Except where otherwise indicated, this work is licensed under a Creative Commons Attribution 4.0 International License
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Rohrmann GF. Baculovirus Molecular Biology [Internet]. 4th edition. Bethesda (MD): National Center for Biotechnology Information (US); 2019.
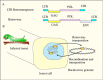
There are a variety of classes of transposable elements (TEs) integrated into the genomes of all cells and they are often major components of cellular genomes. For example, from 0.3% of bacterial genomes up to almost 80% of some vertebrate genomes are composed of these elements (1). This chapter will focus on two types of TEs that were discovered because they were found integrated into the AcMNPV genome. These include a DNA transposon called piggyBac that is of major interest as a vector for engineering cell lines and organisms. In addition to DNA transposons, many TEs are retroelements, which have an RNA intermediate. They often encode a reverse transcriptase, which can convert the RNA of the retroelement into cDNA that is then integrated into the host cell genome. A major category of retroelements includes the retroviruses that are infectious and can spread between organisms. The genome of the retrovirus becomes integrated into a host genome and is called a provirus and contains long terminal repeats (LTR) at either end that encode regulatory elements, a gag gene that encodes a structural (capsid) protein, the polymerase gene that encodes several enzymatic functions, and the envelope gene, env, that provides the virus with the ability to infect other cells (Figure 1A). There are several different categories of retroelements; some lack LTRs, while others lack an env gene, and are normally confined to a cell and are not infectious. In addition, some do not encode a reverse transcriptase, but rely on other retroelements to supply this enzyme. The focus of this chapter is on a group of retroelements found in insects that resemble retroviruses.
A baculovirus-associated errantivirus (retrovirus)
Few polyhedra (FP) mutants are a readily observable baculovirus phenotype that result in reduced numbers of polyhedra and an elevated titer of budded virus. Such mutants often contain an insert in the fp-25k gene (ac61), although the phenotype can result from mutations elsewhere in the genome. In the process of characterizing AcMNPV FP mutants produced after 25 passages in Trichoplusia ni cells, an isolate, (FP-D), was found to contain an integrated retrotransposon that originated from the host genome. It was called transposable element D or TED (2). This element had features of a retrovirus, including long terminal repeats and was demonstrated to express gag, pol, and env-like genes that are capable of being incorporated into virus-like particles (3) (4) (5). Normally retroviruses that integrate into a genome remain integrated and are spread and amplified by the RNA intermediate that is transcribed from the integrated provirus by the host cell RNA polymerase II. In contrast, the TED provirus was found to be unstable and upon excision, a copy of one LTR of about 270 bp remained in the baculovirus genome (6). This instability was probably due to the large size of the TED genome relative to the baculovirus genome. Viruses related to TED have been found in other insects with the retrotransposon called gypsy from Drosophila being the most well studied example.
The insect retroelements that encode an env gene are called the errantiviruses (from Latin errans, to wander). Although similar to retroviruses, they have not been included within the Retroviridae because they are a distinct lineage, and evidence that they are infectious is indirect (see below). Kanga and roo-like retroviruses also encode a related env gene (7). These retrovirus-like elements are often found in multiple copies and are present as apparent complete and truncated or defective genomes. For example, in the Drosophila melanogaster genome there are five different categories of errantiviruses encompassing 78 complete or partial sequences that range from a single full-length copy of gypsy to 18 full length/39 partial copies of the element 297. Other categories include 17.6 (7 full length/5 partial) and idefix (2 full length/5 partial). A fifth category, zam, was not found in this sequence, indicating the variability of errantivirus distribution between D. melanogaster strains (8).
The fp25-locus (Ac61); a remnant of a LINE-1 integration? Fp-25k (Ac61) is present in the genomes of most if not all Alpha-, Beta-, and Gammabaculoviruses. Analysis using the structure prediction program Hhpred (9) indicates that fp25k is related to orf1p of the Line-1 group of retrotransposons with a probability of 99.8%. Orf1p acts as a nucleic acid chaperone and similar to orf1p, fp25k has a coiled-coil domain and a predicted RNA binding motif (10). Deletion or mutations in the fp-25 locus are not lethal, but results in a 'few polyhedra phenotype' (fp) (11, 12). FP mutants are defective in virion occlusion and nucleocapsid envelopment in nuclei and release two- to fivefold more infectious BV than wt in infected Sf9 cells (12, 13). Although the significance the relatedness is compelling, how this gene adapted to baculovirus biology is unclear.
Errantiviruses in Lepidoptera
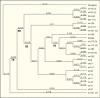
Two of the main cell lines used for baculovirus research were derived from primary explants of pupal ovarian tissue from moths of the family Noctuidae. One is from the fall army worm (Spodoptera frugiperda (Sf)) (14), while the other is derived from the cabbage looper, Trichoplusia ni (Tn) (15). In a survey of the genomes of these cell lines, using degenerate oligomers targeted to a conserved region of the errantivirus reverse transcriptase gene, over 20 different PCR products from each cell line were amplified, cloned, and sequenced. Analysis of these sequences resulted in the identification of over 20 lineages that could be grouped into several major clades (Figure 2). Three of the sequences were identical to the TED errantivirus described above (16). Phylogenetic analyses indicated that most of the Sf and Tn sequences were closely related to each other and to sequences from other Lepidoptera. The next most closely related sequences were from the Drosophila (Diptera). However, there are several sequences from both Sf and Tn that form lineages distinct from the majority of the lepidopteran or dipteran sequences. This research was confirmed for S. frugiperda cells when genome and transcriptome assemblies were characterized (17). Thirteen different errantivirus sequences were identified, nine were similar to the lineages previously identified from partial sequences (16). Most of these elements were closely related to each other and to the TED element from T. ni although two lineages were significantly different. Five of the elements appeared to be transcribed and had relative abundances of 763, 292, 67, 19, and 8. The elements with the highest level of transcripts present (763 and 292) corresponded to sf37 and sf20 that were also the two most abundant sf lineages in the previous study (16).
