Except where otherwise indicated, this work is licensed under a Creative Commons Attribution 4.0 International License
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Rohrmann GF. Baculovirus Molecular Biology [Internet]. 4th edition. Bethesda (MD): National Center for Biotechnology Information (US); 2019.
The discovery of baculoviruses
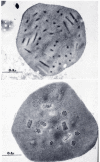
The history of the discovery of baculoviruses is related to the development of the silk industry in China over 5000 years ago. The culture of silkworms spread throughout Asia and reached Japan via Korea by about 300 C.E. arriving in Europe by about 550 C.E. By the 12th century it was established in Italy and Spain spreading to France and England and to Mexico by the 1500s. Silk production has been of major cultural significance in both China and Japan. It was a major item of commerce and in Japan is extensively documented in historic prints (1). Although no longer a major industry, silk production is still practiced symbolically by the Japanese royal family. (For a review of the history of silkworm culture, see (2)). As with any agricultural enterprise, problems were encountered caused by a variety of diseases, and these had to be confronted for the industry to flourish. Even before the germ theory of disease was introduced, a variety of different types of illnesses afflicting silkworms had been described, and methods to mitigate the effects of these diseases had been developed. With the advent of light microscopy, a prominent feature of one of the types of diseases was characterized by the presence of highly refractile occlusion bodies that were symptomatic of the affected insects. These were commonly polyhedron shaped (Figure 1) and led to the naming of the diseases associated with these structures as 'polyhedroses' by the mid-1800s. Although the presence of infectious particles within occlusion bodies had been suggested earlier, it was not until the late 1940s that the presence of rod-shaped virions was convincingly demonstrated by electron microscopy (3). These and other studies demonstrated the crystalline nature of the occlusion body structure. Subsequently, two different types of polyhedrosis diseases were distinguished: those in which the polyhedra developed in nuclei called nuclear polyhedroses (NPVs), and those with occlusion bodies present in the cytoplasm (Figure 2) called cytoplasmic polyhedroses (CPVs) (4). In contrast to the rod-shaped, DNA-containing NPVs, the CPVs have icosahedral capsids and were placed in the Reoviridae (genus Cypovirus), a family of viruses with segmented, double-stranded RNA genomes. A second category of baculovirus characterized by the presence of small, granular, ellipsoidal-shaped occlusion bodies was originally reported in the 1920s (5) was named granulosis viruses (GVs). The division of the baculoviruses into two major groups, the nuclear polyhedrosis viruses (now called nucleopolyhedroviruses (NPV)) (Figure 3) and the granulosis viruses (now called granuloviruses (GVs)) (Figure 4), based on occlusion body morphology defined the major taxonomical divisions of these viruses until the advent of molecular biology.

Figure 1.
Baculovirus occlusion bodies. Scanning EM by K. Hughes and R. B. Addison.
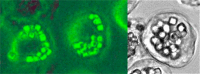
Figure 2.
Comparison of cells infected with a cypovirus (CPV – Reoviridae) and a baculovirus. Left panel: CPV of Orgyia pseudotsugata. Photo by G. S. Beaudreau. Right Panel: AcMNPV infected S. frugiperda cells. From Manji and Friesen (83).
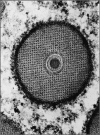
Figure 4.
Cross section of a granulovirus of Plodia interpunctella. From Arnott and Smith (84). Copyright Elsevier 1967. Reproduced with permission via Copyright Clearance Center.
The terminology for these viruses went through a series of names, and it was not until 1973 that a nomenclature that included Borrelinavirus, Bergoldiavirus, Smithiavirus, Moratorvirus and Vagoiavirus in honor of various historic individuals who had done early research on NPVs and GVs was changed and unified into the Baculoviridae (6). The name baculovirus was proposed by Mauro Martignoni, who because of his Italian-Swiss heritage was a Latin scholar. He suggested that they be named baculoviruses (family Baculoviridae) because of the rod-shape of their virions, which is derived from Latin baculum — cane, walking stick, staff. For an excellent biography of Prof. Martignoni, see (7).
The significance of baculoviruses in Nature
Although much of the early interest in baculoviruses was due to the threat they posed to the silk industry, baculoviruses play a major role in the control of natural insect populations. For example, they are a major regulator of gypsy moth populations in North America and in some instances have been shown to be responsible for over 50% of the mortality observed (8). They also are major contributors to the collapse of Douglas-fir tussock moth outbreaks (9). In addition to forest insect populations, they also appear to be important in the natural control of agricultural pests of human food crops, and as a result they may be a substantial contaminant of the human diet. For example, in one study it was found that cabbage purchased from 5 different supermarkets in the Washington D.C. area were all contaminated with baculoviruses to such an extent that each serving (about 100 cm2 of leaf material) would contain up to 108 polyhedra of an NPV pathogenic for the cabbage looper, Trichoplusia ni (10)!
The definition is not in the name: Naming baculoviruses
Baculoviruses are normally named for the initial host from which they were isolated. Consequently, the type NPV species, AcMNPV, was named for its host, the alfalfa looper, Autographa californica (Ac). This naming would be straightforward except that AcMNPV infects a wide variety of lepidopteran insects and its name originated because of its initial association. Consequently, a variety of other virus isolates e.g., Galleria mellonella (GmNPV), Rachiplusia ou (RoNPV), and Plutella xylostella (PlxyNPV), although having unique names, are closely related (~96-98.5% at the amino acid sequence level) variants of AcMNPV. In addition, viruses were originally named by the first letter of the genus and species of their host. However, as more viruses were discovered, some infected different insects that had names with the same first letters. This resulted in different viruses with the same descriptor. Consequently, the first two letters of the genus and species have become the convention, i.e., AcMNPV should really be AucaMNPV. However, since AcMNPV, GmMNPV and RoMNPV have been used so extensively, the original abbreviations have been retained.
What defines a baculovirus?
