NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Certain DNA sequences preferentially adopt multistranded, non-B-form structures under physiological conditions. These include three-stranded DNA triplexes and four-stranded DNA quadruplexes. Several lines of evidence suggest that multiplex structures can form in vivo, either from the addition of oligonucleotides or through the transient formation of single-stranded regions. The consequences of multiplex structures on many DNA-dependent biological processes have been described. In this chapter we will review the effects of different DNA multiplexes on the process of transcription. The influence of parameters such as multiplex type and multiplex formation conditions on different transcription mechanistic steps in organisms spanning from prokaryotes to Xenopus oocytes and mammalian cells will be discussed.
Introduction
Oligopurine/oligopyrimidine-rich DNA sequences have long been known to preferentially adopt multistranded structures quite different from the familiar Watson-Crick base-paired, right-handed, antiparallel-stranded, B-form double-helical structure.1,2 Examples include triple helical DNA (triplexes) and G-quartet-containing quadruplexes (G4), both of which can form under physiological conditions, and once formed, are extremely stable.3,6
Although a considerable amount of information is available about the properties of DNA multiplex structures in vitro, little is known about their existence and biological roles in vivo.7,10 Sequences capable of forming these structures abound in all eukaryotic organisms.11 Examples include the G-rich 3' overhangs on the ends of chromosomes and long oligopurine tracts within the promoter regions of several genes. DNA multiplexes have been invoked as necessary intermediates in many biological processes, including chromosome condensation, recombination, replication, telomere function, and transcriptional control.12,18 Potentially deleterious multiplex structures could also form as a consequence of essential biological processes that use the DNA as a template (e.g., replication and transcription) with removal of these structures then being necessary for viability.19,20 In this chapter, we review the literature to address these questions: (1) can DNA multiplexes such as triplexes and quadruplexes affect the process of transcription, and (2) do DNA multiplexes play a role in transcription regulation in vivo?
DNA Triplexes
As has been well known, certain nucleic acid sequences preferentially adopt a triple-helical structure under the proper conditions.3,4 Triplex structures are characterized by a single polynucleotide strand residing in the former major groove of a homopurine-homopyrimidine duplex (fig. 1A), which are reviewed in Chapter 1 of this book. Two triplex motifs are known. The parallel- or pyrimidine-motif (Py) has a C- or T-rich third strand bound in a parallel orientation with respect to the duplex homopurine strand, while the antiparallel- or purine-motif (Pu) has the opposite orientation and a primarily A- or G-rich third strand. Both types of triplexes utilize Hoogsteen hydrogen bonding between their third strands and purines in their duplex acceptors. The primary base triplets of Py triplexes are T•A•T and C•G•C+, while the base triplets of Pu triplexes are T•A•A, T•A•T, and C•G•G (fig. 1B,C). Py triplexes can occur with RNA being present as any of the three strands, while Pu triplexes only occur with DNA.21,22 Both inter- and intramolecular triplexes have been observed. The former involves a third DNA strand that originates from either a second DNA molecule or from a distal site on the same molecule, while the latter involves homopurine-homopyrimidine sequences immediately adjacent to the duplex acceptor (fig. 2A). Four isomers of intramolecular triplexes can exist dependent on the half-element strand that serves as the third strand (fig. 2B). Intramolecular triplexes are also known as H-DNA or H'-DNA, depending on whether they contain Py or Pu triplexes, respectively.
In theory, a homopurine-homopyrimidine duplex should be capable of forming triplexes of either motif. However, under physiological conditions, cytosine protonation is not favored, and C•G•G is the most stable base triplet in the purine motif. T-rich nucleic acids would be expected, therefore, to form Py triplexes, while G-rich DNAs would form Pu triplexes. The same is true for intramolecular triplexes, with the additional condition that the different isomers are not isoenergetic.23,24 In both intermolecular and intramolecular triplexes, contiguous homopurine-homopyrimidine runs of at least 10 base pairs are required for the duplex acceptor, since shorter triplexes are not very stable under physiological conditions, and even single base interruptions are known to greatly destabilize triplexes.25,27 Triplex formation is kinetically slow compared to duplex annealing.25,28 However, once formed, triplex RNA and DNA are very stable, exhibiting half-lives on the order of days.25,29
DNA Quadruplexes
DNAs (and RNAs) containing guanine tracts will associate in vitro to form four-stranded, right-handed helices known as quadruplexes or tetraplexes.5,6 These G4 nucleic acids are characterized by stacked G-quartet structures, square planar arrays of four guanines, each serving as the donor and acceptor of two Hoogsteen hydrogen bonds. Electronegative carbonyl oxygens line the center of the G-ring, where they interact with a suitably sized monovalent cation, typically Na+ or K+ (fig. 3A). Several isoforms of DNA and RNA quadruplexes have been described by NMR and X-ray crystallographic studies.30,31 The isoforms are characterized by either parallel or cis or trans antiparallel strand orientations and may be composed of either intermolecular or intramolecular or both types of hydrogen bonding (fig. 3B). G-rich nucleic acids can be highly polymorphic, adoption of the exact G4 structure depending on several factors including nucleotide sequence, strand concentration, and the types and concentrations of monovalent, divalent, and polyvalent cations present. Formation of G4 nucleic acids requires one or more polynucleotide strands, each containing one or more runs of two or more contiguous guanosine nucleotides. Four parallel-stranded intermolecular G4 nucleic acids (Fig. 3B, structure 1) require only a single G-tract. However, their strand stoichiometry and very slow formation kinetics lessen the likelihood that this form of G4 nucleic acid often occurs in vivo. More likely in vivo are G4 multiplex species formed from polynucleotides containing multiple G-runs, which have the ability to form intramolecular Hoogsteen hydrogen bonds. These species include purely intramolecular G4' nucleic acids that require only a single DNA or RNA molecule (fig. 3B,2), and G'2 hairpin dimer species that can link two separate polynucleotides (fig. 3B,3,4). Formation of intermolecular species may be rather slow under physiological conditions, though intermediates containing intramolecular G•G Hoogsteen base pairs (e.g., G') form quite rapidly.32 Once formed, each of the G4 species is quite stable, with measured enthalpies approaching -25 kcal/mole of G-quartet.33 Thus, equilibration between different G4 species is glacially slow under physiological conditions, and the thermodynamically favored structure is not necessarily the species that occurs in vivo.
Multiplexes and Transcription
A considerable body of evidence indicates that multiplex nucleic acids may affect transcription. Briefly, transcription requires a start site (+1), usually indicated by an arrow in most schematic representation, to define where transcription begins and in which direction it proceeds. In addition, transcription requires an RNA polymerase, which is the enzyme that catalyzes the template-directed sequential condensation of ribonucleotides to generate a product RNA. In many cases, the RNA polymerase itself does not directly recognize the +1 site but relies on auxiliary proteins for this purpose. In addition, these proteins and/or RNA polymerase do not typically interact directly with the +1 site but rather recognize nearby sequences known as the promoter. Once these proteins and RNA polymerase have assembled on a promoter, addition of ribonucleotides will allow transcription to begin. The process of transcription initiation is shown schematically in Figure 4A. Afterward, the transcribing RNA polymerase can proceed downstream of the +1 site and generate an RNA transcript (wavy line) in a process known as elongation (fig. 4B). Note that transcription is a multistep process: promoter recognition, initiation, elongation, and that the overall rates of transcription depend on the efficiencies of these different steps. These steps are often affected by a class of nucleic acid binding proteins (specific transcription factors), which can greatly modulate transcription.
Multiplex structures are believed to interfere with transcription primarily through two different mechanisms: promoter occlusion and elongation arrest.34 In promoter occlusion (fig. 4C), a DNA multiplex interferes with the binding of a transcription factor to a gene promoter. Note that, for occlusion to occur, the sites of transcription factor binding and multiplex formation need to overlap, and the extent of overlap necessary depending on the transcription factor and multiplex structure used. As shown in this example (fig. 4C), the typical occluded protein is a specific transcription factor that normally stimulates transcription initiation or elongation. However, it is also possible to inhibit transcription of a targeted promoter by occluding a DNA-binding basic transcription factor (e.g., TFIID). Likewise, it is possible to stimulate transcription through protein occlusion, if the occluded protein is a transcriptional repressor. In elongation arrest (fig. 4D), a post-initiation RNA polymerase II has its progress impeded by a downstream-situated multiplex. Note that a multiplex alone usually cannot effectively impede elongation by an RNA polymerase, especially eukaryotic RNA polymerases that normally function in a chromatin environment. Thus, unless the multiplex is located immediately downstream of a transcription pause or termination site, it is usually necessary for the multiplex to direct a subsequent covalent modification of the template (e.g., a cross-link or strand break) that renders it unsuitable for elongation.
Conceivably, there are several other mechanisms by which a multiplex structure might affect transcription. Some are shown schematically in Figure 5. For example, multiplex-forming oligonucleotides could themselves adopt structures that bind proteins involved in transcription (fig. 5A). Note that these could include proteins directly involved in RNA synthesis (e.g., transcription factors) as well as proteins that ultimately modulate their activity (e.g., signal transduction proteins). Given the appropriate sequence homology, multiplex-forming oligonucleotides could bind to RNA transcripts through conventional Watson-Crick base pairing, thereby leading to transcript degradation (and apparent loss) through endogenous RNase H activity (fig. 5B). Alternatively, multiplexes could inhibit transcription through the delivery of nonspecific and specific inhibitors of transcription, instead of through the direct occlusion of stimulatory transcription factors (Fig. 5C). Discerning these possible mechanisms relies on the use of adequate and sufficient controls, including mutagenesis of multiplex-forming sequences, order-of-addition experiments, and physical verification of multiplex structures.
Intermolecular Triplexes and Transcription
Most studies on the modulation of transcription by multiplexes have been done with oligonucleotides and intermolecular triplexes, because of the ease of forming such structures, the variety of controls that can be performed, and the flexibility possible through use of chemically modified triplex-forming oligonucleotides (TFOs). Studies on intermolecular triplexes have been performed both in vitro and in vivo, “in vivo” referring to any living organism, including cultured cells.