Relationships between insect retroviruses and baculoviruses: the env gene
Phylogenetic analyses of the errantivirus env gene and baculovirus genome sequence data resulted in the unexpected observation that the errantivirus env gene is related to the baculovirus envelope fusion protein, or F protein lineage (18, 19) (20). The sequence similarity was most striking in the region that includes the furin cleavage signal (RxxR) and a predicted fusion peptide immediately downstream.
This fusion peptide of about 20 amino acids is highly hydrophobic except for 2 D and 1 K residue (Figure 3). The errantiviruses most likely obtained the F protein from a recombination event that occurred when a retrotransposon integrated into a baculovirus genome (Figure 1). Evidence for such an event is compelling because the errantivirus TED was found integrated into a baculovirus genome as described above (2). Baculoviruses have two different envelope fusion proteins: gp64 and F. GP64 appears to have been recently incorporated into one baculovirus lineage called Group I. Whereas the Group I baculoviruses retain a copy of the F gene, it no longer functions independently as a fusion protein. In contrast, most baculoviruses (Group II) lack gp64 and contain only a copy of the F gene, suggesting that it is the fusion protein in these viruses (21) (see Figure 2, Chapter 2).
In addition to the strong evidence for the capture of a baculovirus envelope fusion protein leading to the evolution of errantiviruses, this phenomenon appears to be a relatively common event in virus evolution and may have occurred a number of times (18, 22). It has not only occurred with elements such as retrotransposons which commonly integrate into DNA, but also has been observed for a variety of other categories of viruses, including members of the Orthomyxoviridae and with the baculovirus gp64 gene described above (21) (see Figure 7, Chapter 2).
Cellular homologs of baculovirus F/errantivirus env proteins
In addition to the relatedness of the baculovirus F and errantivirus env genes, a cellular homolog in the Drosophila genome sequence was also identified (19). However, this protein is not cleaved (see below), does not have membrane fusion activity, and appears to localize to intracellular organelles rather than cell membranes (23). This gene was determined to have entered the Drosophila lineage once, and another time into a mosquito lineage. In Drosophila it is expressed in most tissues analyzed in both adult males and females. It was suggested that it was incorporated into and retained by the insect genome because its expression could protect the host cell from infection by retroviruses or baculoviruses that shared a related env protein. This could be accomplished if the cellular homolog binds to and interferes with the viral receptors on the cell surface or if they act as dominant negative inhibitors in which the endogenous env would complex with and inactivate the viral env protein (7).
Features of baculovirus F and insect retrovirus env proteins: Class I fusion proteins
The baculovirus F and errantivirus env proteins appear to be members of the Class I (24) group of envelope fusion proteins common to many vertebrate viruses. Although, in general, they lack sequence relatedness, it has long been suggested that a number of envelope fusion proteins from a variety of disparate viruses are related. This is based on their requirement for cleavage to be activated and the fact that one of the resulting peptides is membrane associated via a transmembrane domain. In addition, the membrane-associated peptide contains a hydophobic fusion peptide domain downstream of a cleavage site followed by predicted coiled-coil domains that are involved in forming hairpin-like structures that are important in virus-cell fusion (25, 26). Such structures have been characterized in fusion proteins from viruses as diverse as filoviruses, retroviruses, orthomyxoviruses, and paramyxoviruses (24). Evidence suggests that baculovirus F proteins are members of this group (Figure 3), and it has been demonstrated that they require cleavage, most likely by the host cell furin protease, for activation (27, 28). Errantivirus env proteins also have similar features (19) consistent with their being members of this group. Furthermore, gypsy env accumulates at the cell membrane as would be expected for a viral envelope protein and contains a predicted furin cleavage site; it is cleaved when expressed in both Drosophila S2 and Sf9 cells (29). Recently with the determination of structures of a variety of viral fusion proteins, the baculovirus F proteins are predicted to have a high degree of structural similarity to the F proteins of paramxyoviruses by the Hhpred structure prediction program (30) (see Chapter 2).
Additional relationships of insect retroviruses and baculoviruses
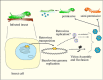
The relatedness of the errantivirus env protein and the baculovirus F homologs may reflect more than a fortuitous recombination event between these two viruses. The errantivirus TED is a mid-repetitive element (about 50 copies/genome) in Trichoplusia ni (6) and is capable of transposition from the insect into the baculovirus genome. A key feature of the relationship that may have led to the capture of a baculovirus F gene by a pre-errantivirus retrotransposon involves the ability of baculoviruses to express genes at very high levels. This feature appears to be due at least in part to the fact that they encode an RNA polymerase (31) capable of high levels of transcription in the context of the virus replication program. This polymerase recognizes a unique promoter sequence (A/G/TTAAG) (32) that is found in the TED LTR as a palindrome. Late in the baculovirus infection, mRNA is expressed from these LTRs at high levels (6). Therefore, integration into a baculovirus genome may reflect a strategy to exploit baculovirus late gene expression to express the integrated retrotransposon/retrovirus genome at high levels. This could result in the production of a mixture of retrovirus particles and occluded baculoviruses containing integrated retroviruses and would provide two methods of escape from an insect with a fatal baculovirus infection: they could either survive by integration into baculovirus genomes, or possibly as infectious virus particles (Figure 4). The evolution of this relationship between a baculovirus and a primordial LTR-type retrotransposon provides a clear pathway, via DNA recombination, for the transposable element to incorporate the baculovirus F homolog into its genome, thereby converting it into a potentially infectious retrovirus (Figure 1B).
Are errantiviruses infectious?
Early on it was noted that retrovirus-like particles and reverse transcriptase activity were present in Drosophila cells (33). Subsequently, gypsy became the most intensively studied retrovirus-like element in D. melanogaster. Indirect evidence suggests that gypsy is infectious for Drosophila (34, 35). These data were obtained by feeding a strain of Drosophila that lacks active gypsy transposition with either purified virus like particles (vlps) from insects with transpositionally active gypsy, or extracts derived from such insects, and then documenting increased levels of transposition in the recipient insects. Similarly, it was observed that gypsy could be transmitted between cells in culture (36).