Genomes and nucleocapsids
Baculoviruses are a very diverse group of viruses with double-stranded, circular, supercoiled genomes, with sizes varying from about 80 to over 180 kb, that encode between 90 and 180 genes. Of these genes, a common set of about 38 homologous, or core genes, has been identified in all baculovirus genomes, and there are probably others that cannot be recognized because of the extent of changes incorporated over time. The genome is packaged in rod-shaped nucleocapsids that are 230–385 nm in length and 40–60 nm in diameter (11, 12). In the most well characterized baculoviruses, the virions are present as two types, occluded virions (ODV) and budded virions (BV). Although these two types of virions are similar in their nucleocapsid structure, they differ in the origin and composition of their envelopes and their roles in the virus life cycle (see Chapter 2).
Occlusion bodies
Members of the Baculoviridae are characterized by their presence in occlusion bodies called polyhedra for NPVs and granules or capsules for GVs (Figure 3, Figure 4). Polyhedra are about 0.6–2 μM in diameter, whereas granules are oval-shaped with diameters of about 0.2–0.4 μM (11). Occlusion bodies are highly stable and can resist most normal environmental conditions thereby allowing virions to remain infectious indefinitely. Evidence suggests that they can survive passage through the gastrointestinal tract of birds, which can facilitate their dispersal (13, 14). The occlusion body consists of a crystalline matrix composed of a protein called polyhedrin in NPVs and granulin in GVs. Although they have different names, these two proteins are closely related.
Recently, nudiviruses, which may be occluded or non-occluded, have been shown to be related to baculoviruses (15). Therefore, in the future occlusion might not be considered an exclusive feature of the Baculoviridae.
Baculovirus hosts
Over the years, baculoviruses have been reported from a variety of different species of invertebrates. However, the only well-documented hosts are Diptera, Hymenoptera, and Lepidoptera. Convincing documentation has been reported for occluded virions resembling NPVs in a caddis fly (Trichoptera) (16) and a shrimp species (17, 18) (see below). An occluded baculovirus-like virus was also reported for a thysanuran, but it did not appear to affect its host and transmission studies failed (19). Baculoviruses have also been reported from Orthoptera (20), but later these were classified as entomopox viruses, and from Coleoptera, but these are normally not occluded and were included with the nudiviruses (21). Reports of infection of other insects, e.g., a coleopteran (22) could not be confirmed (Rohrmann, unpublished). In addition, there is a report of a baculovirus infecting a neuropteran (23). However, the infection occurred under laboratory conditions, where neuropterans were fed on lepidoptera that had died of an NPV infection. Consequently, the neuropterans were likely heavily contaminated from their food source, and although they appeared to die of an NPV infection, they were probably exposed to an unusually high virus dose. Naturally infected Neuroptera have not been documented. An example of the distribution of baculoviruses in insects that has been reported in the literature (24) is shown in Table 1. These numbers should be viewed with caution because many of the reports could be of the same virus infecting different species. However, they do give a good general overview of the likely distribution of baculoviruses. Of particular note is that GVs are confined to the Lepidoptera. In addition, all the hymenopteran hosts belong to a suborder called Symphyta that are comprised of sawflies (named because their ovipositor resembles a saw and in some species is used to cut into plants for egg deposition). Sawflies resemble Lepidoptera; they have herbivorous, caterpillar-like larvae and are distinct from the other hymenopteran suborder, Apocrita, which includes bees, ants, and wasps.
Other defining features of the Baculoviridae: a virus encoded RNA polymerase
In addition to invertebrate hosts, circular, supercoiled double-stranded DNA genomes, rod-shaped enveloped nucleocapsids, and the production of occluded virions, an additional defining feature of baculoviruses is that they encode their own RNA polymerase. The core enzyme is composed of four subunits and functions in the transcription of late promoters that are initiated within a novel sequence element (see Chapter 6). Whereas other DNA viruses of eukaryotes encode their own polymerase, e.g., poxviruses, they replicate in the cytoplasm and therefore do not have access to the host cell transcriptional apparatus located in nuclei. Baculoviruses, in contrast, exploit the host cell transcriptional system for expressing their early genes, but after initiation of DNA replication they are dependent upon their own RNA polymerase for transcription of their late and very late genes. The closely related Nudiviridae also likely encoded their own RNA polymerase related to the baculovirus enzyme.
Baculovirus Diversity
Phylogenetic analysis suggests that large DNA viruses of insects evolved several hundred million years ago along with the first insects (25) (26) and consequently as different insects evolved and proliferated, so did their associated viruses. The recent proliferation of baculovirus genome sequences has greatly expanded our understanding of their diversity and evolution. This has yielded distinct patterns of virus relatedness (Figure 5) in which virus lineages are associated by the host that they infect. The Baculoviridae are divided into four genera (27). The viruses of Lepidoptera are divided into Alpha- and Betabaculoviruses encompassing the NPVs and GVs, respectively, and those infecting Hymenoptera and Diptera are named Gamma- and Deltabaculoviruses, respectively (Table 2). Such patterns of host-associated virus diversity were first observed for small DNA viruses (papilloma and polyoma viruses) of mammals (28) and originally suggested for baculoviruses based on N-terminal polyhedrin and granulin sequences (29). In this process, viruses associate with a host, and as their host becomes genetically isolated and speciates, so does the virus in a process called host-dependent evolution. As more sequence data has become available, this process has been more convincingly demonstrated for baculoviruses (30) and is clearly reflected in Figure 5 in which the major lineages are clustered into clades based on the host insect that they infect. In GVs, host-dependent evolution has been suggested at the level of insect families (31). A major division has been observed in the lepidopteran NPVs that has resulted in the separation of this lineage into two major Groups, I and II (32). These two groups differ significantly in gene content, most notably Group I NPVs use GP64 as their budded virus (BV) fusion protein, whereas Group II NPVs lack gp64 and utilize a protein called F (33). There are also 11 other genes in addition to gp64 that appear to be found only in Group I NPVs (Table 3) (34). It has been suggested that the Group I lineage originated when a NPV variant uniquely containing these genes incorporated gp64 which stimulated their evolution as a distinct lineage (35) (36) (see Chapter 2). Although the list of host insects provides information on baculovirus distribution, sequence analysis of common sets of genes from a wide variety of baculoviruses has provided a picture of the actual extent of their diversity (37).
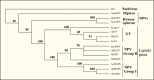
Figure 5.
