NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Strassman JE, Queller DC, Avise JC, et al., editors. In the Light of Evolution: Volume V: Cooperation and Conflict. Washington (DC): National Academies Press (US); 2011.

In the Light of Evolution: Volume V: Cooperation and Conflict.
Show detailsSocial Hymenoptera have played a leading role in development and testing of kin selection theory. Inclusive fitness models, following from Hamilton's rule, successfully predict major life history characteristics, such as biased sex investment ratios and conflict over parentage of male offspring. However, kin selection models poorly predict patterns of caste-biasing nepotism and reproductive skew within groups unless kin recognition constraints or group-level selection is also invoked. These successes and failures mirror the underlying kin recognition mechanisms. With reliable environmental cues, such as the sex of offspring or the origin of male eggs, predictions are supported. When only genetic recognition cues are potentially available, predictions are not supported. Mathematical simulations demonstrate that these differing mechanisms for determining kinship produce very different patterns of behavior. Decisions based on environmental cues for relatedness result in a robust mixture of cooperation and noncooperation depending on whether or not Hamilton's rule is met. In contrast, cooperation evolves under a wider range of conditions and to higher frequencies with genetic kin recognition as shared greenbeard traits. This “excess of niceness” matches the existing patterns in caste bias and reproductive skew; individuals often help others at an apparent cost to their inclusive fitness. The results further imply a potential for greenbeard-type kin recognition to create arbitrary runaway social selection for shared genetic traits. Suggestive examples in social evolution may be alloparental care and unicoloniality in ants. Differences in kin recognition mechanisms also can have consequences for maintenance of advantageous genetic diversity within populations.
Seemingly overtly altruistic behavior, such as individuals accepting sterility, has puzzled evolutionary biologists since the time of Darwin. The first truly predictive framework for how reducing one's own reproduction could be adaptive came from the seminal work of W. D. Hamilton (1964a). His key insight was that fitness is “inclusive” of both an individual's direct reproduction and indirect gains arising through help provided to genetic relatives. Gains in indirect fitness, often labeled as “kin selection,” recast altruism as an ultimately selfish act. Help relatives if the benefit provided (b), prorated by the genetic relatedness of the recipient (r), exceeds the cost to self (c). This is Hamilton's rule: Helping is adaptive if br > c.
More than any other taxonomic group, social Hymenoptera (ants, bees, and wasps) sit at an apparent peak of kin selection, with many species having morphologically sterile workers. Kin selection and applications of Hamilton's rule, however, extend far beyond the evolution of sterile castes to examine many aspects of cooperative (and noncooperative) behavior (Bourke and Franks, 1995). Thus, social insects have had a pivotal role in the development of kin selection theory and its elevation to being the dominant evolutionary paradigm for the study of cooperation and conflict. To date, there have been hundreds of tests of kin selection predictions in social insects (Abbot et al., 2011). However, despite this track record of remarkable utility, kin selection theory has recently become embroiled in controversy. The mathematics of inclusive fitness modeling have been directly challenged (Nowak et al., 2010). The evolution of cooperation is argued as better explained by group-level selection than by nepotism toward kin (Wilson and Wilson, 2007). Finally, the status of social insects as being a paramount example of kin selection has been questioned, with kin selection relegated to being a dissolutive force that primarily selects against cooperation and sociality (Wilson and Hölldobler, 2005). The response to these criticisms from defenders of inclusive fitness modeling and kin selection has been simultaneously vigorous and dismissive (Foster et al., 2006; Lehmann et al., 2007; West et al., 2007c, 2008; Abbot et al., 2011; Herre and Wcislo, 2011; Strassmann et al., 2011b).
The current conceptual maelstrom offers an opportunity for a critical appraisal of the effects of kin selection in the social Hymenoptera. Considering a model or hypothesis as either a failure or success is highly subjective. No single model can be expected to be 100% accurate for all taxonomic groups and in all situations. It is nevertheless fair to categorize a model as failing when a strong majority of studies reject its predictions. Conversely, models that are consistently supported by data are valued for accurately tracking evolutionary outcomes. By such standards, and contrary to its critics, kin selection theory has had major successes. However, contrary to its apologists, kin selection theory also has had some major failures. I will briefly review within the social Hymenoptera two successes, (i) sex investment ratios and (ii) conflict over reproduction by workers (i.e., worker policing), and two failures, (i) caste-biasing nepotism and (ii) reproductive skew theory. I will thereafter consider the mechanism for how kin are recognized as the driving force for the observed pattern of success and failure.
