This publication is provided for historical reference only and the information may be out of date.
This chapter describes methods used to obtain expert input and peer review, identify key questions, conduct literature searches, select and abstract relevant studies, and analyze data.
Expert Input
We owe a major debt of gratitude to the following groups of multidisciplinary experts from around the world who assisted in preparing this report: 10 national advisory panel members and 5 technical experts who helped define the scope and shape the content, 20 peer reviewers representing a variety of backgrounds and viewpoints, 4 scientific authors who provided additional data from their studies, and 15 staff members of the San Antonio Evidence-based Practice Center and the San Antonio Veterans Evidence-based Research, Dissemination, and Implementation Center, a Veterans Affairs Health Service Research and Development Center of Excellence. Their names are listed in Appendix B. Acknowledgments.
Questions Addressed in Evidence Report
Using a modified Delphi process, the national advisory panel identified several important specific questions about garlic that the evidence report should address. These questions and the types of studies that were deemed appropriate to answer them (selection criteria) are given in Table 3.
Table
Table 3. Questions and selection criteria grouped by search strategies.
Literature Search and Selection Methods
Sources and Search Methods
English and non-English citations were identified through February 2000 from 11 electronic databases (Table 4); references of pertinent articles and reviews; manufacturers; and technical experts. An update search that was limited to PubMed was conducted in February 2000. Database searching used maximally sensitive strategies that identified all papers on garlic and on herbal treatments for cardiovascular disease or cancer. The International Bibliographic Information on Dietary Supplements (IBIDS) database was not searched because it did not contain information on botanicals.
Table
Table 4. Electronic sources searched.
Titles, abstracts, and keyword lists of the 11 electronic databases listed in Table 4 were searched using the following terms, which include Latin names for garlic and names of garlic extracts and constituents (".tw." indicates text word searches, "/" indicates keyword searches, and "$" indicates truncated words that facilitate searching all formations and suffixes of particular terms):
Table
Sources and Search Methods.
Selection Processes
At least two independent reviewers scanned the titles and abstracts of all records identified from the search, using selection criteria given in Table 3. Selection criteria that were specified for each formulated question included the types of participants, interventions, control groups, outcomes, and study designs that were deemed appropriate. Cardiovascular-related trials were arbitrarily limited to those that were at least 4 weeks in duration, because the national advisory panel thought that several weeks of garlic administration might be necessary to demonstrate effects on factors such as glucose, blood pressure, and lipids. Figure 2 schematically presents the selection process. Of 1,798 records, reviewers excluded 1,480 with certainty when screening titles and abstracts. Most of these were in vitro studies, involved animals, or did not meet design inclusion criteria. When screening the full text of the remaining 317 records, 178 were excluded. Of the 139 records meeting selection criteria, 49 were records of 45 randomized trials (some trials were reported in multiple publications or records), 17 were records of 13 cancer survey, case-control, or cohort studies, and 73 were reports of adverse effects or results of skin patch tests with garlic. Of note, of the 45 randomized trials meeting eligibility requirements, 34 were cited in MEDLINE, 2 additional trials were identified from EMBASE only, 1 was identified from PHYTODOK, and 8 were identified either by experts or from scanning symposium proceedings.
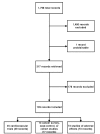
Figure
Figure 2. Flow diagram of selection process.
Data Abstraction Process
Two independent physicians abstracted data from trials that were identified in the efficacy searches. They were not blinded either to study title or to author names. Items that were related to the quality of assessed studies included adequacy of randomization (method and concealment of assignment); whether the trial was single or double blind; whether the intervention and control groups were adequately matched to maintain blinding; cointerventions such as diet, exercise, and cardiovascular medications; and the number of dropouts. Disagreements in abstractions were uncommon (less than 1 percent of items) and were resolved by consensus. No formal reliability testing was done. All abstracted outcome data were verified by a third person with expertise in quantitative data. Abstractions were filed electronically to enable easy updating.