Phylogenetic relatedness of LEF8 from selected baculoviruses. Neighbor joining; bootstrap analysis (1000 reps).
Table 2.
Genera of the Baculoviridae
| Genus | Members |
|---|---|
| Alphabaculovirus | Lepidopteran NPVs |
| Betabaculovirus | Lepidopteran GVs |
| Gammabaculovirus | Hymenopteran NPVs |
| Deltabaculovirus | Dipteran NPVs |
Table 3.
Genes1 found in and unique to most sequenced Group I NPV genomes
| Ac1 (ptp), Ac5, Ac16 (BV-ODV26), Ac27 (iap-1), Ac30, Ac72, Ac73, Ac114, Ac124, Ac128 (gp64), Ac 132, Ac151 (ie2) |
- 1
Genes are designated by their AcMNPV orf number
Multiple versus single nucleocapsids
A prominent feature of the nucleocapsids within polyhedra is their organization into either single or multiple aggregates of nucleocapsids within an envelope (Figure 3). For example, in some NPVs (called multiple or MNPVs) there can be from 1 to 15 nucleocapsids per envelope, with bundles of 5 to 15 predominating. In contrast, strains defined as having a single nucleocapsid per envelope (called SNPVs) rarely show more than one nucleocapsid per envelope (11, 38). It has been suggested that the MNPV phenotype may accelerate the ability of the virus to establish the infection because if multiple nucleocapsids infect a cell simultaneously, some could enter the nuclei and begin replication whereas others could transit the cell and spread the infection elsewhere (this is discussed in Chapter 3). Another benefit of groups of nucleocapsids infecting a cell would be that they could repair damaged DNA via recombination. Fatally damaged genomes could recombine to produce a viable molecule. Because this feature is so distinctive and characteristic of specific isolates, it was incorporated into the early nomenclature such that NPVs were categorized as either MNPV or SNPVs (also previously called multiply or singly embedded virions (MEV and SEV)). In addition, whereas MNPVs and SNPVs were both found in lepidopteran viruses, only SNPVs were observed in other insect orders. GVs were also categorized as singly enveloped; however, multiple GVs, although rare, have been described (39). With the accumulation of DNA sequence data that allowed for the determination of definitive phylogenetic relationships, it was found that the MNPV and SNPV division did not conform to the phylogeny of the viruses. For example, at one point BmNPV was considered the type virus for SNPVs because of its production of predominantly single nucleocapsids (40). However, sequence data indicates that BmNPV is closely related to AcMNPV (both belong to Group I), whereas other MNPVs such as LdMNPV and SeMNPV are more distantly related Group II viruses, which also includes SNPV-type viruses. Drawing conclusions regarding the properties of MNPV and SNPV viruses is fraught with difficulty because their genetic content is often so different that it is not possible to attribute characteristics of a virus to its M- or SNPV phenotype.
Several genes alter the morphotype. For example, AcMNPV deleted for Ac23, a homolog of the F fusion proteins of Group II viruses, shows an elevated percentage of singly enveloped virions (45%) vs. 11–22% for different virus constructs encoding Ac23 (41). It has also been shown that deletion of Ac92, a gene encoding a sulfhydryl oxidase, results in virions that superficially resemble the SNPV phenotype (42). However, they are not infectious, indicating that they are very different from true SNPVs. It was also observed that point mutations in domains II or III of the AcMNPV DNA polymerase gene caused a dramatic change in the ratios of S- and M- NPV types. For example wt AcMNPV showed about 3% S nucleocapsids in infected cell nuclei, whereas a point mutant in domain III or a double mutant in domains II and III resulted in 61 and 89% single nucleocapsids, respectively (43). Therefore, the expression of some baculovirus genes can influence MNPV production and this may be linked to the efficiency of virion replication or virion integrity.
It also appears that the type of cell infected can influence the morphotype. In a report describing the isolation of a virus pathogenic for Bombyx mandarina (BomaNPV S2) (a close relative of BmNPV), it was observed that the virus grew in both Bm5 and T. ni cells. In Bm5 cells, the virus was present as an SNPV. However, in T. ni cells it had an MNPV phenotype (44). Therefore, the viral morphotype may be influenced by the physiology of the cell, how the cell responds to viral infection, or cell specific factors.
In summary, the significance of the MNPV morphotype is complex; a specific causal genetic component has not been identified and although it occurs in most Group I Alphabaculoviruses, MNPVs are also common in Group II viruses and sometimes are found in the Betabaculovirus (GVs). Furthermore, MNPV formation can be influenced by the type of cell infected, suggesting an environmental or physiological influence. Despite agreement that the MNPV and SNPV designation is not a useful taxonomical trait, it continues to be employed, in part for historical continuity, and also because it can be a convenient method for distinguishing different viruses that are pathogenic for the same host, e.g., OpMNPV and OpSNPV which both infect Orgyia pseudotsugata (Figure 3), but are members of Group I and II, respectively. For a detailed review see (45).
Gene content and organization
Although there are major differences in the sizes of baculovirus genomes, a few patterns are evident (Table 4). The viruses of members of the Hymenoptera contain the smallest genomes at a little over 80 kb. In contrast, the GV genomes vary from 101 kb (PlxyGV) up to 178 kb (XecnGV). Group I NPVs cluster around 130 kb, whereas Group II show a much higher degree of diversity, varying from about 130 to almost 170 kb. The small size of the hymenopteran NPV genomes might be attributed to a restricted life cycle confined to replication in insect gut cells (46). Consequently, unlike other baculoviruses that cause systemic infections, genes required for spread throughout the insect, where the virus encounters and replicates in a variety of tissues, might not be necessary. In contrast, the large size of a Group II NPV (LdMNPV), which at 161 kb is about 30 kb larger than many other NPV genomes, can be attributed to a combination of repetitive genes (16 bro [baculovirus repeated orf] genes) that add about half of the additional 30 kb, while genes not found in smaller genomes add most of the remainder. These additional genes encode ribonucleotide reductase subunits and two enhancin genes (47). In other viruses with large genomes (e.g., XecnGV), repeated genes including 10 bro and 4 enhancin genes, comprise up to 17% (30 kb) of the genome (48). Despite the large difference in gene content in GV genomes as reflected in their size range, their genomes are surprisingly collinear (49). In contrast, NPVs even from the same order (Lepidoptera) show a high degree of variation (50).