These four topics are chosen because I believe the underlying theory for the kin selective predictions is sound and that the possibility for kin nepotism to evolve is at least potentially present. This differs from two other cases, where kin selection predictions are suggested to have failed: the haplodiplody and monogamy hypotheses (Nowak et al., 2010). First, cooperative breeding has repeatedly evolved in the haplodiploid Hymenoptera. Haplodiplody creates a genetic asymmetry, such that a female is more related to her full sister (r = 0.75) than she is to her own offspring (r = 0.5). Therefore, if a singly mated mother produces a female-biased offspring sex ratio, it is genetically more advantageous for a daughter to help her mother raise more sisters. However, the balance of evidence from existing species where cooperative breeding is facultative finds that such species are not monogamous, do not predictably bias sex ratios toward females, or both (Bourke and Franks, 1995). Hence, the haplodiploidy hypothesis is not a robust test of kin selection because the required patterns of genetic relatedness likely did not exist in the putative ancestors of eusocial species (Nonacs, 2010). The second example is the “monogamy hypothesis,” where cooperative breeding is predicted to be more likely to evolve in species where family groups are full siblings because of monogamy (Boomsma, 2009). However, a gene-based model for the evolution of cooperation found that helping actually often tended to spread more rapidly through populations with polygamy (Nonacs, 2011). This may be an instance where the underlying kin selection model actually produces erroneous predictions [as postulated by Nowak et al. (2010)].
SUCCESSES: BIASED SEX INVESTMENT RATIOS AND PARENTAGE OF MALES THROUGH WORKER POLICING
Another genetic consequence of the asymmetrical relatedness patterns attributable to haplodiploidy is that with one singly mated queen (i.e., monogynous with monandry), workers are more closely related to full sisters than to their brothers. This preference should drive female investment bias to the point where it is exactly offset by the relative mating advantage of the rare male sex (Trivers and Hare, 1976). Thus, if workers control sex investment ratios, a 3:1 female-to-male investment bias should result. Over 3 decades of evidence has solidly supported the core of the Trivers and Hare hypothesis (Nonacs, 1986; Bourke and Franks, 1995; Chapuis and Keller, 1999; Strassmann and Queller, 2007). Female-biased investment occurs frequently in species where the sister-brother relatedness asymmetry is present and far less often in species where it is absent. Within some species, there are both monogynous, monandrous colonies and others in which the relatedness difference between females and males is reduced or absent because of having multiple laying queens or one queen that uses sperm from multiple males. Extending Trivers' observation predicts that such populations should exhibit split sex ratios, with the monogynous, monandrous colonies favoring females and the others favoring males (Boomsma and Grafen, 1990). A recent review (Meunier et al., 2008) found that within-colony relatedness asymmetries do significantly affect bias in sex investment as predicted by kin selection.
Conflict over male parentage in Hymenoptera can also be present because workers retain the ability to produce haploid eggs in many species, and therefore can produce sons (Bourke and Franks, 1995). Although a worker's sons and nephews (r = 0.375) are more closely related than brothers (r = 0.25), males produced by half sisters are less related (r = 0.125). Thus, when more than 50% of the workers are half sisters, a nonlaying worker is more related to a queen-produced male than to the average worker-produced male. Maximization of inclusive fitness would therefore predict that workers replace (i.e., police) worker-produced eggs with queen-produced eggs (Ratnieks et al., 2006). Policing occurs in all examined species where workers are more related to the queen's male offspring than to the average worker-produced male (Wenseleers and Ratnieks, 2006a). Contrary to expectations generated from the relative genetic value of brothers vs. nephews, however, worker policing also occurs in species where workers are more related to worker-produced males than to queen-produced males (Hammond and Keller, 2004). Thus, worker policing may also often reflect group-level advantages, such as the replacement of less viable worker eggs with more viable queen-laid eggs (Nonacs, 2006a). Despite these notable exceptions, extensive phylogenetic analyses support broad predictions of kin selection theory (Ratnieks et al., 2006; Wenseleers and Ratnieks, 2006b): (i) Worker policing occurs more frequently in species where queen-produced males have higher mean relatedness to workers; (ii) frequencies of worker-produced males correlate to increasing relatedness between worker-produced males and workers; (iii) frequency of worker laying negatively correlates to effectiveness of policing; and (iv) worker policing is less prevalent after queen death in colonies with lower between-worker relatedness [i.e., a reversal of the pattern found in (ii) in the presence of viable queens].
FAILURES: PATRILINE NEPOTISM IN CASTE BIAS AND REPRODUCTIVE SKEW
In the eusocial Hymenoptera, sterile female workers primarily raise the offspring (Bourke and Franks, 1995). These offspring can have a wide range of relatedness to the tending females as a result of queens mating with multiple males or multiple fertile queens in colonies. The former creates patrilines of full sisters within the colony, and the latter can create worker cohorts ranging in relatedness from nieces (if queens are sisters) to totally unrelated. Therefore, any worker cohort that manages to have its full sisters preferentially raised as the future reproductives should significantly increase its fitness. Careful observation, however, has yet to find any significant conflict or favoritism over care giving under a wide range of scenarios. Honey bees (Apis mellifera), whose queens mate with many males, have been studied most extensively for evidence of queen-rearing nepotism, but none has been conclusively demonstrated (Tarpy et al., 2004; Ratnieks et al., 2006). Similar studies in other social Hymenoptera have also failed to find nepotism in colonies with multiply mated females or multiple queens (Keller, 1997; Gamboa, 2004).