Does env play a role in errantivirus infectivity?
The evidence implicating env in errantivirus infectivity is varied. In one study, it was found that a preparation of two monoclonal antibodies against gypsy env mixed with the vlp fraction reduced the number of insertion events in insect feeding experiments (37). In addition, evidence suggests that an integrated Moloney leukemia virus-luciferase construct pseudotyped with a gypsy envelope is infectious for Drosophila cells (38). This suggested that gypsy env is capable of mediating infection of Drosophila cells. Although gypsy may be infectious, its infectivity appears to be very limited. It has been suggested that since they are adapted for integration into the cell genome, they no longer require propagation via infection (39). However, this does not explain why they have retained, conserved, and continue to express an env gene. Since envelope proteins often play major roles in the virulence of other viruses (40), the errantivirus env proteins may allow the viruses to be infectious and spread between different organisms, but this infectivity may be restricted by features of their env proteins. It has also been suggested that gypsy env has fusogenic properties (41).
The invasion and amplification of retroelements
TED, the baculovirus associated errantivirus described above that was originally found integrated into the AcMNPV genome is a particularly useful example of how a virus could spread a transposable element to other species. One can imagine a situation in insects where a baculovirus abortively infects a species that might be semi permissive for virus replication (Figure 4). The infection could result in low levels of baculovirus replication that would allow mRNA expression, cDNA production and transposition of a TE from the invading baculovirus into host cell DNA, but the host would eventually overcome and survive the baculovirus infection. An example of such virus/insect combinations could be AcMNPV pathogenesis in Helicoverpa zea which is over 1000-fold more resistant to fatal infection than Heliothis virescens (42). The primary midgut infection and secondary infection of the tracheal epidermis are similar in the two insects, but H. zea larvae are able to encapsidate infected cells, thereby limiting the infection. Under these conditions, the surviving insects could have been exposed to and possibly parasitized by a TE carried by the invading baculovirus, whereas in contrast, the carrier baculovirus would have been eliminated.
Once a retroelement integrates into a genome, they can be transcribed into RNA, reverse transcribed into cDNA that can integrate elsewhere in the genome. Via this process they can greatly amplify their copy number and the size of the genome of their host organism. Depending on the element, some TEs can transpose at a rate of from 10_3 to 10_5 per element per generation. Consequently, they can be more significant in the production of genetic change that normal nucleotide base changes which are altered at about 10_8–10_9 per nucleotide per generation. Bursts of transpositional activity are thought to have been a major force in the evolution of new species (1). For example, the activation of a gypsy-like retrotransposon in plants of the genus Gossypium, which includes cotton, is responsible for a three-fold difference in the genome size between some species. Similarly, the amplification of several TE families has led to the doubling of the size of the rice genome. These elements often accumulate in heterochromatic regions of the genome. Such regions normally contain repetitive DNA and are transcriptionally inactive and include centromeres and telomeres (43).
What prevents retroelements from amplifying continuously?
The insertion of transposable elements can interrupt genes, alter regulatory regions such as promoters and enhancers, and disrupt patterns of splicing and 3’ transcriptional processing and therefore can be highly mutagenic. Mutations in germ line cells could affect the success of progeny. In somatic cells, mutations could cause localized disruption of cell function, or they could cause more generalized effects if they alter the regulation of functions such as cell division that could lead to the production of oncogenic cells. Consequently, molecular systems have evolved to defend cells against TEs. Therefore, although molecular evidence suggests that amplification of TEs is a major feature of eukaryotic genome evolution, most are eventually silenced by the host. If a lineage survives the invasion and amplification of a TE, at some point its further proliferation is quelled. This is accomplished by several epigenetic mechanisms that involve inherited processes that do not affect the DNA sequence. These include post-transcriptional silencing by RNAi, chromatin modifications by changes in DNA methylation patterns, histone modifications including methylation, and changes in chromatin condensation and packing. It is likely that these processes work cooperatively to both suppress transcription and to eliminate RNA that is expressed.
Suppression of transposable elements by DNA and histone methylation
Although endogenous retroviruses are normally silenced, their active transcription can be detected at different stages during development. For example, a number of retroelements in Drosophila show patterns of spatially regulated expression during embryogenesis (44). DNA methylation involves the addition of a methyl group to the cytocine in CpG sequences. CpG sequences are often found concentrated in ‘islands’ present in promoter regions. Such regions are often characterized by increased histone occupancy with a corresponding reduction in the binding of transcription factors. Methylated DNA can also attract methyl-binding proteins that also can inhibit transcription. It has been suggested that one of the primary roles for this phenomenon is to prevent transcription of TEs thereby protecting the host from endogenous retroviruses, reviewed in (45, 46). Although some insect genomes are highly methylated, the patterns of these modifications may differ from other organisms. It may be associated with non-CpG dinucleotides and may not be focused on mobile sequences that are often heavily methylated in the genomes of vertebrates and plants (47, 48). Transcription can also regulated both positively and negatively by the pattern of histone methylation (49).
Suppression of transposable elements by RNA interference
Three pathways involved in RNA silencing have been identified in both mammals and insects and a major focus of these pathways is the suppression of endogenous transposable elements. These pathways include: i) RNA interference (RNAi) that employs small interfering RNAs (siRNAs) that are derived from exogenous double stranded RNA (dsRNA) and can act as a defense against viral infection by targeting TE RNA for degradation. In addition, siRNAs can also be derived from endogenous sequences and are involved in suppressing the expression of endogenous transposable elements in somatic cells; ii) microRNAs (miRNAs) involve endogenous small RNAs that repress partially complementary mRNAs(50); and iii) Piwi-interacting RNAs (piRNA) that repress transposons in germ line cells and can also activate transcription in heterochromatin which is a gene-poor, highly condensed, DNA-protein complex, reviewed in (51). Therefore, both exogenous and endogenous RNAs can be inactivated and different mechanisms can be involved in germ line and somatic cells.
