From: miRNAs need a Trim Regulation of miRNA Activity by Trim-NHL Proteins

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
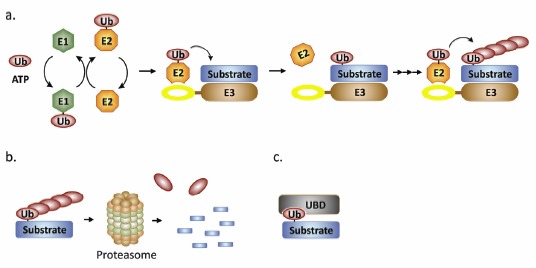
Basic principles of ubiquitination. a) A ubiquitin monomer is covalently linked to an E1 activating enzyme. Interaction of the E1 with an E2 conjugating enzyme results in transfer of the primed ubiquitin to an E2 conjugating enzyme. For RING domain E3 ubiquitin ligases, the E2 interacts directly with the RING. In general, the E3 attracts substrates via additional interaction surfaces such as the coiled-coil or NHL domains. Construction of polyubiquitin chains requires iterative cycles of E2 binding and ubiquitin ligation. Ubiquitin is covalently attached via an isopeptide bond between the C-terminal glycine of ubiquitin and a substrate lysine. See text and references for details. b) Current research is revealing new forms of ubiquitin linkages, in the classical pathway depicted here linear chains of at least four residues are connected at Lys48 of ubiquitin. This linkage is specifically recognized by the 26S proteasome, resulting in ubiquitin release for recycling and proteolytic degradation of the tagged substrate. c) One alternative to the classical pathway is schematically shown. Monoubiquitination can support protein binding with a partner containing a ubiquitin binding domain (UBD), allowing dynamic regulation analogous to protein phosphorylation.
From: miRNAs need a Trim Regulation of miRNA Activity by Trim-NHL Proteins

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.