Table 4.
Genome size and predicted ORF content* of selected baculoviruses
Regulatory content: homologous regions (hrs)
In addition to containing a set of genes encoding proteins required for productive infection, most baculovirus genomes also contain homologous repeated regions. In AcMNPV, hrs are comprised of repeated units of about 70-bp with an imperfect 30-bp palindrome near their center. Hrs are repeated at eight locations in the genome with 2 to 8 repeats at each site (see Chapter 4). They are highly variable, and although they are closely related within a genome, they may show very limited homology between different viruses. For example, in the CpGV genome, tandem repeated sequences are not evident, although a 75-bp imperfect palindrome is present at 13 different locations on the genome (51). In addition, in the TnSNPV (group II) sequence, hrs were not found (52). Hrs have been implicated as both transcriptional enhancers (see Chapter 4) and origins of DNA replication for some baculoviruses (53-58) (see Chapter 5).
Conservation of baculovirus genes: core genes
Despite the diversity in gene content present in different baculovirus genomes, a set of 38 genes that are present in all sequenced baculovirus genomes has been identified (Table 5). Such genes can be very diverse and careful analysis may identify additional core genes. This is particularly true of the dipteran virus that is most distant from the baculoviruses of Lepidoptera and Hymenoptera. Of the conserved genes, about half are virion-associated proteins that are involved in capsid structure, the occlusion-derived virus envelope, and larval infectivity. Most of the others are related to DNA replication or processing, and late or very late transcription. Despite the limited set of genes that baculoviruses share, they do provide insight into some of the major functions required of all baculoviruses such as a common virion structure, the necessity to infect gut cells, and the use of a novel polymerase for the expression of late genes. Furthermore, there are sets of genes specific to, and conserved in, each clade of viruses such as gp64 in Group I NPVs and a number of GV-specific genes (see below). These genes likely reflect major recombination events that altered the properties of the viruses to such an extent that they subsequently developed along distinctive phylogenetic pathways. The observation that different types of NPVs and GVs and other types of DNA viruses (e.g., entomopox, ascovirus, nudivirus) have the potential to simultaneously infect the same insect indicates how via recombination, genes can be transferred between different viruses and between viruses and the host insect.
Table 5.
Conserved genes1 in baculoviruses, nudiviruses, hytrosaviruses, and polydnaviruses
Lepidopteran baculovirus (NPV and GV) core genes
In addition to the core genes found in all baculoviruses, lepidopteran baculoviruses encode an additional set of genes that appear to be present in most of their genomes, and about half are also found in Gammabaculovirus (hymenopteran virus) genomes (59) (Table 6). Homologs or functional analogs of many of these genes are likely present in all baculoviruses, but the relationships cannot be detected due to the extent of divergence of the genes. Since they appear to be so central to the replication of the baculoviruses, the single-stranded DNA binding protein, LEF-3, and the transcriptional activator IE-1 may be examples of gene divergence which prevents their detection in the more distantly related viruses. In contrast, homologs of Ac38, an ADP-ribose phosphatase, belong to a family found in many organisms and would likely be detected outside the lepidopteran baculoviruses, if it were present. It acts as a decapping enzyme in vaccinia virus (60) and its presence suggests that the lepidopteran viruses employ molecular strategies significantly different from the other baculovirus genera lacking orthologs of Ac38. Polyhedrin appears to be an example of possible convergent evolution since the occlusion body protein gene of the dipteran virus appears to encode a novel protein (61).
Table 6.
Additional Core genes present in all Lepidopteran (Alpha- and Beta-) and some Gammabaculoviruses
A few genes are present in most Alphabaculoviruses but are found in some, but not all, Betabaculoviruses (34). The most well-characterized of these is Ac9 (orf1629), which is a homolog of WASP proteins that are involved in movement of the virus based on actin polymerization (see Chapter 3). This would suggest that other baculoviruses have major differences in their pathogenesis.
Granulovirus-specific genes
In addition to their occlusion body morphology, the pathology of GV infection differs from NPVs. For example, their replication is not confined to nuclei as the nucleus and cytoplasm appear to merge during GV infections. The presence of GV specific genes may reflect these differences. Granuloviruses encode a number of genes that are found in most GV genomes. Of these GV genes, 19 appear to be specific to GVs (34), whereas others, although found in all GV genomes, are also found in a few other baculoviruses. Examples of the latter are DNA ligase and helicase-2 homologs that, in addition to GV genomes, are also present in at least one NPV genome (LdMNPV). The helicase-2 homolog is also found in an additional NPV (Maco-B-NPV). There are other GV-specific genes that are found in most, but not all, GV genomes. For additional information or the relatedness of baculovirus genes see (34, 62).
Related viruses: the Nudiviridae
In addition to baculoviruses, there are several groups of viruses pathogenic for invertebrates that have large double-stranded DNA genomes (63). These include nudiviruses, ascoviruses, irridoviruses, and entomopox viruses. However, with the exception of nudiviruses, all these viruses replicate in the cytoplasm or exhibit a combination of both nuclear and cytoplasmic replication (ascoviruses) (reviewed in (64)). Nudiviruses closely resemble baculoviruses including nuclear replication and rod-shaped, enveloped nucleocapsids. Because of the devastation caused to coconut and oil palm trees by the rhinoceros beetle (Oryctes rhinoceros) in the South Pacific in the 1950’s, it was of great interest when a virus that could successfully control this insect was discovered in the 1960’s (65) (66, 67). Subsequently, similar viruses were identified and were categorized as non-occluded baculoviruses (68). They have double-strand, circular DNA genomes varying in size from 97–230 kb (15). As shown in Table 5, nudivirus genomes contain homologs of up to 20 of the 38 core genes conserved in all baculovirus. These include homologs of genes encoding the novel baculovirus RNA polymerase subunits, virion structural proteins, and a set of genes required for infection of gut tissue called per os infectivity factors or PIFs. Therefore, based on morphology and molecular phylogeny, these two groups of viruses clearly share membership in a distinct viral lineage. They were removed from the Baculoviridae in the 1980s because they were all thought to be nonoccluded and were named nudiviruses (latin nudi = naked, bare, uncovered). They also have a somewhat different host range than baculoviruses, having been characterized not only from Lepidoptera and Diptera (69), but also from Coleoptera (the rhinoceros beetle) and Orthoptera (a cricket) (70) and crustacea (Figure 6). Phylogenetic analysis of multiple shared genes shows the separation of the baculovirus and nudivirus into distinct but related lineages (Figure 7) (71).