The Argonautes: proteins with RNAse activity that are critical in RNA interference
Key to the function of interfering RNAs is their interaction with Argonaute proteins (52). Although in Greek mythology the Argonautes were sailors on the ship Argo who accompanied Jason in his search for the Golden Fleece, in molecular biology the term was originally used to describe the shape of the leaves of a mutant of Arabidopsis thaliana, AGO1, because they resembled the squid Argonauta argo (53). The Argonaute proteins facilitate both the processing of some micro RNAs by eliminating the non-active siRNA strand and also use small RNAs as guides to identify and repress complementary transcripts by degradation (via endonuclease activity) or by inhibition of translation. They appear to have evolved from the RNAse H family of endonucleases but have substituted ssRNA for ssDNA as the template to target RNA, reviewed in (52).
Suppression of transposable elements in gonadal cells
A major category of piRNAs (54) (55) include rasiRNAs (repeat associated siRNAs) that are involved in the silencing of transposable elements. PiRNAs map to repetitive elements throughout the Drosophila genome, however a limited number of loci called piRNA clusters appear to match most piRNAs. The transposons in the clusters involved in piRNA production appear to be truncated or defective relicts and are probably not capable of autonomous expression or transposition. It has been suggested that piRNAs are derived from long single stranded precursor RNAs in which a 5' cleavage occurs at a uridine residue. The sequence then becomes incorporated into a Piwi protein complex where a second cleavage occurs generating the specific size. The piRNA then targets the piwi complex to RNA expressed from transposable elements and can interfere with splicing of the retroelement (56). Further evidence for the role of Piwi type proteins in the suppression of transposable elements in gonadal tissue was the observation that a mutation in piwi reduced the repression of gypsy in restricted tissues leading to up to a 150 fold increase in gypsy RNA levels (57, 58) (54). PiRNAs have also been described from Bombyx mori and they appear to include a major subclass of rasiRNAs and are thought to be involved in transposon silencing and development of germ line cells (59, 60) (61). The evolution of piRNA clusters has been examined experimentally by incorporating GFP into silkworm cells via piggyBac transposition and then isolating cell lines. It was observed that these lines amplified piRNAs capable of silencing GFP providing a system to examine a pathway for the development of piRNAs against a new insertion (62). Similar investigations have also been carried out with Drosophila and show that a piRNA complex can be activated against an invading mobile element within a single generation, reviewed in (63). Although it has been thought that suppression of TEs might prevent their accumulation, it has been argued that, in fact, by silencing their activity it promotes their incorporation by preventing the subsequent damage that they can cause. It was also suggested that this was important for genome expansion (64).
In Drosophila; flamenco, a source for piRNAs
A locus called flamenco controls the activity of retroviral elements gypsy (65), idefix, and zam (66). The flamenco locus was mapped to a region that corresponds to a piRNA cluster spanning a region of 179 kb and is comprised of nested transposable elements and fragments including those specific to gypsy, idefix and zam, in addition to other transposable element-specific sequences. Such regions are called piRNA clusters and essentially lack protein coding sequences and are comprised of truncated or damaged copies of TEs that appear to lack the capacity to be mobilized and are concentrated in the pericentromeric or telomeric heterochromatin (58, 67). Other loci that are involved piRNA production have also been described (68, 69) and are reviewed in (54).
Activation of endogenous retroelement sequences during a baculovirus infection
A major question is if the baculovirus infection influences the host cell silencing systems. In an investigation examining the expression of HzSNPV in hemocytes of H. virescens, it was observed that over a hundred genes that were differentially regulated after infection were derived from sequences related to endogenous retroelements. Many of these genes were upregulated from 3 to 11 fold compared to controls (70). Similarly, infection of Antherea pernyi with ApNPV, 12 full-length LTR-retrotransposons significantly changed their levels of expression with 6 being up- and 6 being down-regulated (71). Also in Helicoverpa zea cells reverse transcriptase activity was up-regulated after infection with HearNPV (72). This may be related to the reduction in piRNA population in sf21 cells 12 hpi with SpliNPV (73). This suggests that in these systems virus infection might relax or inhibit the silencing systems present in their host cells thereby facilitating gene expression and possibly amplification of endogenous TEs. This is not unexpected as viruses have been demonstrated to interfere with RNAi systems in a variety of organisms (74). However, it is not clear how the non-specific effects of baculovirus replication might influence this process. For instance, it is well documented that transcription of many RNA polymerase II transcribed genes is turned off as the baculovirus infection progresses (see Chapter 4 and 10) (75-77). It has also been shown that RNA polymerase II transcripts of viral mRNA are degraded late in infection, e.g. see (78). This could interfere with RNAi production if it was carried out by this polymerase. How these characteristics of baculovirus infection might affect the RNA pol II transcripts derived from the integrated provirus such as TED (79) remains to be determined.
Piggybac, a transposon from Trichoplusia ni, that was originally isolated from a baculovirus.
Isolation of piggyBac
The adaptation of cell lines from Trichoplusia ni and Spodoptera frugiperda that were permissive AcMNPV and related viruses established the basis for understanding the molecular events of baculovirus replication. These cell systems supported both budded virus production early in infection and occluded virions later in the replication cycle. Because they were readily visualized, occlusion bodies provided a convenient marker of infection. As investigators became familiar with this system, it was noted that occlusion body mutants occasionally occurred in which there were fewer than the normal number of occlusion bodies produced. These were called ‘few polyhedra’ or FP mutants and were also characterized by elevated levels of budded virus production such that in cell culture they had a selective advantage and could out compete wt virus (80, 81). Characterization of these mutants indicated that they were derived by the insertion of cellular DNA at a specific location in the genome. This locus was eventually determined to be AcMNPV orf61 that encodes a predicted protein of 214 amino acids (25 kDa) and was called the ‘few polyhedra’ or fp gene (11) (82). Subsequently, it was determined that one of the inserts, IFP2 was found in the genome of AcMNPV grown in T. ni cells. It inserted at TTAA sites that were duplicated upon insertion, contained 13-bp inverted terminal repeats and encoded a 594 amino acid transposase that could facilitate its own transposition and it was renamed piggyBac (83) (84). PiggyBac shows precise excision upon transposition such that the TTAA target site is not duplicated and therefore leaves no footprint upon removal (82) (83).