One physician abstracted data about adverse effects. Items that were abstracted included study design (case report, case series, case control, cohort, and controlled trial) and type of specific adverse effect. Several explicit criteria that were aimed at assessing drug adverse effect causality were assessed, such as appropriate temporal relationship, lack of apparent alternative causes, known toxic concentrations of the drug at the time of the appearance of the symptom, disappearance of the symptom with drug discontinuation, dose-response relationship, and reappearance of the symptom if the drug was readministered.
Unpublished Data
We found two randomized trials that were only published in abstract form.46 64 We obtained the full report of one of these trials.64 The one for which we could not obtain a full report is not included in this review.65 It was a crossover trial with 16 participants that compared standardized dehydrated garlic (Kwai® ) with placebo.46 No data prior to the crossover were given in the abstract. Several published studies met selection criteria but did not report critical design features or outcome data. Authors were contacted and requested to provide information regarding randomization procedures and lipid outcomes. Three of the 11 requested authors provided unpublished raw data for lipid outcomes.
Data Analysis Process
Data were synthesized descriptively, emphasizing methodological characteristics of the studies such as populations enrolled, definitions of selection and outcome criteria, sample sizes, adequacy of randomization process, interventions and comparisons, cointerventions, biases in outcome assessment or intervention administration, and study designs. Relationships among clinical outcomes, participant characteristics, and methodological characteristics were examined in evidence tables and graphical summaries such as forest plots.
Primary outcomes in studies were measured with continuous rather than categorical variables. Two methods were used to estimate "effect size" measures for each study. First, we used the standardized mean differences between treatment and comparison group scores. Hedges' g was used to compute the standardized mean difference for each trial:
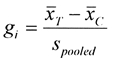
where, for a given trial 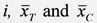 are the mean clinical outcome scores for the treatment group and comparison group, respectively, and spooled
is the pooled standard deviation for the difference between the two means.66
These estimates were adjusted for between-group differences at baseline and for small sample bias.66
Adjusting for baseline differences was accomplished by calculating an effect size at baseline; by definition, it should be zero if study groups were well matched. When a nonzero effect size at baseline was found, outcome effect sizes were adjusted by subtracting the baseline effect size. Second, we used the unstandardized mean differences between treatment and comparison group scores and then adjusted for baseline differences using meta-regression models.
are the mean clinical outcome scores for the treatment group and comparison group, respectively, and spooled
is the pooled standard deviation for the difference between the two means.66
These estimates were adjusted for between-group differences at baseline and for small sample bias.66
Adjusting for baseline differences was accomplished by calculating an effect size at baseline; by definition, it should be zero if study groups were well matched. When a nonzero effect size at baseline was found, outcome effect sizes were adjusted by subtracting the baseline effect size. Second, we used the unstandardized mean differences between treatment and comparison group scores and then adjusted for baseline differences using meta-regression models.
Published reports seldom provided estimates of spooled. One of three strategies was used to estimate spooled when the authors did not directly provide it. First, the individual group variances were used to estimate spooled . If these data were not reported, the pooled variance was back-calculated from either the test statistic or the p-value for differences at followup.67 If neither was possible, a mean variance that was derived from studies of similar size was used. Studies in which the pooled variance was calculated using either of the two latter methods were flagged in the event the magnitude of the effect size resulted in the study being identified as a potential outlier in analyzing heterogeneity.
Placebo-controlled randomized trials with lipid outcomes were quantitatively pooled using a random effects estimator. 66 67 We tried to identify outliers using a standard heterogeneity chi-square test, funnel plot, and Galbraith plot. Studies were considered outliers if the probability for the chi-square value was less than 0.1 and/or the study fell outside of the funnel or Galbraith plot. (A Galbraith plot is a graphical method used to aid in assessing heterogeneity and is particularly useful when the number of studies is small.68 The position of each study along the two axes indicates the weight allocated in the meta-analysis. The vertical axis [a Z statistic equal to the effect size divided by its standard error] gives the contribution of each study to the Q [heterogeneity] statistic. Points outside the confidence bounds are those studies that have major contributions to heterogeneity; in the absence of heterogeneity, all points would be expected to be within the confidence bounds.)