Figure 6.
Polyhedral inclusion body of a nudivirus of the pink shrimp, Penaeus monodon. Arrows indicated nucleocapsids. From Couch (17). Reproduced with permission of Elsevier Limited via Copyright Clearance Center.
Examples of two occluded nudiviruses are a dipteran (crane fly), Tipula oleracea and the pink shrimp Penaeus monodon (Figure 6) both of which were originally thought to be members of the Baculoviridae. However, when their genomes were sequenced, they were both found to be nudiviruses (71). In addition, their occlusion body protein sequences do not appear to be related to baculovirus polyhedrin (29) (71) (72).
Hytrosaviruses
Hytrosaviruses are named for the salivary gland hypertrophy viruses (hypertrophy of salivary gland) and have been characterized from several Diptera including the tsetse fly, the vector for sleeping sickness. They are non-occluded and contain large double-stranded DNA genomes and have a virion morphology similar in both size and appearance to baculoviruses. They appear to infect the salivary gland and although not particularly virulent, they can result in a significant reduction in reproductive fitness (73). Genomes of several hytrosaviruses have been sequenced and found to encode homologs of four baculovirus/nudivirus genes involved in per os infectivity including pifs 1-3 and p74 (15). In addition, they also appear to encode orthologs of baculovirus RNA polymerase subunits including LEF-4, -8, -9, but not p47, along with LEF-5 and VLF-1 (74). Phylogenetic analysis indicates that they are a distinct lineage related to baculovirus and nudiviruses (Figure 7).
A Whispovirus
The white spot syndrome virus (WSSV) belongs to the genus Whispovirus (white spot virus) and causes severe disease outbreaks in cultured penaeid shrimp, particularly in Asia. It is the only member of the Nimaviridae although there may be several more (81). Nimaviridae is derived from Greek nima=thread referring to its flagellar-like structure at the end of the virion. It is a non-occluded, enveloped, rod-shaped virus with a double-stranded DNA genome of up to 300 kb. It is a highly virulent virus and causes major tissue damage; the infection results in white spots of calcium deposited in the shell (82). Although the genome of WSSV encodes homologs of several baculovirus/nudivirus genes involved in per os infectivity including pifs 1-3 and p74 (15) (81), it is unclear if this represents a direct phylogenetic relatedness to the baculovirus/nudivirus lineage, or whether this is an example of a set of genes important in oral infectivity that was transferred between viruses. The lack of RNA polymerase subunits suggests that their molecular biology may be significantly different from the baculovirus/nudivirus lineage.
Related References
- 1.
- Aoki D, Owashi E, Yazaki S. 2001. Ukiyoe VR-Museum. http://www
.biblio.tuat .ac.jp/vr-museum/ukiyoeEng.htm. - 2.
- Steinhaus EA. 1975. Disease in a minor chord. Ohio State University Press, Columbus, Ohio.
- 3.
- Bergold GH. Die isolierung des polyeder-virus and die natur der polyeder. Z Naturforsch. 1947;1947:122–143.
- 4.
- Xeros N. Cytoplasmic polyhedral virus diseases. Nature. 1952;170:1073. [PubMed: 13013322]
- 5.
- Paillot A. Sur une nouvelle maladie du noyau au grasserie des chenilles de P. brassicae et un nouveau groupe de microoganismes parasites. Compt Rend. 1926;182:180–2.
- 6.
- Vago C, Aizawa K, Ignoffo C, Martignoni ME, Tarasevitch L, Tinsley TW. Editorial: Present status of the nomenclature and classification of invertebrate viruses. J Invertebr Pathol. 1974;23:133–4. [PubMed: 4825251]
- 7.
- Arif BM. Society of Invertebrate Pathology Founders' Lecture, 2010. Mauro Emilio Martignoni: a renaissance scientist in the 20th century. J Invertebr Pathol. 2010;107:173–8. [PubMed: 21762697]
- 8.
- Podgwaite JD. 1981. Natural disease within dense gypsy moth populations. In Doane CC, McManus ML (ed), The Gypsy Moth: Research Towards Integrated Pest Management. U.S. Dept. of Agriculture, Washington, D.C.
- 9.
- Thompson CG. 1978. Nuclear polyhedrosis epizootiology. In Brookes MH, Stark RW, Campbell RW (ed), The Douglas-fir tussock moth: a synthesis. U.S. Department of Agriculture, Washington, D.C.
- 10.
- Heimpel AM, Thomas ED, Adams JR, Smith LJ. The presence of nuclear polyhedrosis virus of Trichoplusia ni on cabbage from the market shelf. Environ Entomol. 1973;2:72–75.
- 11.
- Akermann H-W, Smirnoff WA. A morphological investigation of 23 baculoviruses. J Invertebr Pathol. 1983;41:269–280.
- 12.
- Federici BA. 1986. Ultrastructure of baculoviruses, p 61-88. In Granados RR, Federici BA (ed), The biology of baculoviruses, vol I. CRC Press, Boca Raton.
- 13.
- Entwistle PF, Adams PH, Evans HF. Epizootiology of a nuclear polyhedrosis virus in European spruce sawfly (Gilpinia hercyniae): the rate of passage of infective virus through the gut of birds during cage tests. J Invertebr Pathol. 1978;31:307–12. [PubMed: 355558]
- 14.
- Hostetter DL, Bell MR. 1985. Natural dispersal of baculoviruses in the envirionment, p 249-284. In Maramorosch K, Sherman KE (ed), Virol Insecticides for biological control. Academic Press.
- 15.
- Wang Y, Jehle JA. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: New insights on an old topic. J Invertebr Pathol. 2009;101:187–93. [PubMed: 19460388]
- 16.