Up to 98 copies of sequences similar to piggyBac were found in the B. mori genome, although only 5 appeared to be complete (85). A related transposon was also identified in the silkworm genome (86) and named yabusame element after a type of Japanese archery performed while riding a horse. (Perhaps the bow represents the transposase, the arrow the transposon, DNA is the target, and the archer on the horse is the cell?). Related transposons have now been found in many different species from plants, insects and mammals, reviewed in (87).
Although piggyBac demonstrates limited sequence similarity with other transposon superfamilies, its transposase contains DDD acidic residues in its catalytic site suggesting that it belongs to the DDE superfamily of recombinases. In these recombinases, the acidic amino acids DDE or DDD coordinate metal ions required for activity (88).
PiggyBac has potential in many different systems
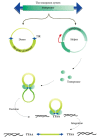
PiggyBac has demonstrated widespread potential as a genetic tool because of its ability to integrate into a variety of heterologous systems including numerous insect species (89), mammals including human (90) (91) and mice (92), planaria (93), the malaria parasite Plasmodium falciparum (94) and the schistosome, Schistosoma mansoni (95). In addition to its widespread host range, it is regarded as having great potential for applications in the production of transgenic animals and in gene therapy. It is normally delivered in a combination of two plasmids; a helper plasmid encoding the transposase and the transfer plasmid containing the transgene flanked by the specific inverted terminal repeats that bind the transposase that subsequently cleaves out the transgene and directs it to and facilitates its integration into the target site (Figure 5). The advantages of piggybac have been reviewed by (92) (96), and elsewhere. They include: i) its ability to be efficiently inserted into germline cells and the levels of insertion can be increased by optimizing the transposase codon preferences to the host species being engineered. In addition, the inserts are stably integrated when the piggyBac transposase is absent and there is a lower level of silencing of piggyBac relative to other transposons such as Sleeping Beauty or Tol2. It also has the potential to be engineered for more specificity of its integration site (97); ii) single copies of the transgene are inserted rather than concatemers that are often produced by other systems. Such multiple inserts can be detrimental because they can result in higher than normal levels of the expression of the insert; iii) the sites of integration can be rapidly identified using inverse PCR allowing rapid analysis of their potential effects at the integration site. iv) PiggyBac allows the integration of inserts of up to 100 kb (98), significantly larger than other vectors such as those derived from retroviruses and adeno associated viruses. These features have led Meir et al (96) to conclude ‘Collectively, piggyBac is currently the most promising DNA transposon for gene and stem cell therapy because of its highly effective transposition activity, large cargo capacity, ability of stably expressing transgene, capability to be molecularly engineered to achieve site-specific gene targeting, and the unique feature of generating foreign DNA-free iPSCs (induced pluripotent stem cells).’
Another DNA Transposon found in a baculovirus genome.
In the course of a genome sequencing project another DNA transposon encoding a transposase and a novel adjacent orf were found integrated into the genome of an NPV isolated from an oak looper, Lambdina fiscellaria (99). The transposase is most closely related (49% amino acid identity) to an orf in Amyelois transitella, the navel orange worm moth, also a lepidopteran. The second orf was also most closely related to an orf in this same insect.
References
- 1.
- Biemont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–4. [PubMed: 17024082]
- 2.
- Miller DW, Miller LK. A virus mutant with an insertion of a copia-like transposable element. Nature (London). 1982;299:562–564. [PubMed: 6289125]
- 3.
- Hajek KL, Friesen PD. Proteolytic processing and assembly of gag and gag-pol proteins of TED, a baculovirus-associated retrotransposon of the gypsy family. J Virol. 1998;72:8718–24. [PMC free article: PMC110286] [PubMed: 9765414]
- 4.
- Lerch RA, Friesen PD. The baculovirus-integrated retrotransposon TED encodes gag and pol proteins that assemble into viruslike particles with reverse transcriptase. J Virol. 1992;66:1590–601. [PMC free article: PMC240890] [PubMed: 1371168]
- 5.
- Ozers MS, Friesen P. The Env-like open reading frame of the baculovirus-integrated retrotransposon TED encodes a retrovirus-like envelope protein. Virology. 1996;226:252–9. [PubMed: 8955045]
- 6.
- Friesen PD, Rice WC, Miller DW, Miller LK. Bidirectional transcription from a solo long terminal repeat of the retrotransposon TED: Symmetrical RNA start sites. Mol Cell Biol. 1986;6:1599–1607. [PMC free article: PMC367686] [PubMed: 3023897]
- 7.
- Malik HS, Henikoff S. Positive Selection of Iris, a Retroviral Envelope-Derived Host Gene in Drosophila melanogaster. PLoS Genet. 2005;1:429–43. [PMC free article: PMC1262188] [PubMed: 16244705]
- 8.
- Kaminker J, Bergman C, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler D, Lewis S, Rubin G, Ashburner M, Celniker S. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002:3. [PMC free article: PMC151186] [PubMed: 12537573]
- 9.
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430:2237–2243. [PubMed: 29258817]
- 10.
- Garretson TA, McCoy JC, Cheng XW. Baculovirus FP25K Localization: Role of the Coiled-Coil Domain. J Virol. 2016;90:9582–9597. [PMC free article: PMC5068512] [PubMed: 27512078]
- 11.
