From: Chapter 18, The Reference Sequence (RefSeq) Database

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
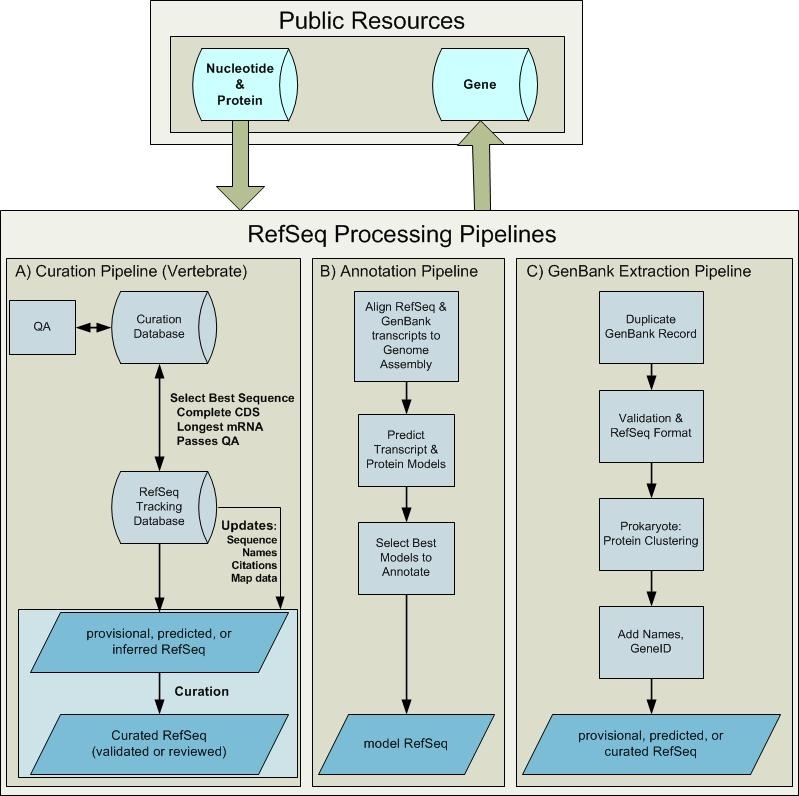
RefSeq Processing Pipelines. Sequence data deposited in the public archival databases is available for RefSeq processing. Processing pipelines include the vertebrate curation pipeline, the computational genome annotation pipeline, and extraction from GenBank. These pipelines generate new and updated RefSeq records that become publicly available in Entrez Nucleotide, Protein, and Gene databases. (A) Once a gene is defined and associated with sufficient sequence information in an internal curation database, it can be pushed into the RefSeq pipeline. The RefSeq process is initiated by selecting the longest mRNA annotated with a complete coding sequence for each locus. This RefSeq record has a status of PROVISIONAL, PREDICTED, or INFERRED. Subsequent curation may result in a sequence or annotation update and a RefSeq status of VALIDATED or REVIEWED. Records are updated if the underlying INSDC submission is updated or if other associated data are updated, including nomenclature, publications, or map location. (B) Available RefSeq and INSDC data are aligned to an assembled genome, ab initio gene prediction that uses the alignment data is performed, and an analysis program integrates all available data to define the annotation models. New MODEL RefSeq records are generated by this pipeline. (C) When a complete, annotated genome becomes available in the INSDC, a set of corresponding RefSeq records are generated by duplicating the GenBank records, followed by validation and addition of cross-references to Gene (via a db_xref citing the GeneID) and more informative and standardized protein names, when available.
From: Chapter 18, The Reference Sequence (RefSeq) Database

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.