Intermolecular triplex effects on transcription have been investigated in vitro for a number of model systems, including prokaryotic, eukaryotic, and various hybrid systems. A list of representative studies is presented in Table 1. Both triplex motifs, purine and pyrimidine, have been explored, as have binding modes that are less well defined. Occlusion of specific transcription factors or general transcription factor/RNA polymerase binding has been proposed and/or reported in many studies.35,39,42,45,47,51,54,56,71 Typical observed results have been in the range of 50% to 105% transcription inhibition when 0.2 to 50 μM TFO was present. Control reactions usually involved oligonucleotides (ODN) that were not capable of triplex formation or templates that lacked TFO binding sites. Some unusual findings include the demonstration that a TFO targeting an upstream stimulatory transcription factor could apparently inhibit transcript appearance through partial hybridization to these transcripts and RNase H-mediated RNA degradation, and that transcription was inhibited when triplexes were located distal to transcription factor binding sites.39,88 Promotion of transcription has also been described in vitro, through the direct delivery of transcription activators by hybrid TFOs.85 Inhibition of transcription elongation has been observed in vitro as well.38,40,49,52,58,60,61,68,72 Typical effects range from 60% to 95% transcription inhibition, depending on several factors including the location of the intermolecular triplex relative to the start site of transcription, the type of RNA polymerase investigated, and whether the TFO was noncovalently bound or whether it directed a covalent modification of the DNA template. Taken together, these data demonstrated that many types of intermolecular triple helices can specifically and effectively inhibit several types of transcription through multiple mechanisms in vitro.
Given the observed successes with intermolecular triplexes in vitro, several research groups have investigated the effects of intermolecular triplexes on transcription in vivo (see Table 1). Both transcription factor occlusion and polymerase elongation mechanisms of triplex action have been investigated in vivo, with reports of efficiencies in excess of -90% reported in some circumstances, depending on oligonucleotide type, delivery method, and target site. Substantial transcription stimulation in vivo mediated by an activation domain peptide/triplex-forming oligonucleotide hybrid has also been reported.85 These findings suggest that intermolecular triplexes appear to be an efficient means of inhibiting specific gene transcription in vivo.
In experiments performed on in vivo targets, researchers encounter complications not found with in vitro experiments, including maintaining oligonucleotide stability in the presence of serum and cellular nucleases, delivering adequate concentrations of TFO to the proper cellular compartment (nucleus), and ensuring that triplex-formation actually occurs. Each of these difficulties has been addressed by a variety of means. Stability questions have been addressed by chemical modifications of the TFO termini and/or its phosphodiester backbone.36,41,42,44,46,47,53,55,62,66,69,70,75,82,84 Delivery difficulties have been surmounted by transfection with cationic lipids, electroporation, microinjection, or synthesis in situ.48,53,55,57,65,67,70,76,80,81,83,85,86 Even triplex formation, which can be highly problematic in an intracellular milieu with its high protein concentrations and surfeit of nonspecific nucleic acid targets, has been overcome by first preforming triplexes on their plasmid targets in vitro and then introducing the entire complex into cells.41,44,46,50,63,64,66,69,71,73,75,78,79,82,87 Note that these ex vivo experiments, although successful at addressing particular aspects of triplex-mediated transcription modulation, do not completely address the overall feasibility of triplexes in vivo. In addition, since very few investigators have actually demonstrated triplex formation in vivo, and oligonucleotides can affect cells through multiple specific and nonspecific mechanisms, most studies supporting triplex effects in vivo are not as compelling as they could be.
Intramolecular Triplexes and Transcription
Oligopurine•oligopyrimidine sequences have long been understood to play an instrumental role in the regulation of transcription for many genes.1 It has also been well known that certain oligopurine•oligopyrimidine sequences, especially those possessing mirror repeats, can form intramolecular triplexes in vitro under conditions of low pH or increased negative superhelicity.3 Thus it has been tempting to speculate that intramolecular triplexes are responsible for the transcriptional regulation observed at these sites. There is some evidence that intramolecular triplexes can form in vivo, albeit under less than physiological conditions in prokaryotic systems.89,90 Additionally, in triplex-specific antibody studies, cross-reactive structures have been identified near the centromeres of chromosomes.12,91 However, most reports in the literature regarding the involvement of H-DNA (or its purine-motif counterpart, H'-DNA) on transcription are only suppositions; few researchers have tested whether these sequences actually form intramolecular triplexes, and most physical studies have been performed in vitro. Nonetheless, a few exemplary studies have been done to investigate the possible role of intramolecular triplex structures on transcriptional regulation. Some are presented in Table 2.
Intramolecular triplexes may affect transcription from two locales: either proximally upstream the transcription start site or at any distance downstream. In the former case, intramolecular triplexes located within gene promoters are believed to arise in response to increased negative superhelical tension, which can result from nearby transcription. Such triplexes could then inhibit subsequent transcription events by displacing necessary transactivating proteins (fig. 4C) or by recruiting repressive proteins (fig. 5C). An alternative view is that the single-stranded region resulting from intramolecular triplex formation could serve as an entry point for RNA polymerase and thus serve as an activator of transcription.105 In the latter case, downstream intramolecular triplexes could arise as a result of processes that locally denature the DNA template (e.g., replication, transcription). These downstream triplexes would then either impede subsequent transcription elongation (fig. 4D) or inhibit transcription elongation by sequestering essential proteins (fig. 5A) or by delivering repressive proteins (fig. 5C).
Promoter-based intramolecular triplex effects on transcription have been reported to be quite variable, with magnitudes ranging from highly stimulatory to no effect to moderately inhibitory.15,92,94,96,97,102,104 More telling have been the results of the corresponding control experiments, which in the majority of studies showed no correlation between intermolecular triplex formation and transcriptional strength.92,94,97,102,104 For downstream intramolecular triplexes, significant inhibitory effects have been consistently reported both in vitro and in vivo, though their exact correlation with a specific triplex structure has been somewhat weak.98,101 All in all, these studies suggested that the transcriptional effects ascribed to relatively short polypurine•polypyrimidine sequences located upstream of many genes is most likely not the result of intramolecular triplex formation, whereas the transcriptional effects observed with very long downstream polypurine•polypyrimidine sequences may well involve some form of intramolecular triplex, especially of the H' variety.
Quadruplexes and Transcription
While quadruplexes, especially of the G-quartet variety, have primarily been invoked as playing a role in the biogenesis of chromosome telomeres, recent studies have suggested that they may also have a role in the transcriptional regulation of certain genes.9,10 G-rich sequences capable of forming quadruplex structures in vitro have been identified in the immunoglobulin switch region, the c-myc promoter, and upstream of the insulin gene.32,106,107 Use of a single-chain antibody fragment probe specific for guanine quadruplexes has led to identification of cross-reactive species in the macronucleus but not the micronucleus of Stylonychia lemnae, suggesting that quadruplexes do exist in vivo.17 Less clear is how such quadruplex structures arise, although arguments concerning the formation of intramolecular triplexes, including local negative superhelical tension in the promoter region, chromatin remodeling, and the consequence of transcription and/or replication events may also apply here.9,19 At present only a few studies directly describe quadruplex effects on transcription (see Table 3). Effects are believed to occur at the level of transcription factor occlusion and/or transcription factor recruitment, and significant effects, both stimulatory and repressive, have been observed.108,111 One major weakness of all these studies is the lack of in vivo characterization of G4 structures, which makes ascribing transcriptional effects to bona fide quadruplexes a bit tenuous.
Conclusions
Do DNA multiplexes affect transcription? From the aforementioned studies, the following conclusions can be made: (1) Some intermolecular triplexes can significantly repress transcription in vitro. However, their effectiveness in vivo often requires triplex preassembly in vitro. (2) Intramolecular triplexes may be responsible for impeding transcription on long, repeated sequences. (3) G-quadruplexes may affect transcription.
Acknowledgments
This work was supported by a grant from the Robert A. Welch Foundation (G-1199), and is dedicated to the memory of Claude Hélène (1938-2003).
References
- 1.
- Wells RD, Collier DA, Hanvey JC. et al. The chemistry and biology of unusual DNA structures adopted by oligopurine•oligopyrimidine sequences. FASEB J. 1988;2:2939–2949. [PubMed: 3053307]
- 2.
- Sinden RR.ed.DNA Structure and Function. San Diego: Academic Press 1994 .
- 3.
- Mirkin SM, Frank-Kamenetskii MD. H-DNA and related structures. Annu Rev Biophys Biomol Struct. 1994;23:541–576. [PubMed: 7919793]
- 4.
- Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu Rev Biochem. 1995;64:65–95. [PubMed: 7574496]
- 5.
- Sen D, Gilbert W. The structure of telomeric DNA:DNA quadruplex formation. Curr Opin Struct Biol. 1991;1:435–438.
- 6.
- Williamson JR. G-quartet structures in telomeric DNA. Annu Rev Biophys Biomol Struct. 1994;23:541–576. [PubMed: 7919797]
- 7.
- Guntaka RV, Varma BR, Weber KT. Triplex-forming oligonucleotides as modulators of gene expression. Int J Biochem Cell Biol. 2003;35:22–31. [PubMed: 12467644]
- 8.
- Zain R, Sun JS. Do natural DNA triple-helical structures occur and function in vivo? Cell Mol Life Sci. 2003;60:862–870. [PubMed: 12827276]
- 9.
- Arthanari H, Bolton PH. Functional and dysfunctional roles of quadruplex DNA in cells. Chem Biol. 2001;8:221–230. [PubMed: 11306347]
- 10.
- Schafer RH, Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2001;56:209–227. [PubMed: 11745112]
- 11.
- Behe MJ. An overabundance of long oligopurine tracts occurs in the genomes of simple and complex eukaryotes. Nucleic Acids Res. 1995;23:689–695. [PMC free article: PMC306739] [PubMed: 7899090]
- 12.
