From: Centrins, a Novel Group of Ca2+-Binding Proteins in Vertebrate Photoreceptor Cells

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
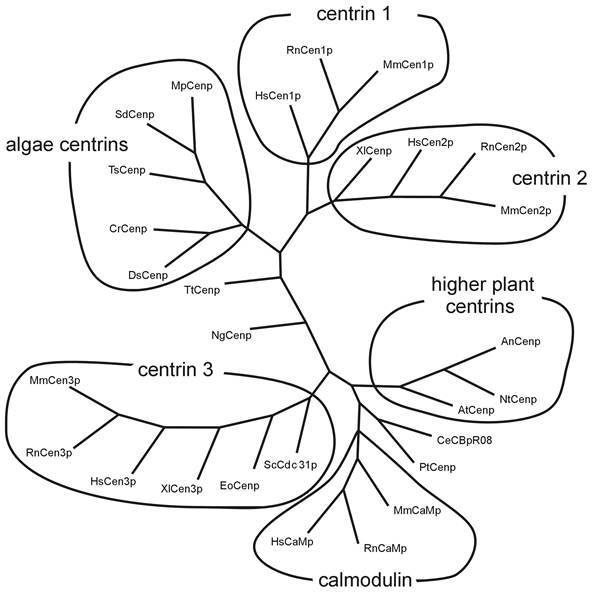
Comparison of centrin isoforms of diverse species. Comparison (using programs: Omiga 2.0, Genedoc 2.5.006 and phylip) of 28 different amino acid sequences of centrins and calmodulins. The phylogram shows a consensus tree which shows the highest frequency of each node of 1000 repetitions. Phylip divides the centrins into subgroups of centrin isoforms 1, 2, 3, algae centrins, higher plant centrins and a group of calmodulin (RnCaMp = rat calmodulin Accsession Number (AN): CAA32120; MmCaMp = mouse calmodulin AN: NP_033920; HsCaMp = human calmodulin AN: BAA08302; NgCenp = Naegleria gruberi centrin AN: AAA75032; XlCenp = Xenopus laevis centrin AN: AAA79194; XlCenp3 = Xenopus laevis centrin 3 AN AAG30507; PtCenp = Paramecium tetrauelia centrin AN: AAB188752; DsCenp = Dunaliella salina centrin AN: AAB67855; HsCen1p, 2p, 3p = human centrins 1, 2, 3 AN: AAC27343, AAH13873, AAH05383; MmCen1p, 2p, 3p = mouse centrins 1, 2, 3 AN: AAD46390, AAD46391, AAH02162; RnCen1p, 2p, 3p = rat centrins AN AAK20385, AAK20386, AAK83217; AtCenp = Arabidopsis thaliana centrin AN: CAB16762, AnCenp = Atriplex nummularia centrin AN: P41210; NtCenp = Nicotiana tabacum centrin AN AAF07221; CrCenp = Chlamydomonas reinhardtii centrin AN CAA41039; SdCenp = Scherffelia dubia centrin AN CAA49153; MpCenp = Micromonas pusilla centrin AN CAA58718; EoCenp = Euplotes octocarinatus centrin AN CAB40791; TsCenp = Tetraselmis striata centrin AN P43646; ScCdc31p = Saccharomyces cerevisiae AN P06704; CeCBpR08 = Caenorhabditis elegans AN P30644; TtCenp = Tetrahymena thermophila AN AAF66602. Tree is not complete.
From: Centrins, a Novel Group of Ca2+-Binding Proteins in Vertebrate Photoreceptor Cells

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.