- Hall DW, Hazard EI. A nuclear polyhedrosis virus of a caddisfly, Neophylax sp. J Invertebr Pathol. 1973;21:323–4.
- 17.
- Couch JA. An enzootic nuclear polyhedrosis virus of pink shrimp: ultrastructure, prevalence, and enhancement. J Invertebrate Pathol. 1974;24:311–331. [PubMed: 4613755]
- 18.
- Couch JA. Free and occluded virus, similar to Baculovirus, in hepatopancreas of pink shrimp. Nature. 1974;247:229–231.
- 19.
- Larsson R. Insect pathological investigations on Swedish Thysaura: A nuclear polyhedrosis virus of the bristletail Dilta hibernica. J Invertebr Pathol. 1984;44:172.
- 20.
- Henry JE, Jutila JW. The isolation of a polyhedrosis virus from a a grasshopper. J Invertebr Pathol. 1966;8:417–418.
- 21.
- Wang Y, Kleespies RG, Ramle MB, Jehle JA. Sequencing of the large dsDNA genome of Oryctes rhinoceros nudivirus using multiple displacement amplification of nanogram amounts of virus DNA. J Virol Methods. 2008;152:106–8. [PubMed: 18598718]
- 22.
- Ryel RB, Cline GB. Isolation and characterization of a nuclear polyhedral inclusion body from the boll weevil, Athonomas grandis. J Alabama Acad Sci. 1970;41:193.
- 23.
- Smith KM, Hills GJ, Rivers CF. Polyhedroses in neuropterous insects. J Insect Pathol. 1959;1:431–437.
- 24.
- Martignoni ME, Iwai PJ. 1986. A catalog of viral diseases of insects, mites, and ticks, Fourth ed. USDA Forest Service PNW-195.
- 25.
- Theze J, Bezier A, Periquet G, Drezen JM, Herniou EA. Paleozoic origin of insect large dsDNA viruses. Proc Natl Acad Sci U S A. 2011;108:15931–5. [PMC free article: PMC3179036] [PubMed: 21911395]
- 26.
- Wu GA, Jun SR, Sims GE, Kim SH. Whole-proteome phylogeny of large dsDNA virus families by an alignment-free method. Proc Natl Acad Sci U S A. 2009;106:12826–31. [PMC free article: PMC2722272] [PubMed: 19553209]
- 27.
- Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, Rohrmann GF, Theilmann DA, Thiem SM, Vlak JM. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch Virol. 2006;151:1257–66. [PubMed: 16648963]
- 28.
- Soeda E, Maruyama T, Arrand J, Griffin B. Host-dependent evolution of three papova viruses. Nature. 1980;285:165–7. [PubMed: 6246442]
- 29.
- Rohrmann G, Pearson M, Bailey T, Becker R, Beaudreau G. N-terminal polyhedrin sequences and occluded Baculovirus evolution. J Mol Evol. 1981;17:329–333. [PubMed: 7026796]
- 30.
- Herniou EA, Olszewski JA, O'Reilly DR, Cory JS. Ancient coevolution of baculoviruses and their insect hosts. J Virol. 2004;78:3244–51. [PMC free article: PMC371050] [PubMed: 15016845]
- 31.
- Bideshi DK, Bigot Y, Federici BA. Molecular characterization and phylogenetic analysis of the Harrisina brillians granulovirus granulin gene. Arch Virol. 2000;145:1933–45. [PubMed: 11043952]
- 32.
- Zanotto P, Kessing B, Maruniak J. Phylogenetic interrelationships among baculoviruses: evolutionary rates and host associations. J Invertebr Pathol. 1993;62:147–64. [PubMed: 8228320]
- 33.
- Pearson MN, Rohrmann GF. Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J Virol. 2002;76:5301–5304. [PMC free article: PMC137044] [PubMed: 11991958]
- 34.
- Miele SA, Garavaglia MJ, Belaich MN, Ghiringhelli PD. Baculovirus: molecular insights on their diversity and conservation. Int J Evol Biol. 2011;2011:379424. [PMC free article: PMC3119482] [PubMed: 21716740]
- 35.
- Herniou EA, Luque T, Chen X, Vlak JM, Winstanley D, Cory JS, O'Reilly DR. Use of whole genome sequence data to infer baculovirus phylogeny. J Virol. 2001;75:8117–26. [PMC free article: PMC115056] [PubMed: 11483757]
- 36.
- Jiang Y, Deng F, Rayner S, Wang H, Hu Z. Evidence of a major role of GP64 in group I alphabaculovirus evolution. Virus Res. 2009;142:85–91. [PubMed: 19428740]
- 37.
- Jehle JA, Lange M, Wang H, Hu Z, Wang Y, Hauschild R. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology. 2006;346:180–193. [PubMed: 16313938]
- 38.
- Hughes KM, Addison RB. Two nuclear polyhedrosis viruses of the Douglas-fir tussock moth. J Invertebr Pathol. 1970;16:196–204.
- 39.
- Falcon LA, Hess RT. Electron microscope observations of multiple occluded virions in the granulosis virus of the codling moth, Cydia pomonella. J Invertebr Pathol. 1985;45:356–359.
- 40.
- Hukuhara T. Genetic variation of polyhedrosis viruses of insects, p 7-11. In (ed),
- 41.
- Yu IL, Bray D, Lin YC, Lung O. Autographa californica multiple nucleopolyhedrovirus ORF 23 null mutant produces occlusion-derived virions with fewer nucleocapsids. J Gen Virol. 2009;90:1499–504. [PubMed: 19264654]
- 42.
- Wu W, Passarelli AL. Autographa californica M nucleopolyhedrovirus Ac92 (ORF92, P33) is required for budded virus production and multiply-enveloped occlusion-derived virus formation. J Virol. in press. [PMC free article: PMC2976406] [PubMed: 20861245]
- 43.
- Feng G, Thumbi DK, de Jong J, Hodgson JJ, Arif BM, Doucet D, Krell PJ. Selection and Characterization of Autographa californica Multiple Nucleopolyhedrovirus (AcMNPV) DNA Polymerase Mutations. J Virol. 2012;86:13576–13588. [PMC free article: PMC3503129] [PubMed: 23035236]
- 44.