If facultatively cooperative groups are more productive than the expected cumulative output of all their individual members, it is possible for reproduction to be shared so that all individuals have higher fitness through cooperation (Nonacs and Hager, 2011). A fitness-maximizing division of the reproduction (i.e., the reproductive skew within the group) can be predicted through an inclusive approach. Groups can be stable if all individuals accrue fitness that is equal to or greater than their expected fitness from reproducing on their own (Nonacs, 2006b). Although there are numerous variants of skew models, all share the general features that predicted skew within groups ought to be affected by the genetic relatedness of the group members, their relative ability to compete for reproductive shares, and the relative productivity of groups vs. solitary individuals (Nonacs and Hager, 2011). In an extensive review of experimental tests of skew theory, none of these predicted relationships were consistently found (Nonacs and Hager, 2011). In 21 of 27 studies, there was no significant correlation between genetic relatedness and skew across groups within populations. In 13 of 18 cases, correlates with competitive ability had no significant effect on skew, and in only 3 of 18 cases did skew significantly correlate to factors likely to cause ecological constraints. Finally, 3 of 4 studies estimating inclusive fitness of group members and solitary individuals found it unlikely that cooperation was favorable for all group members. Individuals of low relatedness would probably gain higher fitness by reproducing solitarily rather than being subordinate group members. Therefore, the overall evidence suggests that individuals rarely modulate reproductive shares in response to the intrinsic characteristics of other group members, such as relatedness (Nonacs, 2006b; Port and Kappeler, 2010; Nonacs and Hager, 2011).
The failure of reproductive skew models to predict the behavioral dynamics between group members within populations contrasts with more accurate predictions at the population or species level. In these comparisons, differences in mean within-group relatedness or differences in environmental constraints for being solitary do accurately predict which population or species should exhibit the greater reproductive skew (Reeve and Keller, 1995; Nonacs and Hager, 2011). For example, if two populations significantly differ in mean within-group relatedness, subordinate individuals in low-relatedness populations stand to gain more fitness by reducing reproductive skew. Thus, there would be greater selective pressure with overall low relatedness to share reproduction more equally, independent of whether individual-level relatedness can be recognized (Reeve and Keller, 1995).
MECHANISMS OF KIN SELECTION
“Failure” is a semantically loaded word and should not imply that there are no evolutionary explanations for the observed outcomes in caste rearing and reproductive skew. However, these explanations invoke elements that are added to kin selection theory and do not follow from it. For example, caste-rearing nepotism would be absent if workers are constrained by an inability to distinguish their own patrilines from others. Without some means of kin recognition, kin selection could not “fail” as a hypothesis because it simply would not be relevant to the phenomenon at hand. Alternately, caste-rearing nepotism could create such within-colony conflict that overall colony productivity or survival is seriously reduced. Hence, conflict would not be selectively advantageous. This alternative explanation, again, does not follow from kin selection theory. No matter the overall cost, successful nepotists would always have a selective advantage relative to nonnepotists within colonies. It is only selection on the across-group level that could favor this ergonomic efficiency explanation for the absence of nepotism. The same two possibilities, constraint or ergonomics, could also explain why reproductive skew models do not adequately predict patterns of cooperative breeding (Nonacs and Hager, 2011). Nevertheless, recent work casts doubt on the constraint hypothesis as a broadly viable explanation. For example, individual ants produce genetically heritable hydrocarbon profiles that could, theoretically, be used to identify kin (van Zweden et al., 2010). However, such markers are readily transferred to create effective nestmate recognition rather than within-nest nepotism. This leaves only the group selection hypothesis as a plausible, if unsatisfying, explanation. It is unsatisfying because it is untestable in many cases; that is, if species A never exhibits caste-rearing nepotism, how can it be shown that it is because such behavior reduces overall colony productivity? Therefore, instead of relegating the solution as only explainable by difficult-to-test group selection, it is useful to reexamine kin selection predictions relative to the mechanistic aspects of exactly how individuals recognize or define other group mates as genetic kin. In essence, kin selection theory may accurately predict the outcomes for caste rearing and reproductive skew, but the predictions themselves may differ from earlier expectations.
There are only two ways that kinship can be assigned, either through recognizing genetic similarity or by using environmental cues that accurately predict genetic similarity. The latter can lead to simple and relatively effective rules-of-thumb for cooperation. For example, in a population where most females mate with only one male, two female wasps maturing at the same time on a nest are likely to be full sisters and can behave according to predictions from Hamilton's rule. However, such rules-of-thumb are not absolutely reliable: The females could also be half sisters, cousins, or unrelated because of multiple mating, multiple foundresses, or nest usurpation. Errors in estimation of relatedness could be common (Nonacs, 2006b).