- Beames B, Summers MD. Location and nucleotide sequence of the 25k protein missing from baculovirus few polyhedra (FP) mutants. Virology. 1989;168:344–353. [PubMed: 2644735]
- 12.
- Harrison RL, Jarvis DL, Summers MD. The role of the AcMNPV 25K gene, "FP25," in baculovirus polh and p10 expression. Virology. 1996;226:34–46. [PubMed: 8941320]
- 13.
- Fraser MJ, Brusca JS, Smith GE, Summers MD. Transposon mediated mutagenesis of a baculovirus. Virology. 1985;145:356–361. [PubMed: 2992159]
- 14.
- Vaughn JL, Goodwin RH, Tompkins GJ, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae). In Vitro. 1977;13:213–217. [PubMed: 68913]
- 15.
- Hink WF. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature. 1970;226:466–467. [PubMed: 16057320]
- 16.
- Menzel T, Rohrmann GF. Diversity of errantivirus (retrovirus) sequences in two cell lines used for baculovirus expression, Spodoptera frugiperda and Trichoplusia ni. Virus Genes. 2008;36:583–586. [PubMed: 18363091]
- 17.
- Geisler C. A new approach for detecting adventitious viruses shows Sf-rhabdovirus-negative Sf-RVN cells are suitable for safe biologicals production. BMC Biotechnol. 2018;18:8. [PMC free article: PMC5803895] [PubMed: 29415704]
- 18.
- Malik HS, Henikoff S, Eickbush TH. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–18. [PubMed: 10984449]
- 19.
- Rohrmann GF, Karplus PA. Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC Evol Biol. 2001;1:1. [PMC free article: PMC29073] [PubMed: 11244578]
- 20.
- Terzian C, Pélisson A, Bucheton A. Evolution and phylogeny of insect endogenous retroviruses. BMC Evol Biol. 2001:1. [PMC free article: PMC57737] [PubMed: 11591216]
- 21.
- Pearson MN, Rohrmann GF. Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J Virol. 2002;76:5301–5304. [PMC free article: PMC137044] [PubMed: 11991958]
- 22.
- Kim FJ, Battini JL, Manel N, Sitbon M. Emergence of vertebrate retroviruses and envelope capture. Virology. 2004;318:183–91. [PubMed: 14972546]
- 23.
- Lung O, Blissard GW. A cellular Drosophila melanogaster protein with similarity to baculovirus F envelope fusion proteins. J Virol. 2005;79:7979–89. [PMC free article: PMC1143756] [PubMed: 15956544]
- 24.
- Harrison SC. Viral membrane fusion. Virology. 2015;479-480:498–507. [PMC free article: PMC4424100] [PubMed: 25866377]
- 25.
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. [PubMed: 11395423]
- 26.
- Skehel J, Wiley D. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. [PubMed: 10966468]
- 27.
- Westenberg M, Wang H, IJkel WF, Goldbach RW, Vlak JM, Zuidema D. Furin is involved in baculovirus envelope fusion protein activation. J Virol. 2002;76:178–84. [PMC free article: PMC135720] [PubMed: 11739683]
- 28.
- Pearson MN, Russell RLQ, Rohrmann GF. Functional analysis of a conserved region of the baculovirus envelope fusion protein, LD130. Virology. 2002;304:81–88. [PubMed: 12490405]
- 29.
- Pearson MN, Rohrmann GF. Conservation of a proteinase cleavage site between an insect retrovirus (gypsy) env protein and a baculovirus envelope fusion protein. Virology. 2004;322:61–68. [PubMed: 15063117]
- 30.
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. [PubMed: 15531603]
- 31.
- Guarino LA, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol. 1998;72:7985–7991. [PMC free article: PMC110134] [PubMed: 9733837]
- 32.
- Rohrmann GF. Polyhedrin structure. J Gen Virol. 1986;67:1499–1513. [PubMed: 3525744]
- 33.
- Heine CW, Kelly DC, Avery RJ. The detection of intracellular retrovirus-like entities in Drosophila melanogaster cell cultures. J Gen Virol. 1980;49:385–95. [PubMed: 6160197]
- 34.
- Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–57. [PubMed: 7958877]
- 35.
- Kim A, Terzian C, Santamaria P, Pelisson A, Prud'homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1994;91:1285–9. [PMC free article: PMC43142] [PubMed: 8108403]
- 36.
- Syomin BV, Fedorova LI, Surkov SA, Ilyin YV. The endogenous Drosophila melanogaster retrovirus gypsy can propagate in Drosophila hydei cells. Mol Gen Genet. 2001;264:588–94. [PubMed: 11212913]
- 37.
- Song SU, Kurkulos M, Boeke JD, Corces VG. Infection of the germ line by retroviral particles produced in the follicle cells: a possible mechanism for the mobilization of the gypsy retroelement of Drosophila. Development. 1997;124:2789–98. [PubMed: 9226450]
- 38.
- Teysset L, Burns JC, Shike H, Sullivan BL, Bucheton A, Terzian C. A Moloney murine leukemia virus-based retroviral vector pseudotyped by the insect retroviral gypsy envelope can infect Drosophila cells. J Virol. 1998;72:853–6. [PMC free article: PMC109448] [PubMed: 9420299]
- 39.
- Pelisson A, Mejlumian L, Robert V, Terzian C, Bucheton A. Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene. Insect Biochem Mol Biol. 2002;32:1249–1256. [PubMed: 12225916]
- 40.
- Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. [PubMed: 10329563]
- 41.
- Misseri Y, Cerutti M, Devauchelle G, Bucheton A, Terzian C. Analysis of the Drosophila gypsy endogenous retrovirus envelope glycoprotein. J Gen Virol. 2004;85:3325–31. [PubMed: 15483247]
- 42.
