (Figure J6) first assumes that a cohort of 3,000 men is subject to prostate cancer screening and 10% are found to be ‘at risk’ due to elevated tPSA, positive DRE and/or other factors. Only these 300 are relevant to the current evidence review. The PCA3 score is then applied to this subset of 300 men, 30 of whom have prostate cancer (10%). Using the example from the previous paragraph, assume a 90% sensitivity for PCA3 testing (cut-off score of 13.3). Among the 30 men with cancer, the PCA3 score will be 13.3 or higher in 27 (90%). Among the 270 men without cancer, PCA3 will be similarly elevated in 184 (68.2%). Among the 211 with a positive PCA3 (27 TP + 184 FP), the positive predictive value (PPV) is 13% (27/211). Another way to express the PPV is via odds. The PPV of 13% is equivalent to odds of 1:6.8 (this could be read as odds of 1 to 6.8 or as a proportion of 1 in 7.8).
This is also called the odds of being affected given a positive result (OAPR). Among the 89 men with a PCA3 score under 13.3, 86 men will not have cancer, resulting in a negative predictive value (NPV) of 96.6%. Another way to understand the NPV is to create odds of having cancer among those with a negative test result. This is called the odds of being affected given a negative result (OANR); in this example, 1:30.
Table J3 provides a summary of positive and negative predictive values for both PCA3 scores and tPSA at seven different sensitivities ranging from 50% to 95%, each with three different cancer rates (5%, 10% and 15%). Within each of the groups with the same sensitivity, the PPV increase while the NPV decreases. Between groups, increasing sensitivity is associated with higher PPV and lower NPV rates, at the same cancer rate. When comparing across the table (PCA3 versus tPSA), both the PPV and NPV are slightly lower.
Table J3
Positive and negative predictive values for PCA3 and tPSA testing at seven selected test sensitivities, each with three different rates of prostate cancer.
Up to this point, all tables and figures were designed to assist health care providers understand each test's performance, evaluate the trade-offs at selected cut-off levels, and compare performance between tests. This is of less interest to an individual male who does not have a PCA3 (or tPSA) level at or above a cut-off level, but instead has a ‘patient –specific’ measurement that could be interpreted for that individual. Table J4 provides this information at select PCA3 and tPSA levels. There are, however, too many possibilities to provide a complete listing, but this type of information could easily be part of a computerized report. Such information could also be tailored to include other relevant risk factors such as family history of prostate cancer or race. The table allows for prior risks of between 5% and 30%. As an example, consider a male with a relatively low PCA3 score of 10. Where his prior risk to be low at 5% (e.g., 50 y.o. white male, no family history and his initial screening test results to be only slightly elevated with a negative DRE), then the PCA 3 score would reduce his risk to 1:106. However, if his PCA3 score more elevated, at 45, his risk would be slightly increased to 1:14. In general, a stronger marker is more able to modify the initial risks. This can be seen by scanning the first column of risks. PCA3 risks vary more than do the corresponding tPSA-related risks, even though the ranges of values are similar (footnote, Table J4).
Table J4
Patient-specific prostate cancer risks by an individual's prior risk of prostate cancer (1:N) and by PCA3 score or tPSA elevation.
References
- 1.
- Punglia RS, D'Amico AV, Catalona WJ, et al. Effect of verification bias on screening for prostate cancer by measurement of prostate-specific antigen. N Engl J Med. 2003 Jul 24;349(4):335–42. [PubMed: 12878740]
- 2.
- Thompson IM, Ankerst DP, Chi C, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/ml or lower. JAMA. 2005 Jul 6;294(1):66–70. [PubMed: 15998892]
- 3.
- Begg CB, Greenes RA. Assessment of diagnostic tests when disease verification is subject to selection bias. Biometrics. 1983 Mar;39(1):207–15. [PubMed: 6871349]
- 4.
- de Groot JA, Janssen KJ, Zwinderman AH, et al. Correcting for partial verification bias: a comparison of methods. Annals of epidemiology. 2011 Feb;21(2):139–48. [PubMed: 21109454]
- 5.
- de Groot JA, Janssen KJ, Zwinderman AH, et al. Multiple imputation to correct for partial verification bias revisited. Stat Med. 2008 Dec 10;27(28):5880–9. [PubMed: 18752256]
- 6.
- Gupta A, Roehrborn CG. Verification and incorporation biases in studies assessing screening tests: prostate-specific antigen as an example. Urology. 2004 Jul;64(1):106–11. [PubMed: 15245945]
- 7.
- Rosman AS, Korsten MA. Effect of verification bias on the sensitivity of fecal occult blood testing: a meta-analysis. Journal of general internal medicine. 2010 Nov;25(11):1211–21. [PMC free article: PMC2947648] [PubMed: 20499198]
- 8.