In contrast to using environmental cues, individuals could recognize kin through a shared phenotypic trait that uniquely represents the presence of identical alleles. To function as an effective kin selective mechanism, genetic recognition must have three elements: expressed phenotypic trait(s), an ability to recognize traits in other individuals, and the proclivity to direct aid toward those others if a suitable opportunity arises (Gardner and West, 2010). These three elements can either be genetically linked as a single “greenbeard” system or be a set of genetic markers monitored by a single perception locus that triggers cooperative behavior (Lacy and Sherman, 1983). In the second scenario, recognition and action need not be linked to any cue alleles. Although the basic premise of greenbeard kin selection is sound, functioning greenbeards have been thought likely to be rare for several reasons (Gardner and West, 2010). A greenbeard system must always co-occur within individuals. For example, if the willingness to aid becomes decoupled from the phenotypic trait, a “falsebeard” individual would result. Such individuals would receive benefits from other greenbeards but would never provide benefits (and thus never incur any costs for being helpful). Under a wide range of conditions, cheating falsebeards could be at a selective advantage over true greenbeards and prevent widescale cooperation (Gardner and West, 2010). A second problem is that successful greenbeard genetic systems would encounter Crozier's paradox (Crozier, 1986). As the greenbeard system increases in frequency, it becomes less useful for identifying kinship (Rousset and Roze, 2007). As it approaches fixation, it can no longer function for kin nepotism because almost all individuals in the population would be viewed as identically related. A final problem for a greenbeard system, but not for phenotype matching, is that individuals are either highly related at a locus (both have identical alleles) or totally unrelated. Thus, if the rest of the genome is Hamiltonian in the sense of estimating relatedness relative to environmental cues, this could lead to different estimates of r as well as intragenomic and interlocus conflict. Assuming that disproportionally more loci are Hamiltonian in driving behavior, this is predicted to lead to suppression of any greenbeard favoritism (Helanterä and Bargum, 2007); however, an alternative argument is supported by Gardner and West (2010).
Despite the theoretical objections, more examples of greenbeard behavior have been found recently in hydrozoans (Grosberg et al., 1985), slime molds (Queller et al., 2003), yeast (Smukalla et al., 2008), and ants (Keller, 2007). The most compelling example is in side-blotch lizards (Uta stansburiana), where blue-throated males preferentially establish territories next to each other (Sinervo et al., 2006). These aggregations form with respect to the blue-throat trait and are not predicted by whole-genome relatedness. Neighboring blue-throats are more successful at mate guarding against larger aggressive orange males. However, these benefits accrue only to blue males not next to an orange male; blue males adjacent to orange males suffer a cost. Possibly in reaction to a perception that such a greenbeard system could not be evolutionarily stable, Sinervo et al. (2006) insightfully comment, “A proximate explanation for kin altruism is not that kin share a fractional number of genes; rather, kin altruists share key genes for signal, self-recognition, and donation behavior” (p. 7376). In this context, it is seen that kin nepotism through genetic recognition differs from existing models of greenbeard nepotism not in process but simply in the number of genes involved. This view argues that kin selection can result from a collaboration of multiple greenbeards sharing a common interest.
SIMULATING DIFFERENT PATHWAYS FOR THE EVOLUTION OF COOPERATION
Although the evolutionary dynamics of single greenbeard systems have been extensively examined (Gardner and West, 2010), multiple greenbeards evolving synchronously have not received similar attention. I consider this latter situation by simulating the evolution of helping behavior under three scenarios:
- Help is allocated so as to maximize inclusive fitness as predicted by Hamilton's rule.
- Help is allocated relative to recognized shared alleles, with the helper and helped having the same greenbeard allele(s). In this scenario, falsebeard mutants can also arise that induce cooperation from greenbeard alleles but never extend help.
- A mixed system exists where some loci favor cooperation as predicted by Hamilton's rule and others behave as greenbeards or falsebeards. This tests the degree to which interlocus conflict can suppress or mask greenbeard effects.
Phenotype matching (Lacy and Sherman, 1983) is a hybrid version of greenbeard and Hamiltonian kin recognition, where a number of cues act as greenbeards but there is only one recognition locus. This locus collates matches from all the cues to generate an estimate of r that is used in Hamilton's rule to determine if cooperation occurs. I do not specifically evaluate phenotype matching, although scenario (i) can be viewed as a version of the model that is completely accurate for estimating relatedness. The model is written in TrueBasic.