- Xu YP, Gu LZ, Lou YH, Cheng RL, Xu HJ, Wang WB, Zhang CX. A baculovirus isolated from the wild silkworm encompasses the host ranges of Bombyx mori NPV and Autographa californica MNPV in cultured cells. J Gen Virol. 2012;93:2480–9. [PubMed: 22855783]
- 45.
- Rohrmann GF. Baculovirus nucleocapsid aggregation (MNPV vs SNPV): an evolutionary strategy, or a product of replication conditions? Virus Genes. 2014;49:351–7. [PubMed: 25224849]
- 46.
- Lauzon HA, Garcia-Maruniak A, Zanotto PM, Clemente JC, Herniou EA, Lucarotti CJ, Arif BM, Maruniak JE. Genomic comparison of Neodiprion sertifer and Neodiprion lecontei nucleopolyhedroviruses and identification of potential hymenopteran baculovirus-specific open reading frames. J Gen Virol. 2006;87:1477–89. [PubMed: 16690912]
- 47.
- Kuzio J, Pearson MN, Harwood SH, Funk CJ, Evans JT, Slavicek J, Rohrmann GF. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. [PubMed: 9887315]
- 48.
- Hayakawa T, Ko R, Okano K, Seong S, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. [PubMed: 10502508]
- 49.
- Lange M, Jehle JA. The genome of the Cryptophlebia leucotreta granulovirus. Virology. 2003;317:220–36. [PubMed: 14698662]
- 50.
- Hayakawa T, Rohrmann GF, Hashimoto Y. Patterns of genome organization and content in lepidopteran baculoviruses. Virology. 2000;278:1–12. [PubMed: 11112474]
- 51.
- Luque T, Finch R, Crook N, O'Reilly DR, Winstanley D. The complete sequence of the Cydia pomonella granulovirus genome. J Gen Virol. 2001;82:2531–47. [PubMed: 11562546]
- 52.
- Willis LG, Siepp R, Stewart TM, Erlandson MA, Theilmann DA. Sequence analysis of the complete genome of Trichoplusia ni single nucleopolyhedrovirus and the identification of a baculoviral photolyase gene. Virology. 2005;338:209–26. [PubMed: 15951000]
- 53.
- Guarino LA, Gonzalez MA, Summers MD. Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J Virol. 1986;60:224–229. [PMC free article: PMC253920] [PubMed: 16789259]
- 54.
- Guarino LA, Summers MD. Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J Virol. 1986;60:215–223. [PMC free article: PMC253919] [PubMed: 16789258]
- 55.
- Leisy DJ, Rohrmann GF. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology. 1993;196:722–730. [PubMed: 8372444]
- 56.
- Pearson MN, Bjornson RM, Pearson GD, Rohrmann GF. The Autographa californica baculovirus genome: Evidence for multiple replication origins. Science. 1992;257:1382–1384. [PubMed: 1529337]
- 57.
- Pearson MN, Bjornson RM, Ahrens C, Rohrmann GF. Identification and characterization of a putative origin of DNA replication in the genome of a baculovirus pathogenic for Orgyia pseudotsugata. Virology. 1993;197:715–725. [PubMed: 8249294]
- 58.
- Pearson MN, Rohrmann GF. Lymantria dispar nuclear polyhedrosis virus homologous regions: characterization of their ability to function as replication origins. J Virol. 1995;69:213–221. [PMC free article: PMC188566] [PubMed: 7983712]
- 59.
- Arif B, Escasa S, Pavlik L. Biology and genomics of viruses within the genus Gammabaculovirus. Viruses. 2011;3:2214–22. [PMC free article: PMC3230848] [PubMed: 22163341]
- 60.
- Parrish S, Resch W, Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci U S A. 2007;104:2139–44. [PMC free article: PMC1793903] [PubMed: 17283339]
- 61.
- Perera O, Green TB, Stevens SM Jr, White S, Becnel JJ. Proteins associated with Culex nigripalpus nucleopolyhedrovirus occluded virions. J Virol. 2007;81:4585–90. [PMC free article: PMC1900190] [PubMed: 17301145]
- 62.
- Garavaglia MJ, Miele SA, Iserte JA, Belaich MN, Ghiringhelli PD. Ac53, ac78, ac101 and ac103 are newly discovered core genes in the family Baculoviridae. J Virol. 2012;86:12069–79. [PMC free article: PMC3486445] [PubMed: 22933288]
- 63.
- Williams T, Bergoin M, van Oers MM. Diversity of large DNA viruses of invertebrates. J Invertebr Pathol. 2017;147:4–22. [PubMed: 27592378]
- 64.
- Cheng X, Wan X, Xue J, Moore RC. Ascovirus and its evolution. Virolog Sinica. 2007;22:137–147.
- 65.
- Huger AM. A virus disease of the Indian rhinoceros beetle, Oryctes rhinoceros(Linnaeus), caused by a new type of insect virus, Rhabdionvirus oryctes gen. n., sp. n. J Invertebr Pathol. 1966;8:38–51. [PubMed: 5905536]
- 66.
- Ramle M, Wahid MB, Norman K, Glare TR, Jackson TA. The incidence and use of Oryctes virus for control of rhinoceros beetle in oil palm plantations in Malaysia. J Invertebr Pathol. 2005;89:85–90. [PubMed: 16039309]
- 67.
- Huger AM. The Oryctes virus: its detection, identification, and implementation in biological control of the coconut palm rhinoceros beetle, Oryctes rhinoceros (Coleoptera: Scarabaeidae). J Invertebr Pathol. 2005;89:78–84. [PubMed: 16039308]
- 68.
- Granados RR, Nguyen T, Cato B. An insect cell line persistently infected with a baculovirus-like particle. Intervirology. 1978;10:309–17. [PubMed: 681147]
- 69.
- Unckless RL. A DNA virus of Drosophila. PLoS One. 2011;6:e26564. [PMC free article: PMC3203887] [PubMed: 22053195]
- 70.