- Agazie YM, Burkholder GD, Lee JS. Triplex DNA in the nucleus: Direct binding of triplex-specific antibodies and their effects on transcription, replication and cell growth. Biochem J. 1996;316:461–466. [PMC free article: PMC1217372] [PubMed: 8687388]
- 13.
- Rooney SM, Moore PD. Antiparallel, intramolecular triplex DNA stimulates homologous recombination in human cells. Proc Natl Acad Sci USA. 1995;92:2141–2144. [PMC free article: PMC42439] [PubMed: 7892237]
- 14.
- Bianchi A, Wells RD, Heintz NH. et al. Sequences near the origin of replication of the DHFR locus in Chinese hamster ovary cells adopt left-handed Z-DNA and triplex structures. J Biol Chem. 1990;21789-21796 [PubMed: 2254331]
- 15.
- Kohwi Y, Kohwi-Shigamatsu T. Altered gene expression correlates with DNA structure. Genes Dev. 1991;5:2547–2554. [PubMed: 1752443]
- 16.
- Dempsey LA, Sun H, Hanakahi LA. et al. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D: A role for G-G pairing in immunoglobulin switch recombination. J Biol Chem. 1999;274:1066–1071. [PubMed: 9873052]
- 17.
- Schaffitzel C, Berger I, Postberg J. et al. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lamnae macronuclei. Proc Natl Acad Sci USA. 2001;98:8572–8577. [PMC free article: PMC37477] [PubMed: 11438689]
- 18.
- Catasti P, Chen X, Moyzis RK. et al. Structure-function correlations of the insulin-linked polymorphic region. J Mol Biol. 1996;264:534–545. [PubMed: 8969303]
- 19.
- Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. [PMC free article: PMC148410] [PubMed: 10198430]
- 20.
- Fry M, Loeb LA. Human Werner's syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. [PubMed: 10212265]
- 21.
- Roberts RW, Crothers DM. Stabilities and properties of double and triple helices: Dramatic effects of RNA and DNA backbone composition. Science. 1992;258:1463–1466. [PubMed: 1279808]
- 22.
- Semerad CL, Maher LJ. Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res. 1994;22:5321–5325. [PMC free article: PMC332077] [PubMed: 7529405]
- 23.
- Htun H, Dahlberg JE. Topology and formation of triple-stranded H-DNA. Science. 1989;243:1571–1576. [PubMed: 2648571]
- 24.
- Kohwi Y, Kohwi-Shigematsu T. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc Natl Acad Sci USA. 1988;85:3781–3785. [PMC free article: PMC280302] [PubMed: 3375241]
- 25.
- Cheng AJ, Van Dyke MW. Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1993;21:5630–5635. [PMC free article: PMC310527] [PubMed: 8284208]
- 26.
- Cheng AJ, Van Dyke MW>. Oligodeoxyribonucleotide length and sequence effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1994;22:4742–4747. [PMC free article: PMC308526] [PubMed: 7984426]
- 27.
- Orson FM, Klysik J, Bergstrom DE. et al. Triple helix formation: binding avidity of acridine-conjugated AG motif third strands containing natural, modified and surrogate bases opposed to pyrimidine interruptions in a polypurine target. Nucleic Acids Res. 1999;27:810–816. [PMC free article: PMC148251] [PubMed: 9889277]
- 28.
- Paes HM, Fox KR. Kinetic studies on the formation of intermolecular triple helices. Nucleic Acids Res. 1997;25:3269–3274. [PMC free article: PMC146877] [PubMed: 9241240]
- 29.
- Hoyne PR, Gacy AM, McMurray CT. et al. Stabilities of intrastrand pyrimidine motif DNA and RNA triple helices. Nucleic Acids Res. 2000;28:770–775. [PMC free article: PMC102562] [PubMed: 10637329]
- 30.
- Simonsson T. G-quadruplex DNA structures — variations on a theme. Biol Chem. 2001;382:621–628. [PubMed: 11405224]
- 31.
- Deng J, Xiong Y, Sundaralingam M. X-ray analysis of an RNA tetraplex (UGGGGU)(4) with divalent Sr(2+) ions at subatomic resolution (0.61 A). Proc Natl Acad Sci USA. 2001;98:13665–13670. [PMC free article: PMC61098] [PubMed: 11707581]
- 32.
- Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. [PubMed: 2320109]
- 33.
- Lu M, Guo Q, Kallenbach NR. Thermodynamics of G-tetraplex formation by telomeric DNAs. Biochemistry. 1993;32:598–601. [PubMed: 8422371]
- 34.
- Praseuth D, Guieysse AL, Helene C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim Biophys Acta. 1999;1489:181–206. [PubMed: 10807007]
- 35.
- Cooney M, Czernuszewicz G, Postel EH. et al. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. [PubMed: 3293213]
- 36.
- Orson FM, Thomas DW, McShan WM. et al. Oligonucleotide inhibition of IL2R alpha mRNA transcription by promoter region collinear triplex formation in lymphocytes. Nucleic Acids Res. 1991;19:3435–3441. [PMC free article: PMC328345] [PubMed: 2062658]
- 37.
- Postel EH, Flint SJ, Kessler DJ. et al. Evidence that a triplex-forming oligodeoxyribonucleotide binds to the c-myc promoter in HeLa cells, thereby reducing c-myc mRNA levels. Proc Natl Acad Sci USA. 1991;88:8227–8231. [PMC free article: PMC52480] [PubMed: 1896473]
- 38.
- Young SL, Krawczyk SH, Matteucci MD. et al. Triple helix formation inhibits transcription elongation in vitro. Proc Natl Acad Sci USA. 1991;88:10023–10026. [PMC free article: PMC52859] [PubMed: 1946419]
- 39.
- Maher LJ, Dervan PB, Wold B. Analysis of promoter-specific repression by triple-helical DNA complexes in a eukaryotic cell-free transcription system. Biochemistry. 1992;31:70–81. [PubMed: 1731886]
- 40.
- Duval-Valentin G, Thuong NT, Helene C. Specific inhibition of transcription by triple helix-forming oligonucleotides. Proc Natl Acad Sci USA. 1992;89:504–508. [PMC free article: PMC48267] [PubMed: 1731320]
- 41.
- Grigoriev M, Praseuth D, Robin P. et al. A triple helix-forming oligonucleotide-intercalator conjugate acts as a transcriptional repressor via inhibition of NF kappa B binding to interleukin-2 receptor alpha-regulatory sequence. J Biol Chem. 1992;267:3389–3395. [PubMed: 1737792]
- 42.
- McShan WM, Rossen RD, Laughter AH. et al. Inhibition of transcription of HIV-1 in infected human cells by oligodeoxynucleotides designed to form DNA triple helices. J Biol Chem. 1992;267:5712–5721. [PubMed: 1544943]
- 43.
- Maher LJ. Inhibition of T7 RNA polymerase initiation by triple-helical DNA complexes: A model for artificial gene repression. Biochemistry. 1992;31:7587–7594. [PubMed: 1510945]
- 44.
- Grigoriev M, Praseuth D, Guieysse AL. et al. Inhibition of gene expression by triple helix-directed DNA cross-linking at specific sites. Proc Natl Acad Sci USA. 1993;90:3501–3505. [PMC free article: PMC46328] [PubMed: 8475098]
- 45.
- Skoog JU, Maher LJ. Repression of bacteriophage promoters by DNA and RNA oligonucleotides. Nucleic Acids Res. 1993;21:2131–2138. [PMC free article: PMC309475] [PubMed: 8502553]
- 46.
- Grigoriev M, Praseuth D, Guieysse AL. et al. Inhibition of interleukin-2 receptor alpha-subunit gene expression by oligonucleotide-directed triple helix formation. CR Acad Sci III. 1993;316:492–495. [PubMed: 8221232]
- 47.
- Ing NH, Beekman JM, Kessler DJ. et al. In vivo transcription of a progesterone-responsive gene is specifically inhibited by a triplex-forming oligonucleotide. Nucleic Acids Res. 1993;21:2789–2796. [PMC free article: PMC309654] [PubMed: 8332487]
- 48.
- Roy C. Inhibition of gene transcription by purine rich triplex forming oligodeoxyribonucleotides. Nucleic Acids Res. 1993;21:2845–2852. [PMC free article: PMC309666] [PubMed: 7687346]
- 49.
- Rando RF, DePaolis L, Durland RH. et al. Inhibition of T7 and T3 RNA polymerase directed transcription elongation in vitro. Nucleic Acids Res. 1994;22:678–685. [PMC free article: PMC307860] [PubMed: 8127717]
- 50.
- Degols G, Clarenc JP, Lebleu B. et al. Reversible inhibition of gene expression by a psoralen functionalized triple helix forming oligonucleotide in intact cells. J Biol Chem. 1994;269:16933–16937. [PubMed: 8207016]
- 51.
- Mayfield C, Ebbinghaus S, Gee J. et al. Triplex formation by the human Ha-ras promoter inhibits Sp1 binding and in vitro transcription. J Biol Chem. 1994;269:18232–18238. [PubMed: 8027084]
- 52.
- Xodo L, Alunni-Fabbroni M, Manzini G. et al. Pyrimidine phosphorothioate oligonucleotides form triple-stranded helices and promote transcription inhibition. Nucleic Acids Res. 1994;22:3322–3330. [PMC free article: PMC523725] [PubMed: 8078767]
- 53.
- Neurath MF, Max EE, Strober W. Pax5 (BSAP) regulates the murine immunoglobulin 3' alpha enhancer by suppressing binding of NF-alpha P, a protein that controls heavy chain transcription. Proc Natl Acad Sci USA. 1995;92:5336–5340. [PMC free article: PMC41689] [PubMed: 7777508]
- 54.
- Kim HG, Miller DM. Inhibition of in vitro transcription by a triplex-forming oligonucleotide targeted to human c-myc P2 promoter. Biochemistry. 1995;34:8165–8171. [PubMed: 7794930]
- 55.