All simulations assumed a diploid population and started with 500 mothers, each mating with two males. All mothers produced two offspring, a potential helper and helped offspring that could be full siblings or half siblings. All offspring genotypes were randomly determined with respect to father and which allele was contributed by each parent. Loci were not linked and segregated independently. Only offspring and no mothers, helpers, or helped survived to the next generation. For the first two scenarios, all offspring had 1–15 loci (depending on the simulation) at which helping behavior could independently evolve. All alleles at these loci were initially “null” and neutral with respect to whether help was offered. Helping alleles were randomly mutated into the population at the rate of 0.01 mutations per offspring. If no cooperation occurred, both offspring mated twice and produced two offspring of their own, which were added to the pool from which the next generation's mothers and fathers were chosen. Thus, the inclusive fitness of noncooperation (IFNC) equaled 2(0.5) + 2r, where r is the relatedness of the sibling's future offspring calculated at the level of the entire genome. If cooperation occurred, the helper did not reproduce and its sibling produced 2–12 offspring (b) depending on the conditions of the simulations (Table 3.1). This results in IFC = rb. A Hamiltonian allele would vote “yes” if IFC > IFNC and “no” otherwise. (Note that to be completely consonant with Hamilton's rule, the number of offspring raised by the helped offspring without a helper ought to be subtracted from both IF values. For mathematical simplicity, I allow this value to cancel out during calculations.) I assume that Hamiltonian alleles are completely accurate in discriminating full sibs and half sibs. This could occur as a modified greenbeard system where a single recognition/action system simultaneously monitors multiple phenotypic or environmental cues (unspecified in this model) to estimate genetic relatedness reliably. Unlike a single greenbeard, the components would not have to be linked to each other. Similarly, I assume that although b varies stochastically, the level is always recognized. Thus, Hamiltonian alleles vote identically across all loci, giving either a 100% yes or no vote for helping among themselves. Therefore, depending on whether overall environments were good, moderate, or poor for producing benefits from cooperation, Hamiltonian alleles would favor helping in ∼50%, 17%, or 0% of the possible cases (Table 3.1).
TABLE 3.1
Payoffs for Helping Behavior.
In contrast, at each greenbeard locus, there are four different possible greenbeard alleles (each producing a hypothetical unique phenotypic cue) that can mutate into the population. For each greenbeard allele, a matching falsebeard allele can also mutate into the population. A greenbeard allele in a helper could vote yes if the helped sib had identical greenbeard or matching falsebeard allele(s) at the same locus. It would vote yes, however, only if a helped individual would be expected to transmit more greenbeard alleles to the offspring generation than the combined reproduction of noncooperating helpers and helped (Table 3.1). It would vote no if noncooperation was expected to transmit more alleles. Thus, the vote of a greenbeard locus depended on b and the heterozygosity or homozygosity of the helper and helped with respect to greenbeard and matching falsebeard alleles. Different greenbeard alleles at the same locus do not help each other and would vote no. Falsebeard alleles in a helper always vote no. Thus, across greenbeard loci, there could be a mixture of yes and no votes and probability of helping occurs relative to the proportion of yes votes (e.g., 12 yes votes and 3 no votes would result in an 80% probability of helping).
In the third scenario, five loci evolved cooperation as Hamiltonian and one to five loci evolved as greenbeards, both as described above (resulting in a range of a 5:1 advantage for Hamiltonian loci to 5:5 equal weighting). Helping occurs with a probability that is the proportion of yes votes across all votes by Hamiltonian and greenbeard loci. All scenarios were simulated for 2,500 generations with 20 replicates. Random mutation was present for the first 2,000 generations to allow all possible alleles to enter the population. For the last 500 generations, greenbeard alleles were prevented from mutating into the population. Therefore, at the end of the simulations, the frequency of greenbeard alleles was more reflective of their relative selective advantage (i.e., they could be selectively eliminated from populations).
RESULTS
The invasion of helping is charted as the proportion of mothers (out of 500) that produced pairs of offspring that decided to cooperate. Hamiltonian alleles rapidly invade a noncooperative population when cooperation is at least sometimes advantageous (Fig. 3.1). The resulting level of sociality is commensurate with how often IFC > IFNC (e.g., 50%, 17%, or 0%). The number of loci that could be involved has no effect on the rate of spread of helping behavior. Overall, the system consistently evolves rapidly to maximize inclusive fitness.
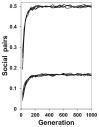
FIGURE 3.1
Increase in sociality with Hamiltonian alleles that help if IFC > IFNC for full or half sibs, as measured by the proportion of offspring pairs that help each other. The upper set of lines is for simulations where helping often produces large benefits (more...)
Sociality also often evolves with only greenbeard loci but differs from Hamiltonian patterns. Unlike with Hamiltonian alleles, a significant fraction of the population can exhibit cooperation under conditions where helping never produced higher inclusive fitness at a genome level (Fig. 3.2A). Although this level of cooperation was often enhanced through continual input of greenbeard alleles by mutation, greenbeards continued to persist for many generations in the population when their entry by mutation was turned off (Fig. 3.2). Moreover, higher levels of cooperation evolved and greenbeard alleles persisted longer in the absence of mutation as the number of greenbeard loci increased across all levels of b.
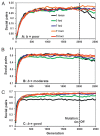
FIGURE 3.2
Proportion of social pairs with loci that favor helping if individuals share the same greenbeard alleles or matching falsebeards. Simulations are where helping provides poor benefits (A, b = 2–6), moderate benefits (B, b = 3–9), or large (more...)