- Ankerst DP, Groskopf J, Day JR, et al. Predicting prostate cancer risk through incorporation of prostate cancer gene 3. J Urol. 2008 Oct;180(4):1303–8. discussion 08. [PubMed: 18707724]
- 9.
- Auprich M, Augustin H, Budaus L, et al. A comparative performance analysis of total prostate-specific antigen, percentage free prostate-specific antigen, prostate-specific antigen velocity and urinary prostate cancer gene 3 in the first, second and third repeat prostate biopsy. BJU Int. 2012 Jun;109(11):1627–35. [PubMed: 21939492]
- 10.
- Bollito E, De Luca S, Cicilano M, et al. Prostate cancer gene 3 urine assay cutoff in diagnosis of prostate cancer: a validation study on an Italian patient population undergoing first and repeat biopsy. Analytical and quantitative cytology and histology / the International Academy of Cytology [and] American Society of Cytology. 2012 Apr;34(2):96–104. [PubMed: 22611765]
- 11.
- Hessels D, van Gils MP, van Hooij O, et al. Predictive value of PCA3 in urinary sediments in determining clinico-pathological characteristics of prostate cancer. Prostate. 2010 Jan 1;70(1):10–6. [PubMed: 19708043]
- 12.
- Nyberg M, Ulmert D, Lindgren A, et al. PCA3 as a diagnostic marker for prostate cancer: a validation study on a Swedish patient population. Scand J Urol Nephrol. 2010 Dec;44(6):378–83. [PubMed: 20961267]
- 13.
- Ochiai A, Okihara K, Kamoi K, et al. Prostate cancer gene 3 urine assay for prostate cancer in Japanese men undergoing prostate biopsy. Int J Urol. 2011 Mar;18(3):200–5. [PubMed: 21332814]
- 14.
- Ferro M, Bruzzese D, Perdona S, et al. Predicting prostate biopsy outcome: prostate health index (phi) and prostate cancer antigen 3 (PCA3) are useful biomarkers. Clin Chim Acta. 2012 Aug 16;413(15-16):1274–8. [PubMed: 22542564]
- 15.
- Perdona S, Cavadas V, Di Lorenzo G, et al. Prostate cancer detection in the “grey area” of prostate-specific antigen below 10 ng/ml: head-to-head comparison of the updated PCPT calculator and Chun's nomogram, two risk estimators incorporating prostate cancer antigen 3. European urology. 2011 Jan;59(1):81–7. [PubMed: 20947244]
- 16.
- Deras IL, Aubin SM, Blase A, et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol. 2008 Apr;179(4):1587–92. [PubMed: 18295257]
- 17.
- Goode RR, Marshall SJ, Duff M, et al. Use of PCA3 in detecting prostate cancer in initial and repeat prostate biopsy patients. Prostate. 2012 May 14 [PubMed: 22585386]
- 18.
- Rigau M, Morote J, Mir MC, et al. PSGR and PCA3 as biomarkers for the detection of prostate cancer in urine. Prostate. 2010 Dec 1;70(16):1760–7. [PubMed: 20672322]
- 19.
- Schilling D, Hennenlotter J, Munz M, et al. Interpretation of the prostate cancer gene 3 in reference to the individual clinical background: implications for daily practice. Urol Int. 2010;85(2):159–65. [PubMed: 20424427]
- 20.
- Wu AK, Reese AC, Cooperberg MR, et al. Utility of PCA3 in patients undergoing repeat biopsy for prostate cancer. Prostate Cancer Prostatic Dis. 2012 Mar;15(1):100–5. [PubMed: 22042252]
- Introduction: Partial Verification Bias for Total PSA (tPSA)
- Literature Regarding Partial Verification Bias for tPSA elevations
- Review of Methodology Specific to the PCA3/tPSA Comparison
- Interpretative Analysis for PCA3
- Interpretative Analysis for tPSA
- Interpretative Analysis Comparing PCA3 and tPSA Performance
- References
- Detailed Description of PCA3 and Total PSA Interpretive Analysis - PCA3 Testing ...Detailed Description of PCA3 and Total PSA Interpretive Analysis - PCA3 Testing for the Diagnosis and Management of Prostate Cancer
- Exact Search Strings - Interventions To Improve Cardiovascular Risk Factors in P...Exact Search Strings - Interventions To Improve Cardiovascular Risk Factors in People With Serious Mental Illness
- Pry3p [Saccharomyces cerevisiae S288C]Pry3p [Saccharomyces cerevisiae S288C]gi|6322383|ref|NP_012457.1|Protein
Your browsing activity is empty.
Activity recording is turned off.
See more...