- Tu GC, Cao QN, Israel Y. Inhibition of gene expression by triple helix formation in hepatoma cells. J Biol Chem. 1995;270:28402–28407. [PubMed: 7499344]
- 56.
- Kovacs A, Kandala JC, Weber KT. et al. Triple helix-forming oligonucleotide corresponding to the polypyrimidine sequence in the rat alpha 1(I) collagen promoter specifically inhibits factor binding and transcription. J Biol Chem. 1996;271:1805–1812. [PubMed: 8576186]
- 57.
- Porumb H, Gousset H, Letellier R. et al. Temporary ex vivo inhibition of the expression of the human oncogene HER2 (NEU) by a triple helix-forming oligonucleotide. Cancer Res. 1996;56:515–522. [PubMed: 8564964]
- 58.
- Escude C, Giovannangeli C, Sun JS. et al. Stable triple helices formed by oligonucleotide N3'→P5' phosphoramidates inhibit transcription elongation. Proc Natl Acad Sci USA. 1996;93:4365–4369. [PMC free article: PMC39543] [PubMed: 8633072]
- 59.
- Kochetkova M, Shannon MF. DNA triplex formation selectively inhibits granulocyte-macrophage colony-stimulating factor gene expression in human T cells. J Biol Chem. 1996;271:14438–14444. [PubMed: 8662666]
- 60.
- Giovannangeli C, Perrouault L, Escude C. et al. Specific inhibition of in vitro transcription elongation by triplex-forming oligonucleotide-intercalator conjugates targeted to HIV proviral DNA. Biochemistry. 1996;35:10539–10548. [PubMed: 8756710]
- 61.
- Giovannangeli C, Perrouault L, Escude C. et al. Efficient inhibition of transcription elongation in vitro by oligonucleotide phosphoramidates targeted to proviral HIV DNA. J Mol Biol. 1996;261:386–398. [PubMed: 8780781]
- 62.
- Aggarwal BB, Schwarz L, Hogan ME. et al. Triple helix-forming oligodeoxyribonucleotides targeted to the human tumor necrosis factor (TNF) gene inhibit TNF production and block the TNF-dependent growth of human glioblastoma tumor cells. Cancer Res. 1996;56:5156–5164. [PubMed: 8912851]
- 63.
- Musso M, Wang JC, Van DykeMW. In vivo persistence of DNA triple helices containing psoralen-conjugated oligodeoxyribonucleotides. Nucleic Acids Res. 1996;24:4924–4932. [PMC free article: PMC146337] [PubMed: 9016662]
- 64.
- Alunni-Fabbroni M, Pirulli D, Manzini G. et al. (A,G)-oligonucleotides form extraordinary stable triple helices with a critical R.Y sequence of the murine c-Ki-ras promoter and inhibit transcription in transfected NIH 3T3 cells. Biochemistry. 1996;35:16361–16369. [PubMed: 8973212]
- 65.
- Shevelev A, Burfeind P, Schulze E. et al. Potential triple helix-mediated inhibition of IGF-I gene expression significantly reduces tumorigenicity of glioblastoma in an animal model. Cancer Gene Ther. 1997;4:105–112. [PubMed: 9080119]
- 66.
- Joseph J, Kandala JC, Veerapanane D. et al. Antiparallel polypurine phosphorothioate oligonucleotides form stable triplexes with the rat alpha1(I) collagen gene promoter and inhibit transcription in cultured rat fibroblasts. Nucleic Acids Res. 1997;25:2182–2188. [PMC free article: PMC146703] [PubMed: 9153319]
- 67.
- Rininsland F, Johnson TR, Chernicky CL. et al. Suppression of insulin-like growth factor type I receptor by a triple-helix strategy inhibits IGF-I transcription and tumorigenic potential of rat C6 glioblastoma cells. Proc Natl Acad Sci USA. 1997;94:5854–5859. [PMC free article: PMC20870] [PubMed: 9159164]
- 68.
- Wang Z, Rana TM. DNA damage-dependent transcriptional arrest and termination of RNA polymerase II elongation complexes in DNA template containing HIV-1 promoter. Proc Natl Acad Sci USA. 1997;94:6688–6693. [PMC free article: PMC21219] [PubMed: 9192626]
- 69.
- Kim HG, Reddoch JF, Mayfield C. et al. Inhibition of transcription of the human c-myc protooncogene by intermolecular triplex. Biochemistry. 1998;37:2299–2304. [PubMed: 9485376]
- 70.
- Kim HG, Miller DM. A novel triplex-forming oligonucleotide targeted to human cyclin D1 (bcl-1, proto-oncogene) promoter inhibits transcription in HeLa cells. Biochemistry. 1998;37:2666–2672. [PubMed: 9485417]
- 71.
- Nakanishi M, Weber KT, Guntaka RV. Triple helix formation with the promoter of human alpha1(I) procollagen gene by an antiparallel triplex-forming oligodeoxyribonucleotide. Nucleic Acids Res. 1998;26:5218–5222. [PMC free article: PMC147955] [PubMed: 9801322]
- 72.
- Ebbinghaus SW, Fortinberry H, Gamper HB. Inhibition of transcription elongation in the HER-2/ neu coding sequence by triplex-directed covalent modification of the template strand. Biochemistry. 1999;38:619–628. [PubMed: 9888801]
- 73.
- Kuznetsova S, Ait-Si-Ali S, Nagibneva I. et al. Gene activation by triplex-forming oligonucleotide coupled to the activating domain of protein VP16. Nucleic Acids Res. 1999;27:3995–4000. [PMC free article: PMC148666] [PubMed: 10497263]
- 74.
- Ritchie S, Boyd FM, Wong J. et al. Transcription of the human c-Src promoter is dependent on Sp1, a novel pyrimidine binding factor SPy, and can be inhibited by triplex-forming oligonucleotides. J Biol Chem. 2000;275:847–854. [PubMed: 10625617]
- 75.
- Bailey C, Weeks DL. Understanding oligonucleotide-mediated inhibition of gene expression in Xenopus laevis oocytes. Nucleic Acids Res. 2000;28:1154–1161. [PMC free article: PMC102614] [PubMed: 10666457]
- 76.
- Faria M, Wood CD, Perrouault L. et al. Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc Natl Acad Sci USA. 2000;97:3862–3867. [PMC free article: PMC18107] [PubMed: 10760257]
- 77.
- Catapano CV, McGuffie EM, Pacheco D. et al. Inhibition of gene expression and cell proliferation by triple helix-forming oligonucleotides directed to the c-myc gene. Biochemistry. 2000;39:5126–5138. [PubMed: 10819980]
- 78.
- Intody Z, Perkins BD, Wilson JH. et al. Blocking transcription of the human rhodopsin gene by triplex-mediated DNA photocrosslinking. Nucleic Acids Res. 2000;28:4283–4290. [PMC free article: PMC113126] [PubMed: 11058128]
- 79.
- Shen C, Buck A, Mehrke G. et al. Triplex forming oligonucleotide targeted to 3'UTR downregulates the expression of the bcl-2 proto-oncogene in HeLa cells. Nucleic Acids Res. 2001;29:622–628. [PMC free article: PMC30398] [PubMed: 11160882]
- 80.
- Cogoi S, Rapozzi V, Quadrifoglio F. et al. Anti-gene effect in live cells of AG motif triplex-forming oligonucleotides containing an increasing number of phosphorothioate linkages. Biochemistry. 2001;40:1135–1143. [PubMed: 11170438]
- 81.
- Faria M, Wood CD, White MR. et al. Transcription inhibition induced by modified triple helix-forming oligonucleotides: A quantitative assay for evaluation in cells. J Mol Biol. 2001;306:15–24. [PubMed: 11178890]
- 82.
- Stutz AM, Hoeck J, Natt F. et al. Inhibition of interleukin-4- and CD40-induced IgE germline gene promoter activity by 2'-aminoethoxy-modified triplex-forming oligonucleotides. J Biol Chem. 2001;276:11759–11765. [PubMed: 11278649]
- 83.
- Besch R, Giovannangeli C, Kammerbauer C. et al. Specific inhibition of ICAM-1 expression mediated by gene targeting with triplex-forming oligonucleotides. J Biol Chem. 2002;277:32473–32479. [PubMed: 12080053]
- 84.
- Rapozzi V, Cogoi S, Spessotto P. et al. Antigene effect in K562 cells of a PEG-conjugated triplex-forming oligonucleotide targeted to the bcr/abl oncogene. Biochemistry. 2002;41:502–510. [PubMed: 11781088]
- 85.
- Stanojevic D, Young RA. A highly potent artificial transcription factor. Biochemistry. 2002;41:7209–7216. [PubMed: 12044151]
- 86.
- Carbone GM, McGuffie EM, Collier A. et al. Selective inhibition of transcription of the Ets2 gene in prostate cancer cells by a triplex-forming oligonucleotide. Nucleic Acids Res. 2003;31:833–843. [PMC free article: PMC149218] [PubMed: 12560478]
- 87.
- Ziemba AJ, Reed MW, Raney KD. et al. A bis-alkylating triplex forming oligonucleotide inhibits intracellular reporter gene expression and prevents triplex unwinding due to helicase activity. Biochemistry. 2003;42:5013–5024. [PubMed: 12718544]
- 88.
- Praseuth D, Guieysse AL, Itkes AV. et al. Unexpected effect of an anti-human immunodeficiency virus intermolecular triplex-forming oligonucleotide in an in vitro transcription system due to RNase H-induced cleavage of the RNA transcript. Antisense Res Dev. 1993;3:33–44. [PubMed: 8388279]
- 89.
- Kohwi Y, Malkhosyan SR, Kohwi-Shigematsu T. Intramolecular dG•dG•dC triplex detected in Escherichia coli cells. J Mol Biol. 1992;223:817–822. [PubMed: 1538396]
- 90.