At most loci, a single greenbeard allele was strongly numerically dominant in frequency no matter the level of b or the number of loci involved. Thus, Crozier's paradox (Crozier, 1986) often occurs at individual loci, where one greenbeard allele rises to high frequency at the expense of all other possible alleles. Although falsebeard alleles occasionally reached high frequency, they did not predominate at the majority of loci for any combination of b and loci number. This result is somewhat surprising because falsebeards contribute to producing nonhelping phenotypes by always voting against cooperation. Thus, conditions where cooperation was never beneficial at a whole-genome level, IFNC ≥ IFC (Table 3.1), might have been expected to tip the balance of selection toward a falsebeard allele, but this happened only at a minority of loci. As more loci are interacting or b becomes moderate or good, allele populations are almost uniformly dominated by a single greenbeard allele.
Across most individual simulation trials, there appeared to be little interaction between greenbeard and falsebeard alleles at a given locus. Occasionally, an increase in the frequency of a matching falsebeard could tip the balance of selection from one greenbeard allele to another and lead to a replacement as the most frequent allele in the population.
The high frequency of greenbeard alleles under many conditions results in more cooperative behavior between siblings than predicted by Hamilton's rule for any given b value. For example, even with a large mean payoff to cooperation, applying Hamilton's rule predicts that only ∼50% of the helpers' decisions ought to be to cooperate (Fig. 3.1). Compared with this prediction, cooperation through greenbeards results in many “mistakes” (Fig. 3.3). Most of the errors are of individuals being helped when IFC < IFNC predicts no helping.
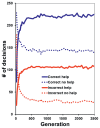
FIGURE 3.3
Outcome of decisions made by individuals with greenbeard loci. “Correct” and “incorrect” are determined for 500 pairings relative to whether IFC > IFNC at the whole-genome level holds with half (r = 0.125) or full (more...)
The results also show that there can be interlocus conflict across Hamiltonian and greenbeard loci. These two regions of the simulated genomes can disagree as to whether cooperation is advantageous. The resulting levels of sociality appear to be an approximate balance between the relative number of loci that select for Hamiltonian or greenbeard levels of cooperation (Fig. 3.4). Thus, cooperation sometimes occurs under conditions where combinations of sibling relatedness and benefit predict it should not; however, cooperation also sometimes does not happen in the presence of shared greenbeard loci across offspring.
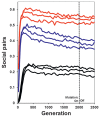
FIGURE 3.4
Proportion of social pairs when individuals have both Hamiltonian and greenbeard loci. The three sets of lines represent conditions with good (top series, b = 4–12), moderate (middle series, b = 3–9), or poor (bottom series, b = 2–6) (more...)
DISCUSSION
Kin Recognition Mechanism and the Expression of Cooperation
In predicting patterns of life history traits, robust theory must combine with a realistic mechanism for action. The distribution of success and apparent failure in past kin selection predictions for social insects illustrates the importance of this connection. Thus, genetic mechanisms for identifying kin (e.g., a hypothetical series of greenbeard loci) are less likely to maximize an actor's inclusive fitness than environmental cues that provide only a probability level of sharing genotypes. That kin selection theory is accurate only with probabilistic environmental cues helps to explain patterns of behavior within social insects. For example, the sex of a given offspring may be hidden for some time through development, but it must eventually become recognizable (Nonacs and Carlin, 1990). This gives workers a clear cue for biasing investment toward females and away from males. Similarly, having multiple matrilines or patrilines within a single colony may give out an unmistakable diversity signal (van Zweden et al., 2010). Therefore, kin selection models for optimal sex ratio investment have usable proximate cues and function well in predicting ultimate allocation patterns at both the colony and population levels.
Conflict over and suppression of worker laying similarly have an available proximate mechanism. Although both workers and queens can produce male eggs, an increasing body of evidence shows that queen eggs are recognizably different from worker eggs in morphology and chemical signatures (Ratnieks et al., 2006; van Zweden et al., 2009; Meunier et al., 2010). As a result of nutritional differences, queens may always be able to imbue their eggs with specific signals that cannot be faked by workers. Thus, workers can discriminate between eggs and maximize their inclusive fitness through manipulating which individuals will produce the males.
In comparison, there is almost no supportive evidence for within-colony nepotism in the production of new queens (Keller, 1997; Gamboa, 2004; Tarpy et al., 2004; Ratnieks et al., 2006). This is despite a potentially huge boost in inclusive fitness for eusocial Hymenoptera (e.g., the replacement of an unrelated female, r = 0, or half sister, r = 0.25, with a full sister, r = 0.75). From a mechanistic perspective, however, the only cues that workers could use to behave nepotistically would be through recognizing one or more shared alleles. Similarly, reproductive skew models require that group members estimate kinship in apportioning reproductive shares. Although unstated in the models, this assumes a type of greenbeard kin recognition and may be why they fail to predict skew across groups within populations. There is no obvious nongenetic cue that would, for instance, be able to differentiate emerging adults on the same nest as full sisters rather than cousins. However, evolved patterns of reproductive sharing at population or species levels ought to reflect the expected mean levels of relatedness or ecological constraints, regardless of any ability for individual-level recognition of genetic relatedness. Hence, skew models are relatively more successful at predicting population-level differences in reproductive sharing (Nonacs and Hager, 2011).