- Trudeau D, Washburn JO, Volkman LE. Central role of hemocytes in Autographa californica M nucleopolyhedrovirus pathogenesis in Heliothis virescens and Helicoverpa zea. J Virol. 2001;75:996–1003. [PMC free article: PMC113996] [PubMed: 11134313]
- 43.
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–85. [PubMed: 17363976]
- 44.
- Ding D, Lipshitz HD. Spatially regulated expression of retrovirus-like transposons during Drosophila melanogaster embryogenesis. Genet Res. 1994;64:167–81. [PubMed: 7698641]
- 45.
- Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329–47. [PubMed: 18818875]
- 46.
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–90. [PubMed: 17708532]
- 47.
- Field LM, Lyko F, Mandrioli M, Prantera G. DNA methylation in insects. Insect Mol Biol. 2004;13:109–15. [PubMed: 15056357]
- 48.
- Schaefer M, Lyko F. DNA methylation with a sting: an active DNA methylation system in the honeybee. Bioessays. 2007;29:208–11. [PubMed: 17295216]
- 49.
- Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. [PMC free article: PMC2857722] [PubMed: 20210320]
- 50.
- Grassmann R, Jeang KT. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta. 2008 [PMC free article: PMC2641032] [PubMed: 18549828]
- 51.
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–81. [PMC free article: PMC2953241] [PubMed: 18403677]
- 52.
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. [PubMed: 18073770]
- 53.
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. Embo J. 1998;17:170–80. [PMC free article: PMC1170368] [PubMed: 9427751]
- 54.
- Senti KA, Brennecke J. The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet. 2010;26:499–509. [PMC free article: PMC4988489] [PubMed: 20934772]
- 55.
- Leslie M. The immune system's compact genomic counterpart. Science. 2013;339:25–7. [PubMed: 23288523]
- 56.
- Teixeira FK, Okuniewska M, Malone CD, Coux RX, Rio DC, Lehmann R. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature. 2017;552:268–272. [PMC free article: PMC5933846] [PubMed: 29211718]
- 57.
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–21. [PMC free article: PMC1470774] [PubMed: 15082550]
- 58.
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. [PubMed: 17346786]
- 59.
- Kawaoka S, Hayashi N, Katsuma S, Kishino H, Kohara Y, Mita K, Shimada T. Bombyx small RNAs: genomic defense system against transposons in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38:1058–65. [PubMed: 18801438]
- 60.
- Kawaoka S, Minami K, Katsuma S, Mita K, Shimada T. Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem Biophys Res Commun. 2008;367:755–60. [PubMed: 18191035]
- 61.
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA. 2009;15:1258–64. [PMC free article: PMC2704083] [PubMed: 19460866]
- 62.
- Kawaoka S, Mitsutake H, Kiuchi T, Kobayashi M, Yoshikawa M, Suzuki Y, Sugano S, Shimada T, Kobayashi J, Tomari Y, Katsuma S. A role for transcription from a piRNA cluster in de novo piRNA production. RNA. 2011;18:265–73. [PMC free article: PMC3264913] [PubMed: 22194309]
- 63.
- Levine MT, Malik HS. Learning to protect your genome on the fly. Cell. 2011;147:1440–1. [PMC free article: PMC3725141] [PubMed: 22196722]
- 64.
- Fedoroff NV. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–67. [PubMed: 23145453]
- 65.
- Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, Corces VG. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. Embo J. 1994;13:4401–11. [PMC free article: PMC395367] [PubMed: 7925283]
- 66.
- Desset S, Meignin C, Dastugue B, Vaury C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–9. [PMC free article: PMC1462594] [PubMed: 12807771]
- 67.
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. [PubMed: 17975059]
- 68.
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–9. [PubMed: 19812547]
- 69.
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, Seitz H, Zamore PD, Weng Z, Theurkauf WE. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–49. [PMC free article: PMC2770713] [PubMed: 19732946]
- 70.
- Breitenbach JE, Shelby KS, Popham HJR. Baculovirus Induced Transcripts in Hemocytes from the Larvae of Heliothis virescens. Viruses. 2011;3:2047–2064. [PMC free article: PMC3230841] [PubMed: 22163334]
- 71.
- Feng M, Ren F, Zhou Y, Zhang N, Lu Q, Swevers L, Sun J. Correlation in Expression between LTR Retrotransposons and Potential Host Cis-Targets during Infection of Antherea pernyi with ApNPV Baculovirus. Viruses. 2019:11. [PMC free article: PMC6563192] [PubMed: 31064084]
- 72.
- Nguyen Q, Chan LC, Nielsen LK, Reid S. Genome scale analysis of differential mRNA expression of Helicoverpa zea insect cells infected with a H. armigera baculovirus. Virology. 2013;444:158–70. [PubMed: 23827436]
- 73.
- Kharbanda N, Jalali SK, Ojha R, Bhatnagar RK. Temporal expression profiling of novel Spodoptera litura nucleopolyhedrovirus-encoded microRNAs upon infection of Sf21 cells. J Gen Virol. 2015;96:688–700. [PubMed: 25481752]
- 74.
- Berkhout B, Haasnoot J. The interplay between virus infection and the cellular RNA interference machinery. FEBS Lett. 2006;580:2896–902. [PMC free article: PMC7094296] [PubMed: 16563388]
- 75.
- Ooi BG, Miller LK. Regulation of host RNA levels during baculovirus infection. Virology. 1988;166:515–523. [PubMed: 2459844]
- 76.
- Nobiron I, O'Reilly DR, Olszewski JA. Autographa californica nucleopolyhedrovirus infection of Spodoptera frugiperda cells: a global analysis of host gene regulation during infection, using a differential display approach. J Gen Virol. 2003;84:3029–39. [PubMed: 14573808]
- 77.
- Katsuma S, Mita K, Shimada T. ERK- and JNK-dependent signaling pathways contribute to Bombyx mori nucleopolyhedrovirus infection. J Virol. 2007;81:13700–9. [PMC free article: PMC2168829] [PubMed: 17913811]
- 78.