- Ussery DW, Sinden RR. Environmental influences on the in vivo level of intramolecular triplex DNA in Escherichia coli. Biochemistry. 1993;32:6206–6213. [PubMed: 8512930]
- 91.
- Ohno M, Fukagawa T, Lee JS. et al. Triplex-forming DNAs in the human interphase nucleus visualized in situ by polypurine/polypyrimidine DNA probes and antitriplex antibodies. Chromosoma. 2002;111:201–213. [PubMed: 12355210]
- 92.
- Glaser RL, Thomas GH, Siegfried E. et al. Optimal heat-induced expression of the Drosophila hsp26 gene requires a promoter sequence containing (CT)n•(GA)n repeats. J Mol Biol. 1990;211:751–761. [PubMed: 2313697]
- 93.
- Sarkar PS, Brahmachari SK. Intramolecular triplex potential sequence within a gene down regulates its expression in vivo. Nucleic Acids Res. 1992;20:5713–5718. [PMC free article: PMC334407] [PubMed: 1454535]
- 94.
- Raghu G, Tevosian S, Anant S. et al. Transcriptional activity of the homopurine-homopyrimidine repeat of the c-Ki-ras promoter is independent of its H-forming potential. Nucleic Acids Res. 1994;22:3271–3279. [PMC free article: PMC523718] [PubMed: 8078760]
- 95.
- Duval-Valentin G, de BizemontT, Takasugi M. et al. Triple-helix specific ligands stabilize H-DNA conformation. J Mol Biol. 1995;247:847–858. [PubMed: 7723037]
- 96.
- Xu G, Goodridge AG. Characterization of a polypyrimidine/polypurine tract in the promoter of the gene for the chicken malic enzyme. J Biol Chem. 1996;271:16008–16019. [PubMed: 8663263]
- 97.
- Becker NA, Maher LJ. Characterization of a polypurine/polypyrimidine sequence upstream of the mouse metallothionein-I gene. Nucleic Acids Res. 1998;26:1951–1958. [PMC free article: PMC147503] [PubMed: 9518488]
- 98.
- Ohshima K, Montermini L, Wells RD. et al. Inhibitory effects of expanded GAA•TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J Biol Chem. 1998;273:14588–14595. [PubMed: 9603975]
- 99.
- Grabczyk E, Usdin K. The GAA•TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acid Res. 2000;28:2815–2822. [PMC free article: PMC102661] [PubMed: 10908340]
- 100.
- Grabczyk E, Usdin K. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acid Res. 2000;28:4930–4937. [PMC free article: PMC115239] [PubMed: 11121484]
- 101.
- Sakamoto N, Ohshima K, Montermini L. et al. Sticky DNA, a self-associated complex formed at long GAA•TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J Biol Chem. 2001;276:27171–27177. [PubMed: 11340071]
- 102.
- Rustighi A, Tessari MA, Vascotto F. et al. A polypyrimidine/polypurine tract within the Hmga2 minimal promoter: A common feature of many growth-related genes. Biochemistry. 2002;41:1229–1240. [PubMed: 11802722]
- 103.
- Hoyne PR, Maher LJ. Functional studies of potential intrastrand triplex elements in the Escherichia coli genome. J Mol Biol. 2002;318:373–386. [PubMed: 12051844]
- 104.
- Lu Q, Teare JM, Granok H. et al. The capacity to form H-DNA cannot substitute for GAGA factor binding to a (CT)n•(GA)n regulatory site. Nucleic Acids Res. 2003;31:2483–2494. [PMC free article: PMC156050] [PubMed: 12736297]
- 105.
- Weintraub H. High-resolution mapping of S1- and DNase I-hypersensitive sites in chromatin. Mol Cell Biol. 1985;5:1538–1539. [PMC free article: PMC366889] [PubMed: 2993871]
- 106.
- Hammond-Kosack MC, Kilpatrick MW, Docherty K. Analysis of DNA structure in the human insulin gene-linked polymorphic region in vivo. J Mol Endocrinol. 1992;9:221–225. [PubMed: 1476609]
- 107.
- Katahira M, Fukuda H, Kawasumi H. et al. Intramolecular quadruplex formation of the G-rich strand of the mouse hypervariable minisatellite Pc-1. Biochem Biophys Res Commun. 1999;264:327–333. [PubMed: 10529363]
- 108.
- Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. [PMC free article: PMC147388] [PubMed: 9469822]
- 109.
- Lew A, Rutter WJ, Kennedy GC. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc Natl Acad Sci USA. 2000:12508–12512. [PMC free article: PMC18794] [PubMed: 11070077]
- 110.
- Kuzmine I, Gottlieb PA, Martin CT. Structure in nascent RNA leads to termination of slippage transcription by T7 RNA polymerase. Nucleic Acids Res. 2001;29:2601–2606. [PMC free article: PMC55752] [PubMed: 11410669]
- 111.
- Simonsson T, Henriksson M. C-myc suppression in Burkitt's lymphoma cells. Biochem Biophys Res Comm. 2002;290:11–15. [PubMed: 11779125]
- 112.
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. [PMC free article: PMC129314] [PubMed: 12195017]
Figures

Figure 1
Triplex nucleic acids. A) Schematic representation of an intermolecular triplex. The third strand (black) is seen residing in the major groove of duplex DNA. Note that this third strand may be part of a larger molecule (dotted line extensions). B) Base triplets and strand orientations in the pyrimidine motif. C) Base triplets and strand orientations in the purine motif.
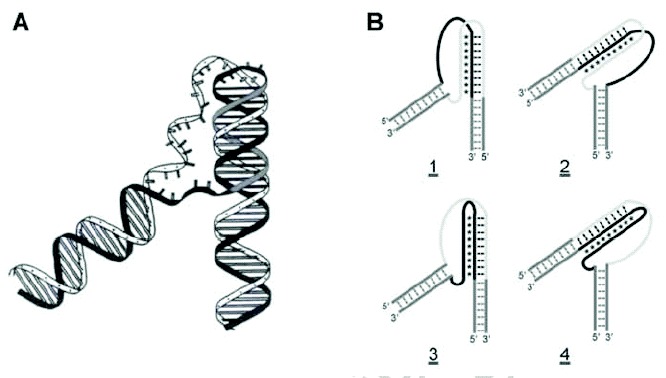
Figure 2
Intramolecular triplexes. A) Schematic representation of an intramolecular triplex. The third strand (gray) is shown residing in the major groove of duplex DNA. B) Schematic representations of H-DNA isomers H-y3 (1) and H-y5 (2), and H'-DNA isomers H-r3 (3) and H-r5 (4). In H(H') DNA nomenclature, the nature of the third strand is identified as either pyrimidine (y) or purine (r) and originating from either the 3' or 5' end of the corresponding oligopyrimidine or oligopurine strand. The purine-rich strand is shown in black, while the pyrimidine-rich strand is shown in light gray.
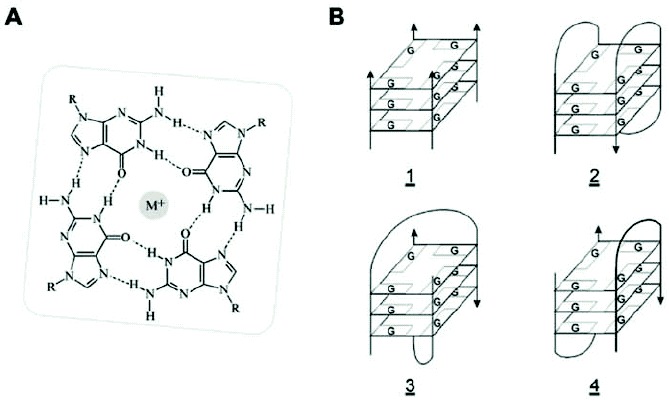
Figure 3
G4 nucleic acids. A) Chemical structure of a Hoogsteen hydrogen-bonded G-quartet. M+ represents a monovalent cation, typically Na+ or K+. B) Schematic representations of a parallel-strand intermolecular tetraplex (G4, 1), a monomeric intramolecular quadruplex (G4', 2), and two forms of dimeric hairpin quadruplexes (G'2, 3,4). Both RNA and DNA and mixtures of these nucleic acids can form G4 structures. A G' structure contains only Hoogsteen hydrogen-bonded G•G base pairs and would correspond to a single hairpin (indicated in bold) in representation 4.
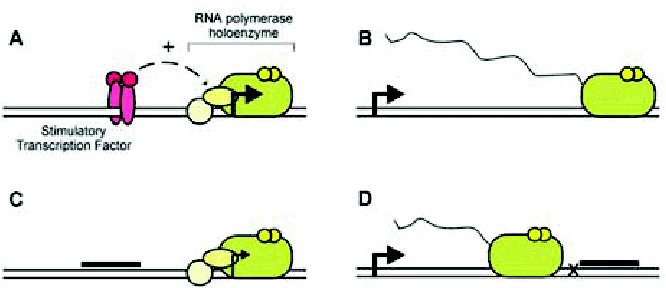
Figure 4
Transcription regulation by multiplexes. Schematic representations of transcription and its inhibition by multiplexes. A) A distal homodimeric specific transcription factor (left, dark gray or red) positively affects the function of basic transcription factors (center, light gray or yellow) and RNA polymerase II (right, medium gray or green) bound proximally to the start site of transcription, thereby promoting initiation (rightward bold arrow). B) An elongating RNA polymerase II proceeds downstream from the start site of transcription, synthesizing an RNA transcript (tail) complementary to the DNA template strand. C) A promoter-bound multiplex (bold line) occludes the binding of a specific transcription factor, thereby diminishing transcription initiation. D) A multiplex located downstream of the transcription start site positions a covalent DNA crosslink (X) that impedes elongation by RNA polymerase II.