An interesting result from the simulation models is that greenbeard nepotism results in seemingly overly cooperative populations (Fig. 3.3), which is exactly how caste nepotism and reproductive skew appear to fail. In both cases, individuals are too nice. In caste determination, matrilines do not discriminate across females in how they are treated. Close kin, distant kin, and nonkin are equally helped. Reproductive skew on wasp nests tends to be very high, indicating that subordinates willingly cooperate to their apparent fitness detriment (Nonacs et al., 2006). Most paradoxically, fertile unrelated wasps join (and are allowed to join) groups with no differential treatment (Queller et al., 2000; Leadbeater et al., 2010). Relatedness has no predictive role in establishing aggression patterns or dominance hierarchies on nests (Nonacs et al., 2006; Leadbeater et al., 2010).
Runaway Social Selection Through Greenbeard Nepotism
The significance of greenbeard kin nepotism in evolutionary biology is controversial (Gardner and West, 2010; Leigh, 2010). However, in the models presented here, two of the three major objections to greenbeards often fail to prevent the evolution of helping. A falsebeard cheating genotype that accepts cooperation but does not reciprocate rarely destabilizes a greenbeard kin recognition. The inability of a falsebeard to predominate may result from several simultaneous processes. First, positive kin assortment into sibling pairs often imposes a cost onto cheating. Although there is the probability of drawing benefits from unrelated greenbeards, there can be the relatively greater chance of not helping siblings that share identical falsebeard alleles when such help would increase allele frequencies. A second process occurs as a greenbeard allele reaches higher frequencies. At this point, more of the helper/helped pairings involve homozygous individuals, which increases the net gain for helping (Table 3.1). This may generate positive feedback in more helping and continued increases in allele frequency. Finally, when greenbeard alleles predominate at multiple loci, they will increase commonality of interest. This could result in greenbeard alleles forming a “voting block” as regards cooperation and depress the selective advantage of a falsebeard at any individual locus. Altogether, the results are strongly suggestive that models of greenbeard recognition systems based on single-locus dynamics (Gardner and West, 2010) may predict very different outcomes from multilocus situations. These ramifications need to be explored more extensively in future work. Also, the models here assume idealized versions of Hamiltonian and greenbeard alleles in that all alleles “know” the payoff for helping, know the cost for abandoning direct reproduction, and properly identify shared alleles or correctly measure r. The effects of mistakes in these estimates could be valuable to consider (Nonacs, 2006b).
The second objection is that greenbeards will sometimes function to the detriment of the much larger nongreenbeard genome, and therefore are expected to be suppressed (Helanterä and Bargum, 2007). However, suppression is only partial in this model. Shared greenbeard alleles increase in populations and raise levels of cooperation proportional to their abundance relative to Hamiltonian loci (Fig. 3.4). This supports the view that greenbeard alleles are not intragenomic “outlaws” but are under similar selection pressures as the rest of the genome (Gardner and West, 2010).
An effect of greenbeard nepotism is that it quickly increases the frequency of arbitrary phenotypic traits in social settings in what is very much a “runaway” process (i.e., in the absence of social interactions, the traits convey no benefit to their bearer). This outcome is relevant to West-Eberhard's (1983) proposal that parents might allocate resources biased toward offspring with particular traits that have no effect other than being “attractive.” If such attractive traits are greenbeards, their bearers would benefit from the genetic correlation between trait and preference as in runaway sexual selection. The process could be further enhanced by the feedback between the commonness of an allele and increased payoffs for cooperation. Such runaway social selection is proposed for bright coloration and plumage in young birds, which appears to function in attracting parental care (Lyon et al., 1994; Ligon and Hill, 2010) but, interestingly, not in identifying kin (Shizuka and Lyon, 2010). Moreover, rapidly reaching Crozier's paradox (i.e., fixation at the loci involved) may leave no trace in present-day behavior of past genetically based favoritism across offspring. This suggests that some morphological and behavioral traits in social species without clear adaptive value may have evolved through greenbeard nepotism.
In this suite of traits arising from greenbeard nepotism could be the initial evolution of cooperative breeding itself. Specifically, greenbeard traits can produce low levels of social behavior under conditions where Hamilton's rule would predict no cooperation (Fig. 3.2). If, for example, offspring dispersal is limited, positive kin assortment would result among neighboring individuals. This would create preconditions where greenbeard alleles producing alloparental behavior could be selectively favored. If alloparental care is as simple as feeding a nearby hungry mouth (Jamieson, 1989), a greenbeard trait for feeding a “mouth like mine” could evolve even if it did not appear to increase kin selective inclusive fitness (Emlen et al., 1991; Komdeur, 1996). Once social groups are established, more elaborated forms of cooperation could evolve with enhanced benefits provided to helped individuals that are potentially in line with Hamilton's rule.