- Blissard GW, Rohrmann GF. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology. 1989;170:537–555. [PubMed: 2658304]
- 79.
- Friesen PD, Nissen MS. Gene organization and transcription of TED, a Lepidopteran retrotransposon integrated within the baculovirus genome. Mol Cell Biol. 1990;10:3067–3077. [PMC free article: PMC360671] [PubMed: 1692964]
- 80.
- Fraser MJ, Hink WF. The isolation and characterization of the MP and FP variants of Galleria mellonella nuclear polyhedrosis virus. Virology. 1982;117:366–378. [PubMed: 18635122]
- 81.
- Fraser MJ, Smith GE, Summers MD. Aquisition of host cell DNA sequences by Baculoviruses; Relationships between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis virus. J Virol. 1983;47:287–300. [PMC free article: PMC255260] [PubMed: 16789244]
- 82.
- Cary LC. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–69. M. G, Corsaro BG, Wang H, Rosen E, Fraser MJ. [PubMed: 2549707]
- 83.
- Fraser MJ, Cary L, Boonvisudhi K, Wang HG. Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology. 1995;211:397–407. [PubMed: 7645244]
- 84.
- Elick TA, Bauser CA, Fraser MJ. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica. 1996;98:33–41. [PubMed: 8765680]
- 85.
- Xu HF, Xia QY, Liu C, Cheng TC, Zhao P, Duan J, Zha XF, Liu SP. Identification and characterization of piggyBac-like elements in the genome of domesticated silkworm, Bombyx mori. Mol Genet Genomics. 2006;276:31–40. [PubMed: 16685528]
- 86.
- Daimon T, Mitsuhiro M, Katsuma S, Abe H, Mita K, Shimada T. Recent transposition of yabusame, a novel piggyBac-like transposable element in the genome of the silkworm, Bombyx mori. Genome. 2010;53:585–93. [PubMed: 20725145]
- 87.
- Yusa K. 2015. piggyBac Transposon. Microbiol Spectr 3:MDNA3-0028-2014. [PubMed: 26104701]
- 88.
- Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27:1097–109. [PMC free article: PMC2323262] [PubMed: 18354502]
- 89.
- Handler AM. Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochem Mol Biol. 2002;32:1211–20. [PubMed: 12225912]
- 90.
- Wilson MH, Coates CJ, George AL Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–45. [PubMed: 17164785]
- 91.
- Tipanee J, VandenDriessche T, Chuah MK. Transposons: Moving Forward from Preclinical Studies to Clinical Trials. Hum Gene Ther. 2017;28:1087–1104. [PubMed: 28920716]
- 92.
- Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–83. [PubMed: 16096065]
- 93.
- Gonzalez-Estevez C, Momose T, Gehring WJ, Salo E. Transgenic planarian lines obtained by electroporation using transposon-derived vectors and an eye-specific GFP marker. Proc Natl Acad Sci U S A. 2003;100:14046–51. [PMC free article: PMC283543] [PubMed: 14615580]
- 94.
- Balu B, Shoue DA, Fraser MJ Jr, Adams JH. High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proc Natl Acad Sci U S A. 2005;102:16391–6. [PMC free article: PMC1275597] [PubMed: 16260745]
- 95.
- Morales ME, Mann VH, Kines KJ, Gobert GN, Fraser MJ Jr, Kalinna BH, Correnti JM, Pearce EJ, Brindley PJ. piggyBac transposon mediated transgenesis of the human blood fluke, Schistosoma mansoni. FASEB J. 2007;21:3479–89. [PubMed: 17586730]
- 96.
- Meir YJ, Wu SC. Transposon-based vector systems for gene therapy clinical trials: challenges and considerations. Chang Gung Med J. 2011;34:565–79. [PubMed: 22196059]
- 97.
- Kettlun C, Galvan DL, George AL Jr, Kaja A, Wilson MH. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol Ther. 2011;19:1636–44. [PMC free article: PMC3182353] [PubMed: 21730970]
- 98.
- Li MA, Turner DJ, Ning Z, Yusa K, Liang Q, Eckert S, Rad L, Fitzgerald TW, Craig NL, Bradley A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2012;39:e148. [PMC free article: PMC3239208] [PubMed: 21948799]
- 99.
- Rohrmann GF, Erlandson MA, Theilmann DA. Genome Sequence of an Alphabaculovirus Isolated from the Oak Looper, Lambdina fiscellaria, Contains a Putative 2-Kilobase-Pair Transposable Element Encoding a Transposase and a FLYWCH Domain-Containing Protein. Genome Announc. 2015:3. [PMC free article: PMC4447894] [PubMed: 26021909]
- A baculovirus-associated errantivirus (retrovirus)
- Errantiviruses in Lepidoptera
- Relationships between insect retroviruses and baculoviruses: the env gene
- Cellular homologs of baculovirus F/errantivirus env proteins
- Features of baculovirus F and insect retrovirus env proteins: Class I fusion proteins
- Additional relationships of insect retroviruses and baculoviruses
- Are errantiviruses infectious?
- Does env play a role in errantivirus infectivity?
- The invasion and amplification of retroelements
- What prevents retroelements from amplifying continuously?
- Suppression of transposable elements by DNA and histone methylation
- Suppression of transposable elements by RNA interference
- The Argonautes: proteins with RNAse activity that are critical in RNA interference
- Suppression of transposable elements in gonadal cells
- In Drosophila; flamenco, a source for piRNAs
- Activation of endogenous retroelement sequences during a baculovirus infection
- Piggybac, a transposon from Trichoplusia ni, that was originally isolated from a baculovirus.
- Another DNA Transposon found in a baculovirus genome.
- References
- Baculoviruses, retroviruses, DNA transposons (piggyBac), and insect cells - Bacu...Baculoviruses, retroviruses, DNA transposons (piggyBac), and insect cells - Baculovirus Molecular Biology
Your browsing activity is empty.
Activity recording is turned off.
See more...