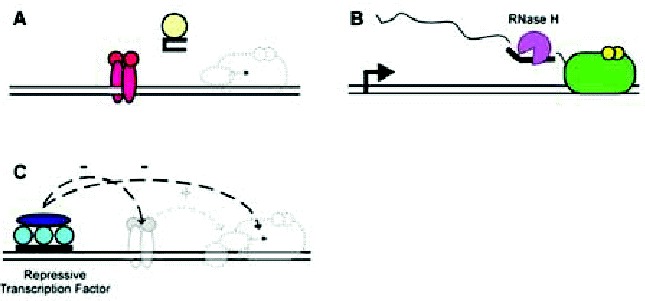
Figure 5
Alternative mechanisms for transcription regulation by multiplexes. Schematic representations of transcription regulation by multiplexes and multiplex-forming oligonucleotides. A) A multiplex-forming oligonucleotide folds into a structure that serves as an aptamer, which competes for promoter binding by a general transcription factor (center, light gray or yellow). B) A multiplex-forming oligonucleotide binds to an RNA transcript and directs its degradation by RNase H. C) An upstream promoter multiplex binds proteins that actively interfere with the function of specific and general transcription factors.
Tables
Table 1Intermolecular triplexes and transcription
| Targeta | Multiplexb | Triplex Proofc | Transcription Assayd | Maxium Effecte | Controlsf | Ref. |
|---|---|---|---|---|---|---|
| human c-myc P1 promoter,linear plasmid | 27mer GAT PO TFO,“parallel” | EMSA, DNase I FP | in vitro, HeLa NE, c-myc P2 runoff | -95% @ 160 nM TFO | complement ODN | 35 |
| human IL2Rα promoter, SRE, chromosome | 28mer PO TFO, 3'- amine parallel | REPA | in vivo, PBMC cells , northern, media TFO | -55% @ 15 μM TFO | 31mer GT ODN/c-myc or β-actin mRNA | 36 |
| human c-myc P1 promoter, chromosome | 27mer GAT PO TFO, Pu3 | EMSA, chromatin DNase I hyper sensitivity | in vivo, HeLa cells, northern, media TFO | -90% @ 125 μM TFO | complement ODN/β-actin mRNA | 37 |
| syn., + 180 G-free cassette Ad2 MLP, linear plasmid | 15mer MT PO TFO, 3'-C*, Py3 | DMS FP | in vitro, Jurkat NE, G-free, pre. 3plx | -90% @ 10 μM TFO, covalent | MT TFO noncovalent, scrambled ODN covalent | 38 |
| syn., 1-5 copies, Sp1 site, Ad E4 core promoter, supercoiled plasmid | 21mer T5meC PO TFO, Py3 | n.s.(prior work) | in vitro, Drosophila Kc cell NE, primer ext., pre. 3plx | -90% @ 205 μM TFO, pre. 3plx, basal transcription (-Sp1) | scrambled ODN/ftz promoter (co-transcribed) | 39 |
| E. coli bla gene, +22, DNA fragment | 13mer T5meC PO TFO, 5'- psoralen, Py3 | DNase I FP /DNase I FP | in vitro, E. coli RNA pol holoenzyme, runoff, pre.3plx | -80% @ 50 μM TFO, covalent | unmod. TFO, nonspecific pso-16-mer (-20% each)/tetR gene | 40 |
| human IL-2Rα , enhancer, NF-κ B site, plasmid | 15mer T5meC PO TFO, 3'- acridine, Py3 | EMSA, / REPA, DNase I FP | in vivo, HSB2 Not cells, CAT assay, pre. 3plx | -90% @ 10 μM TFO | HIV LTR CAT | 41 |
| HIV-1 promoter, Sp1 and +1 sites, plasmid | 31 & 38mer GT PO TFO, 3'-amine, parallel | EMSA, CD | in vitro, HeLa NE, smear; in vivo, U937/ HIV-1 cells, media TFO | -80% (in vitro) & -90% (in vivo)@ 10 μM TFO | scrambled ODN | 42 |
| syn., f 10 promoter, +1, linear plasmid | 21mer T5meC PO TFO Py3; 22mer GT GU PO TFOs, Pu3 | DNase I & MPE FP | in vitro, T7 pol., runoff | -92% @ 1 μM T5meC; - 99% @ 1 mM GT TFO | scrambled ODN / wt f 10 promoter (co-transcribed) | 43 |
| humanIL-2Rα , enhancer, NF-κB site, plasmid | 15mer T5meC PO TFO, 3'- acridine, Py3 | DNase I FP | in vitro, C8166 NE, primer extension, pre. 3plx; in vivo, HSB2 cells, CAT assay, pre. 3plx | -80% @ 1 μM TFO covalent (in vitro) -80% @ 0.2 μM TFO, covalent (in vivo) | mutant 3plx site / HIV LTR | 44 |
| syn., f 10 promoter, +1, linear plasmid | 21mer T5meC PO TFO, Py3; 22mer GT GU PO TFOs, Pu3 | n.s. (prior work) | in vitro, T7 pol., runoff, pre. 3plx | -90% @ 1 μM T5meC, GT, GU TFO | wt f 10 promoter (co-transcribed) | 45 |
| human IL-2Rα , enhancer, NF-κB site, plasmid | 15mer T5meC PO TFO, 3'- acridine, Py3 | DNase I FP | in vivo, HSB2 cells, CAT assay, pre. 3plx? | -90% @ 5 μM TFO | unmod. TFO, mutant mod. ODN / mutant 3plx site | 46 |
| syn., PRE site, Lov core promoter, plasmid | 38mer GT PO TFO,3'- cholesterol, Pu3 | EMSA, DNase I FP | in vitro, HeLa NE, G-free, pre. 3plx; in vivo, CV-1 cells, CAT assay, media TFO | -105% @ 200 nM TFO (in vitro); @ 20 μM TFO, (in vivo) | (in vitro) mut. ODN (-50% @ 200 nM) / mut. promoter, Ad2 MLP (in vivo) (co-transcribed); mut. ODN (-15% @ 20 μM) / SV2CAT | 47 |
| human 6-16 promoter, IRE, plasmid syn. +63 or +103, linear plasmid | 21mer AGT PO TFO, Pu3 | EMSA, DNase I & Cu-phen. FP | in vivo, HeLa cells, CAT, co-transfect | -99% @ 1.8 μM TFO, (-50% for controls) | GA ODN / SV40 or minimal TK promoter | 48 |
| syn. +63 or +103, linear plasmid | 53mer GT PO TFO, 3' -amino, Pu3 | EMSA | in vitro, T3 or T7 RNA pol., truncated transcript, pre. 3plx | -80% @ 0.2 μM TFO, (-30% @ 4 μM reverse ODN) | reverse ODN | 49 |
| syn., +204, plasmid | 15mer T5meC PO TFO, 5'-psoralen, Py3 | PCR / PCR | in vivo, HeLa or XP2Y0(SV) cells, β -gal assay, pre. 3plx | -75% @ 1.8 μM TFO (24h HeLa), -85% (72h XP) | mut. ODN | 50 |
| human H-ras promoter, -8, linear plasmid | 21mer GC PO TFO, Pu3 | EMSA, DNase I FP | in vitro, HeLa NE, runoff, pre. 3plx | -50% @ 15 μM TFO | reverse ODN / CMV promoter | 51 |
| syn., +46,linear plasmid | 11mer CT PO PS TFO, Py3 | Tm, CD | in vitro, T7 RNA pol., runoff, pre. 3plx | -95% @ 5 μM PO TFO | 11mer CT ODN, -TFO | 52 |
| IgH 3' a enhancer, Pax5 site, plasmid & chromosomal | 41mer GT PO TFO, 3'- amine, Pu3 | EMSA / in vivo DMS FP | in vivo, CH12.LX.A2 cells, luciferase, northern, (co)transfect TFO | +160% @ 25 μg TFO | mut. ODN/ mut. enhancer | 53 |
| human c-myc, P2 promoter, MAZ & E2F sites, linear plasmid human ALDH2 downstream promoter, chromosome | 21mer GT PO, PS, & PO/PS TFO, Pu3 | EMSA, DNase I FP | in vitro, HeLa NE, runoff, pre. 3plx | -80% @ 20 μM TFO | reverse ODN / CMV promoter | 54 |
| human ALDH2 downstream promoter, chromosome | 21mer GT PO, PS, & PO/PS TFO, Pu3 | EMSA | in vivo, HepG2 cells, RT-PCR, transfect TFO | -90% @ 600 nM PS TFO | albumin mRNA | 55 |
| rat α 1(I) collagen promoter, -138, fragment or plasmid | 30mer GA PO TFO, parallel | EMSA | in vitro, RCF NE, runoff, pre. 3plx; vitro); in vivo, RCF cells, CAT, post transfect TFO | -95% @ 250 μM TFO, (in vitro);-60% @ 1 μM TFO (in vivo) | 20mer AGCT ODN / mut. & CMV IE promoter | 56 |
| human HER2 core promoter, chromosome | 28mer GAC PO TFO, Pu3 | EMSA, FTIR | in vivo, MCF7 cells, ELISA, northern, transfect TFO | -28% protein, -49% mRNA @ 0.22 μM TFO, 6 h post transfect. | mut., complement, scrambled / EGFR protein, GAPDH mRNA | 57 |
| HIV1 nef gene, linear plasmid | 11mer TGC PN TFO, Py3 | REPA, Tm | in vitro, HeLa NE, truncated transcript, pre. 3plx | -60% @ 2 μM TFO | PO & other modif. TFOs | 58 |
| human GM-CSF promoter, NF-κ B site, plasmid & chromosome | 15mer GT PO TFO, Pu3 | EMSA, DNase I FP | in vivo, Jurkat cells, luciferase, RNase protect, ELISA, media TFO | -70% @ 2.4 μM TFO | random GT ODN / IL3 promoter, mRNA | 59 |
| HIV1 nef gene, linear plasmid | 15mer TG5meC PO TFO, 5'-acridine, Py3, stabilized with BePI | Tm | in vitro, HeLa NE or SP6 or T7 RNA pol., truncated transcript, pre. 3plx | -77% @ 10 μM TFO (pol II), -60% @ 10 μM TFO + 25 μM BEPI (SP6) | unsub. TFO & random ODN | 60 |
| HIV1 nef gene, linear plasmid | 16mer TC or T5meC PN TFO, Py3 | n.s. | in vitro, HeLa NE or SP6 RNA pol., truncated transcript, pre. 3plx | -77% @ 1 μM TC TFO (pol II), -60% @ 10 mM T5meC TFO (SP6) | titration | 61 |
| human TNF gene, 3rd intron, chromosome | 27mer GT PO TFO, 3'- cholesterol, Pu3 | EMSA | in vivo, THP-1 cells, TNF bioassay, RT -PCR, media TFO | -75% @ 0.6 μM TFO | other GT ODNs / IL-β mRNA | 62 |
| syn., +400, plasmid | 19mer GT PO TFO, 5'- psoralen, Pu3 | REPA, PCR/ REPA, PCR | in vivo, HeLa cells, luciferase, pre. 3plx | -79% @ 0.6 μM TFO, covalent | -pre. 3plx & -crosslink / β -gal | 63 |
| murine c-Ki-ras promoter, -290, plasmid | 20mer GA PO TFO, Pu3 | EMSA, Tm, CD | in vivo, NIH 3T3 cells, CAT, pre. 3plx | -90% @ 1 μg TFO (-40% @ 1 μg non- specific ODN) | non-specific ODN | 64 |