Another trait that could result through runaway social selection is the phenomenon within ants of unicoloniality. Unicoloniality occurs when adjacent nests show atypically low or no aggression toward each other and is commonly associated with invasive ant species, where supercolonies can arise that extend over thousands of kilometers (Helanterä et al., 2009). Unicolonial associations are genetically homogeneous, with the majority of ants having low relatedness to each other. The reduction in intraspecific aggression appears to be attributable to a loss of genetic diversity at recognition loci (Suarez et al., 2008). Although unicolonial behavior may be enhanced by genetic bottlenecks, it is also present in situ in native habitats and apparently can evolve within large populations (Pedersen et al., 2006; Wang et al., 2010). In other words, unicoloniality can be selectively favored and is not simply always a byproduct of genetic bottlenecks from introductions. Interestingly, unicoloniality could be categorized as workers showing excessive cooperation by accepting and raising unrelated individuals, exactly as predicted by the model of greenbeard cooperation. The suggestion would be that if greenbeard alleles arise in a species at their recognition loci, it would be possible for such alleles to sweep through a population, carrying unicoloniality in their wake. This would lead to high genetic similarity at recognition loci in populations where within- and across-nest relatedness is almost identical [which is indeed observed in unicolonial ants (Brandt et al., 2009)]. Finally, it is a suggestive coincidence that one of the known greenbeard allele systems (Gp-9 in fire ants) appears to have arisen in an exotic species as that species was simultaneously evolving a unicolonial population social structure (Keller, 2007).
Genetic Diversity, Relatedness, and Social Heterosis
Controversies about kin selection often concern the best methods to model the effects of genetic relatedness (Wilson and Hölldobler, 2005; Foster et al., 2006; Lehmann et al., 2007; West et al., 2007c; Wilson and Wilson, 2007; Nowak et al., 2010). This tends to obscure the more salient point of how important the level of relatedness is for favoring cooperation (Nonacs, 2011). The dynamics of social groups can be dominated by either their genetic relatedness or their genetic diversity, because both bring evolutionarily selective advantages. High relatedness means that group benefits will tend to be exclusively shared by alleles identical by descent. It becomes relatively less important which individuals are the reproducers and which individuals are the helpers (Nonacs and Hager, 2011). In contrast, higher genetic diversity can create social heterosis, or the ability to exploit a wider range of resources more effectively, and thus increase total group benefits (Nonacs and Kapheim, 2007). Although more restrictive on the possible skew between reproducers and helpers, within-group outcomes become less important relative to across-group competition. The evolutionarily inescapable point is, however, that nepotism, as predicated on Hamilton's rule, must come at the expense of genetic diversity. Similarly, selection for genetic diversity may require behavioral biases against closer genetic relatives. Relatedness and diversity have an unavoidable tradeoff between them.
The model presented here gives no advantage to genetic diversity; therefore, cooperation evolves to maximize population-level genetic similarity. Alleles go to high frequency or fixation acting either as greenbeards or to maximize genome-level inclusive fitness. Nevertheless, one might expect that the diversity/ relatedness tradeoff could be different for greenbeard or Hamiltonian alleles. Any natural system where the benefits of cooperation are primarily directed to close relatives would strongly select against genetic diversity. In contrast, cooperation could evolve with considerably less of a tradeoff with a greenbeard kin recognition mechanism. The benefits of cooperation would not necessarily only flow to the closest relatives. As argued above in the case of ant unicoloniality, selection for nepotism based on greenbeard similarity could potentially affect only a limited part of the genome. Social heterosis could simultaneously select for genetic diversity at the remainder of the genome, with the result being a patchwork genome of regions of low and high genetic diversity (Nonacs and Kapheim, 2007). It is difficult to imagine how such opposing selective processes could simultaneously operate when one or more traits are being selected relative to their probabilities of being shared. Kin nepotism following Hamilton's rule will always work to reduce genetic diversity. It is the dynamic evolutionary consequences of selection for kin vs. selection for genetic diversity that should draw the future attention of both theoreticians and experimentalists.
ACKNOWLEDGMENTS
I thank J. Field, D. C. Queller, T. Wang, and an anonymous reviewer for helpful comments on the manuscript.
- Kinship, Greenbeards, and Runaway Social Selection in the Evolution of Social In...Kinship, Greenbeards, and Runaway Social Selection in the Evolution of Social Insect Cooperation - In the Light of Evolution
- Genomic Imprinting and the Evolutionary Psychology of Human Kinship - In the Lig...Genomic Imprinting and the Evolutionary Psychology of Human Kinship - In the Light of Evolution
- LOC130061904 [Homo sapiens]LOC130061904 [Homo sapiens]Gene ID:130061904Gene
- LOC127888357 [Homo sapiens]LOC127888357 [Homo sapiens]Gene ID:127888357Gene
Your browsing activity is empty.
Activity recording is turned off.
See more...