Standardized mean difference effect sizes were converted to clinical laboratory units to aid in interpreting effect size standard deviation units. As noted above, the effect size statistic is calculated by dividing the difference between group means by the pooled standard deviation of the two groups. Because both numerator and denominator are expressed in original units (e.g., milligrams per deciliter [mg/dL]), the units cancel out and the effect size is "unitless." Effect sizes can be back-converted to a value with the original unit, for example, mg/dL, by multiplying the effect size value by a standard deviation value. The statistical significance of the values (effect sizes or converted values) do not change. However, the magnitude of the "converted effect" will vary up or down depending on the magnitude of the standard deviation used. Because these converted clinical units are based on a common standard deviation across all studies, individual study values do not always agree with author-reported results.
Lacking population standard deviation values, we chose to use the "average" standard deviation value for the pooled studies within each group for conversions. Two "averages" were examined: a weighted pooled standard deviation across studies (weighted by sample size) and the median pooled standard deviation. When the two values were substantially different (representing skewness), the median value was chosen. When the values were similar, the weighted pooled standard deviation value was used. The actual weighted average standard deviations that were used in conversions of lipid analyses were total cholesterol: 40.8 mg/dL; low-density lipoprotein level (LDL): 29.1 mg/dL; high-density lipoprotein level (HDL): 11.4 mg/dL; and triglycerides: 85.9 mg/dL.
The following are the rationale for pooling lipid studies: (1) multiple small- to moderate-size studies were available; (2) similar control groups were used; (3) lipid outcomes were measured using similar parameters (e.g., serum total cholesterol) at similar followup times (i.e., 4 to 6 weeks, 8 to 12 weeks, and 20 to 24 weeks); and (4) actual numeric results were often reported. No quantitative summary analysis of blood pressure, glucose, or thrombotic outcomes was performed. Although several studies measured blood pressure, few had a priori hypotheses about blood pressure, and numeric results commonly were not reported, raising the possibility of publication bias in the studies that did report numeric outcomes. Few studies reported glucose or thrombotic outcomes; those that did reported these in several different manners (e.g., fibrinolytic activity and platelet adhesiveness).
Sensitivity and Subgroup Analysis
Different preparations of garlic, including combination preparations, were used in the trials. Subgroup analyses were conducted for trials that used similar dried standardized preparations of garlic and enrolled participants with hypercholesterolemia. Analyses with and without the studies that evaluated the combination preparation of garlic and ginkgo or garlic and hawthorn compared with placebo were conducted because ginkgo is not known to affect lipid parameters. The study that evaluated a garlic and fish oil combination was not pooled with other studies because of possible independent effects of fish oil on lipid parameters. Subgroup analysis based on "doses" of garlic supplements was not conducted because of limited variability of dosing among trials.
Presentation of Results
For this report, results of the standardized mean difference analyses are presented with overall results converted back to original "mg/dL" units; the results and conclusions of the standardized mean difference analyses do not differ substantially from the unstandardized mean difference analyses. Results for total cholesterol, HDL, LDL, and triglyceride levels are all presented in mg/dL. To convert cholesterol, LDL, and HDL values from mg/dL to millimoles per deciliter (mmol/dL), divide by 38.7. To convert triglyceride values from mg/dL to mmol/dL, divide by 88.2. Results were presented in clinical laboratory units to aid interpretation. The magnitude of values depends entirely on the value of the standard deviation. For this report, we chose to use a weighted average standard deviation across all studies at baseline (see the preceding "Data Analysis Process" section). Different values for the common standard deviation would change the magnitude of clinical laboratory unit results (would not change the magnitude of the effect size units), but not the statistical significance.
Publication Details
Copyright
Publisher
Agency for Healthcare Research and Quality (US), Rockville (MD)
NLM Citation
Mulrow C, Lawrence V, Ackermann R, et al. Garlic: Effects on Cardiovascular Risks and Disease, Protective Effects Against Cancer, and Clinical Adverse Effects. Rockville (MD): Agency for Healthcare Research and Quality (US); 2000 Oct. (Evidence Reports/Technology Assessments, No. 20.) 2, Methodology.