| human IGF-I gene, 1st exon, chromosome | RNA cont. 23 nt. GA sequence, Pu3? | n.s. for RNA | in vivo, C6 cells, stable clones, northern, in situ TFO | -95% | complement RNA | 65 |
| rat α 1(I) collagen promoter, -141, plasmid | 18mer GA PS TFO, Pu3 | EMSA, DNase I FP | in vivo, 2TK cells, CAT, pre. 3plx | -80% @ 50 μg TFO | 18mer ACGT ODN | 66 |
| rat IGF-IR gene, 3' (+4504), chromosome | RNA cont. 24 nt. GA sequence, Pu3? | n.s. | in vivo, C6(t1) cells, stable clones, northern, in situ TFO | -75%?, (similar inhibition IGF-I mRNA) | complement RNA / β -actin | 67 |
| syn., +185, linear plasmid | 15mer TG5meC PO TFO, Py3 | n.s. | in vitro, HeLa NE, runoff | -90% @ 10 μM TFO, crosslink | -UV | 68 |
| human, c-myc, P1 promoter PuF site & P2 promoter MAZ site, plasmid | 26 & 23mer GAT PS TFOs, Pu3 | EMSA | in vivo, HeLa cells, luciferase, Do pre. 3plx | 90% @ 100 μM, both TFOs | reverse ODNs | 69 |
| human cyclin D1 promoter, Sp1, chromosome | 18mer GT PS TFO, Pu3 | EMSA, DNase I FP | in vivo, HeLa cells, luciferase, transfect TFO | -60% @ 10 μM TFO | reverse ODN/ CMV luc | 70 |
| human α 1(I) collagen promoter, -140, fragment or plasmid | 30mer GAT PO TFO, Pu3 | EMSA | in vitro, HeLa NE, runoff, pre. 3plx; in vivo, CK-Y cells, GFP, pre. 3plx | -75% @ 200 ng TFO, (in vitro); -55% @ 30 nM TFO (in vivo) | mut. & control? ODN / CMV IE promoter | 71 |
| human HER2,+205, linear plasmid | 23mer PPGAT PO TFO, 5'-pam, Pu3 | EMSA | in vitro, HeLa NE, truncated transcript, pre. 3plx | -80% @ 1 μM TFO | titration, diff. TFOs | 72 |
| syn., -40, minimal c-fos promoter, plasmid | 20mer GA PO TFO, 5' -or 3' -peptide, Pu3 | EMSA, co-migrate | in vivo, NIH 3T3 cells, luciferase, pre. 3plx | +250% @ 1:1 TFO:plasmid (+10% @ 2:1) | titration, unmod. TFO | 73 |
| human c-src promoter, Sp1 & SPY sites, plasmid | 11mer GA PO TFO, Pu3 | n.s. | in vivo, 10T1/2 cells, CAT, pre. 3plx | -70% @ 500:1 TFO:plasmid | GA & ACGT ODNs | 74 |
| CAT gene, +578, plasmid | 19mer GAT PD TFO, Pu3 | n.s. | in vivo, Xenopus oocyte, CAT, northern, pre. 3plx | -90% @ 180 μM TFO | +/- TFO / Cyclin B1 mRNA | 75 |
| syn., +113, plasmid & chromosome | 15mer TG5meC PN TFO, 5'-acridine or psoralen, Py3 | EMSA / REPA | in vivo, P4 cells or nuclei, luciferase, northern, co-transfect TFO or steptolysin permeabilize | -70% @ 0.5 μM TFO (5' acridine, cell, plasmid); -50% @ 0.5 μM TFO (5' psoralen, nuclei, chromosome | mut. & reverse ODN / mut. site | 76 |
| human c-myc, P2 promoter, MAZ site, chromosome | 23mer GT PO TFO, 3'-amine, Pu3 | EMSA, DNase I FP | in vivo, CEM cells, northern, RT-PCR, Not media TFO | -63% @ 20 μM TFO | GA & scrambled ODNs / GAPDH mRNA | 77 |
| human rhodopsin gene, 1st or 2nd intron, plasmid | 18 or 28mer GA(T or C) PO TFO, 5'-psoralen & 3'-amine, Pu3 | n.s. (prior) | in vivo, HT1080 cells, IFM, pre. 3plx | -90% @ 1 μM, both TFOs | -TFO, -UV | 78 |
| human bcl-2 3' UTR, + 1946, plasmid | 18mer GAT PO(PS ends) TFO, 3'- amine, Pu3 | EMSA, co-migrate | in vivo, Tet-On HeLa, western, pre. 3plx | -90% @ 12.5:1 TFO:plasmid | random, scrambled ODN/ actin protein | 79 |
| murine Ki-ras promoter, plasmid | 20mer, PO/PS GA TFO, Pu3 | EMSA, DMS & DNase I FP | in vivo, 293 cells, CAT, co-transfect TFO | -75% @ 5 μM TFO | -TFO | 80 |
| syn., HIV-1PPT, +113, plasmid | 15mer TG5meC PN TFO, Py3 | n.s. / REPA | in vivo, P4 cells, single-cell luciferase, microinject TFO | -85% @ 32 μM TFO | reverse ODN, Renilla luciferase | 81 |
| human IgE promoter, STAT6 & Pu.1/NF-κ B sites, plasmid | 21mer TC AE TFO, Py3 | EMSA | in vivo, DG75 cells, luciferase, pre. 3plx | -80% @ 200:1 TFO: plasmid, (-20% mut. AE ODN) | random PO ODN, mut. AE ODN | 82 |
| human ICAM-1 gene, 3rd intron, chromosome | 16mer GT PO TFO, 3'- or 5'-psoralen, Py3 | EMSA / capture- PCR | in vivo, A431 cells, FACS, northern, transfect TFO | -60% @ 3 μM TFO (northern), +/- UV? | scrambled ODN, HLA-DR expression/ GAPDH mRNA | 83 |
| human bcr promoter, +1, chromosome | 13mer GA PO TFO, 3'-PEG, Pu3 | EMSA, DMS FP | in vivo, K562 cells, RT-PCR, media TFO | -35% @ 15 μM TFO, (random ODN, -15%) | G-rich PEG'd random ODN /abl mRNA | 84 |
| syn., 5 sites upstream core Ad E4 promoter, linear plasmid | 22mer, GT PO TFO, 3' -peptide, Pu3 | co-migrate | in vitro, HeLa NE, runoff, pre. 3plx; in vivo, HeLa cells, CAT, co-transfect TFO | +1000% @ 2.5 nM TFO in vitro (+100% @ 50 nM?); +3000% @ 50 nM TFO in vivo | GAL4 E4 promoter | 85 |
| human Ets2 promoter, Sp1, plasmid & chromosome | 25mer GT PO TFO, Pu3 | EMSA | in vivo, DU145 cells, luciferase, RT-PCR, northern, co-transfect TFO | -80% @ 250 nM TFO (plasmid), -62% @ 400 nM TFO (chromosome) | scrambled ODN, c-src reporters | 86 |
| human HER2, core promoter, plasmid | 25mer PPGAT PO TFO, 5'- /3'-pam, Pu3 | EMSA, alkylation/ Southern | in vivo, HeLa cells, luciferase, pre. 3plx | 70% @ 2 mM TFO (5' & 3' pam) | different modified TFOs | 87 |
a Target information is indicated as gene name, site, and whether located on an extrachromosomal DNA (e.g., plasmid) or on a chromosome. Syn., synthetic target. Ad, adenovirus. MLP, major late promoter.
b Triplex information is indicated as length, base composition (in order of abundance), backbone composition, modifications (indicated by 5'- or 3'-), auxiliary compounds (“stabilized with...”), and triplex motif. Bases include N6-methyl-8-oxo-adenine (M), 5-methylcytidine (5meC), and pyrazolopyrimidine guanine (ppG). Backbone compositions include phosphodiester (PO), phosphorothioate (PS), N3'-P5' phosphoramidate (PN), 2'-aminoethoxyribose (AE), and N,N-diethylethylenediamine phosphoramidate (PD). Modifications include N4,N4-ethano-2'-deoxycytidine (C*), phenylacetate mustard (pam), polyethylene glycol (PEG). Motifs include purine (Pu3), pyrimidine (Py3), and parallel. Additional abbreviations include triplex-forming oligonucleotide (TFO), containing (cont.), nucleotide (nt.).
c Assays used to demonstrate triplex formation include electrophoretic mobility shift assays (EMSA), footprinting (FP) with enzymes like DNase I or with chemicals including dimethyl sulfate (DMS), copper-1,10 phenanthroline (Cu-phen.), and methidiumpropyl-EDTA-iron (MPE), circular dichroism (CD), melting temperature (Tm), Fourier transform infrared spectroscopy (FTIR), and restriction endonuclease protection assays (REPA). Assays shown after a slash (e.g., PCR/PCR) indicate those assays performed after transcription has occurred.
d Transcription assay information is presented as extract (e.g., nuclear, NE) or polymerase (pol.), template, and assay type for in vitro studies and cell type, assay type, and TFO administration method for in vivo studies. In vitro assays include runoff, G-free cassette, primer extension, and truncated transcript. In vivo assays include RNA-based (e.g., northern, reverse transcriptase PCR [RT-PCT]), immunological (e.g., enzyme-linked immunosorption [ELISA], immunofluorescence microscopy [IFM], fluorescence-activated cell sorting [FACS]), and enzymatic activity assays (e.g., chloramphenicol acetyltransferase [CAT], β-galactosidase [β-gal.], luciferase [luc.]). Preformed triplexes (pre. 3plx).
e Maximum effect of triplex on transcription. Minus and plus signs indicate triplex-dependent transcription inhibition and stimulation, respectively. Unexpected or unusual findings are indicated in parentheses. Mut., mutant.
f Controls used to demonstrate specificity of triplex effects on transcription. Controls involving oligonucleotides are shown before the slash, while controls involving targets are shown after (e.g., scrambled ODN/mutant promoter). Additional abbreviations include: unmodified (unmod.), cytomegalovirus immediate-early promoter (CMV IE), and unsubstituted (unsub.).
Table 2Intermolecular triplexes and transcription
| Targeta | Multiplexb | Triplex Proofc | Transcription Assayd | Maxium Effecte | Controlsf | Ref. |
|---|---|---|---|---|---|---|
| Drosophila hsp26 promoter -89, chromosome | 25 bp mirror repeat, H | oligo hybrid., DEPC FP, S1 nuclease/ DEPC FP | in vivo, Drosophila, hsp26-lacZ transgene, β-gal | -67% del. H, +2% mut. H-, -68% H'+ | del. H, mut. H-, H' | 92 |
| syn., promoter, plasmid | (G• C)n, H' | CAA FP | in vivo, LTK-cells, TK CAT, direct & competitions | +970% (G)30, +90% (G)18, +50% (G)35 | number of repeats | 15 |
| syn., +120, plasmid | 38 bp mirror repeat, H or H'? | chloroquine 2D gel electro- phoresis | in vivo, E. coli, lacZ, β-gal | -80% | mut. H- | 93 |
| murine c-Ki-ras promoter, plasmid | 27 bp mirror repeat, H | CAA FP | in vivo, HepG2 cells, CAT | -50% mut. H-, -50% mut. H+ | mut. H-, multi mut. H+ | 94 |
| syn., +17, plasmid | 69 bp mirror repeat, stabilized with BePI, Hy-5 | CAA FP, mung-bean nuclease | in vivo, E. coli, viability | -53% (-74% @ 5 μM BePI) | H- / tetR | 95 |
| chicken malic enzyme promoter, plasmid | 49 bp Py/ Pu, H' | S1 & P1 nuclease | in vivo, chicken hepatocytes, CAT | +900%, (+0% reversed orientation) | H-, H+ reversed | 96 |
| murine metallothionein-1 promoter, plasmid | 128 bp, H-y3 | CAA, DEPC, DMS FP | in vivo, NIH 3T3 cells, luciferase | no effect, basal or Cd2+ -induced | H-, reverse orientation | 97 |
| syn., human frataxin 1st intron GAA repeat, +100? plasmid | (GAA• TTC)n repeats, H'? | n.s. | in vivo, COS-7 cells, RNase Not protection, β-gal | -91% @ n = 230 repeats | number of repeats, reverse orientation | 98 |
| syn., human frataxin 1st intron GAA repeat, +100? plasmid | (GAA• TTC)n repeats, H'? | DEPC (ss DNA tested!) | in vitro, T7 RNA pol., runoff | -90% @ n = 88 repeats | number of repeats | 99 |
| syn., human frataxin 1st intron GAA repeat, +100? plasmid | (GAA)88, H'? | n.s. | in vitro, T7 RNA pol., runoff | -90% (-25% @ 2.5 μM complement [TTC]7 ODN) | (GGA)7 ODN / (CUG)88 template | 100 |
| syn.,human frataxin 1st intron GAA repeat, +100? plasmid | (GAA)150, (multi-H'(sticky DNA) | gel mobility/ gel mobility | in vitro, T7 or SP6 RNA pol. or HeLa NE, runoff | -99% r[UUC]150, -95% co-transcribed control | number of repeats, orientation / co-transcribed control template | 101 |
| human HMGA2 promoter, -25, plasmid | 60 bp Py/Pu, H? | S1 nuclease, EMSA | in vivo, NIH 3T3, luciferase | +400% H+ supercoiled, (+200% mut. H-) | mut. H-, linear or supercoiled/ CMV β-gal | 102 |
| syn., E. coli PIT sequences, promoter & downstream, plasmid | 33 nt multimers, H' | n.s. | in vivo, E. coli, lacZ, β-gal | -0% (promoter PIT); +300% (downstream 2 x PIT), mRNA translation effect | PIT-, mut. PIT | 103 |
| Drosophila hsp26 promoter, chromosome | 25 bp mirror repeat, H | DMS, kethoxal, CMCT, KMnO4 & UV FP / REPA | in vivo, Drosophila, hsp26-lacZ transgene, β-gal | -0% H-, -40% mut. H+ | mut. H- & multi-mut. H+ | 104 |
a Target information is indicated as gene name, site, and whether located on an extrachromosomal DNA (e.g., plasmid) or on a chromosome. Syn., synthetic target.
b Triplex information is indicated as length of the triplex-forming region, sequence of repeating element, auxiliary compounds (“stabilized with...”), and triplex motif (H or H'). BePI, (3-methoxy- 7H-8-methyl-11-[(3'-amino-propyl) amino] benzo[e]pyrido [4,3-b]indole.
c Assays used to demonstrate triplex formation include electrophoretic mobility shift assays (EMSA), oligonucleotide hybridization, footprinting (FP) with enzymes like S1 nuclease or with chemicals including chloroacetaldehyde (CAA), 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho- p-toluene sulfonate (CMCT), diethylpyrocarbonate (DEPC), and dimethyl sulfate (DMS), or with ultraviolet radiation (UV) and restriction endonuclease protection assays (REPA). Assays indicated after a slash (e.g., UV FP / REPA) indicate those assays performed after transcription has occurred or in vivo.
d Transcription assay information is presented as extract (e.g., nuclear, NE) or polymerase (e.g., T7 RNA pol.), template, assay type (e.g., runoff) for in vitro studies, and cell type, template, and assay type for in vivo studies. In vivo assays include RNA-based (e.g., RNase protection) and enzymatic activity assays (e.g., chloramphenicol acetyltransferase [CAT], β-galactosidase [β-gal.], luciferase [luc]).
e Maximum effect of triplex on transcription. Minus and plus sign preceding percentage effect indicate triplex-dependent transcription inhibition and stimulation, respectively. Hand H+ refer to mutations that either disrupt or promote H(H')-DNA formation. Del., deletion. Mut., mutation. Unexpected or unusual findings are indicated in parentheses.
f Controls used to demonstrate specificity of triplex effects on transcription. Controls involving template DNA are shown before the slash, while controls involving alternative targets are shown after (e.g., H- / tetR).
Table 3Quadruplexes and transcription
| Targeta | Multiplexb | Quad. Proofc | Transcription Assayd | Maxmium Effecte | Controlsf | Ref. |
|---|---|---|---|---|---|---|
| human insulin gene, -363, plasmid | (G9A2T2C)n, intra- or inter-molecular G4 | EMSA, DMS FP | in vivo, HeLa cells, luciferase | -80% with Pur-1 | mut. G4 -, multi-mut. G4 +, Pur-1 expression | 109 |
| syn. | G4 RNA? | n.s. | in vitro, T7 RNA pol. + GTP only, PAGE | +100% termination @ 13-14 nt. | 7-deaza- GTP | 110 |
| human c-myc promoter, chromosome | 22mer G18A2T2, G4' | n.s. | in vivo, Ramos cells, proliferation, G4' ODN, media | -70% @ 100 nM G4 ODN, (-55% @ 100 nM ss G-ODN) | random ODN | 111 |
| human c-myc promoter, -115 P1, plasmid or chromosome | 27mer G20A4T3, stimulated by TMPyP4, “chair” G4' | DMS FP, TMPyP4 UV FP, Taq DNA pol. stop | in vivo, HeLa cells, luciferase or Ramos cells, RT-PCR | +200% chair mut. | TmPyP2, alternative G4 - mut., CA46 cells | 112 |
a Target information is indicated as gene name, site, and whether located on an extrachromosomal DNA (e.g., plasmid) or on a chromosome. Syn., synthetic target.
b Quadruplex information is indicated as length of the quadruplex-forming region, composition or sequence, auxiliary compounds (“stabilized with...”), and quadruplex form. TMPyP4, cationic porphyrin.
c Assays used to demonstrate quadruplex formation include electrophoretic mobility shift assays (EMSA), footprinting (FP) with dimethyl sulfate (DMS) or ultraviolet radiation (UV), and polymerase (pol.) stop assays. Data not shown (n.s.).
d Transcription assay information is presented as assay type for in vitro studies and as cell type, template, and assay type for in vivo studies. PAGE, polyacrylamide gel electrophoresis. RT-PCR, reverse transcriptase PCR.
e Maximum effect of quadruplex on transcription. Minus and plus sign preceding percentage effect indicate quadruplex-dependent transcription inhibition and stimulation, respectively. Abbreviations include nucleotide (nt.), single-stranded (ss), and mutation (mut.). Unexpected or unusual findings are indicated in parentheses.
f Controls used to demonstrate specificity of triplex effects on transcription. G4- and G4+ refer to mutations that either disrupt or promote G4 formation.