- Wang Y, Kleespies RG, Huger AM, Jehle JA. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J Virol. 2007;81:5395–406. [PMC free article: PMC1900193] [PubMed: 17360757]
- 71.
- Bezier A, Theze J, Gavory F, Gaillard J, Poulain J, Drezen JM, Herniou EA. The genome of the nucleopolyhedrosis-causing virus from Tipula oleracea sheds new light on the Nudiviridae family. J Virol. 2015;89:3008–25. [PMC free article: PMC4337555] [PubMed: 25540386]
- 72.
- Yang YT, Lee DY, Wang Y, Hu JM, Li WH, Leu JH, Chang GD, Ke HM, Kang ST, Lin SS, Kou GH, Lo CF. The genome and occlusion bodies of marine Penaeus monodon nudivirus (PmNV, also known as MBV and PemoNPV) suggest that it should be assigned to a new nudivirus genus that is distinct from the terrestrial nudiviruses. BMC Genomics. 2014;15:628. [PMC free article: PMC4132918] [PubMed: 25063321]
- 73.
- Abd-Alla AM, Vlak JM, Bergoin M, Maruniak JE, Parker A, Burand JP, Jehle JA, Boucias DG. Hytrosaviridae: a proposal for classification and nomenclature of a new insect virus family. Arch Virol. 2009;154:909–18. [PubMed: 19458899]
- 74.
- Jehle JA, Abd-Alla AM, Wang Y. Phylogeny and evolution of Hytrosaviridae. J Invertebr Pathol. 2013;112:S62–7. [PubMed: 22841640]
- 75.
- Drezen JM, Leobold M, Bezier A, Huguet E, Volkoff AN, Herniou EA. Endogenous viruses of parasitic wasps: variations on a common theme. Curr Opin Virol. 2017;25:41–48. [PubMed: 28728099]
- 76.
- Bezier A, Annaheim M, Herbiniere J, Wetterwald C, Gyapay G, Bernard-Samain S, Wincker P, Roditi I, Heller M, Belghazi M, Pfister-Wilhem R, Periquet G, Dupuy C, Huguet E, Volkoff AN, Lanzrein B, Drezen JM. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science. 2009;323:926–30. [PubMed: 19213916]
- 77.
- Bezier A, Herbiniere J, Lanzrein B, Drezen JM. Polydnavirus hidden face: the genes producing virus particles of parasitic wasps. J Invertebr Pathol. 2009;101:194–203. [PubMed: 19460382]
- 78.
- Burke GR, Strand MR. Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor Bracovirus. J Virol. 2012;86:3293–306. [PMC free article: PMC3302316] [PubMed: 22238295]
- 79.
- Burke GR, Thomas SA, Eum JH, Strand MR. Mutualistic polydnaviruses share essential replication gene functions with pathogenic ancestors. PLoS Pathog. 2013;9:e1003348. [PMC free article: PMC3649998] [PubMed: 23671417]
- 80.
- Volkoff AN, Jouan V, Urbach S, Samain S, Bergoin M, Wincker P, Demettre E, Cousserans F, Provost B, Coulibaly F, Legeai F, Beliveau C, Cusson M, Gyapay G, Drezen JM. Analysis of virion structural components reveals vestiges of the ancestral ichnovirus genome. PLoS Pathog. 2010;6:e1000923. [PMC free article: PMC2877734] [PubMed: 20523890]
- 81.
- Kawato S, Shitara A, Wang Y, Nozaki R, Kondo H, Hirono I. 2018. Crustacean Genome Exploration Reveals the Evolutionary Origin of White Spot Syndrome Virus. J Virol doi: 10.1128/JVI.01144-18:2018 Nov 7. pii: JVI.01144-18. [PMC free article: PMC6340018] [PubMed: 30404800] [CrossRef]
- 82.
- Leu JH, Yang F, Zhang X, Xu X, Kou GH, Lo CF. Whispovirus. Curr Top Microbiol Immunol. 2009;328:197–227. [PubMed: 19216439]
- 83.
- Volkman LE, Summers MD. Nuclear polyhedrosis virus detection: relative capabilities of clones developed from Trichoplusia ni ovarian cell line TN-368 to serve as indicator cells in a plaque assay. J Virol. 1975;16:1630–7. [PMC free article: PMC355772] [PubMed: 1104895]
- 84.
- Arnott HJ, Smith KM. An ultrastructural study of the development of a granulosis virus in the cells of the moth Plodia interpunctella (Hbn.). J Ultrastruct Res. 1967;21:251–68. [PubMed: 5587787]
- 85.
- Hyink O, Dellow RA, Olsen MJ, Caradoc-Davies KM, Drake K, Herniou EA, Cory JS, O'Reilly DR, Ward VK. Whole genome analysis of the Epiphyas postvittana nucleopolyhedrovirus. J Gen Virol. 2002;83:957–71. [PubMed: 11907346]
- 86.
- Fan Q, Li S, Wang L, Zhang B, Ye B, Zhao Z, Cui L. The genome sequence of the multinucleocapsid nucleopolyhedrovirus of the Chinese oak silkworm Antheraea pernyi. Virology. 2007;366:304–15. [PubMed: 17540430]
- 87.
- Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. [PubMed: 8030224]
- 88.
- Nakai M, Goto C, Kang W, Shikata M, Luque T, Kunimi Y. Genome sequence and organization of a nucleopolyhedrovirus isolated from the smaller tea tortrix, Adoxophyes honmai. Virology. 2003;316:171–83. [PubMed: 14599801]
- 89.
- Ijkel WFJ, van Strien EA, Jeldens JGM, Broer R, Zuidema D, Goldbach RW, Vlak JM. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J Gen Virol. 1999;80:3289–3304. [PubMed: 10567663]
- 90.
- Jakubowska AK, Peters SA, Ziemnicka J, Vlak JM, van Oers MM. Genome sequence of an enhancin gene-rich nucleopolyhedrovirus (NPV) from Agrotis segetum: collinearity with Spodoptera exigua multiple NPV. J Gen Virol. 2006;87:537–51. [PubMed: 16476975]
- 91.
- Xiao H, Qi Y. Genome sequence of Leucania seperata nucleopolyhedrovirus. Virus Genes. 2007;35:845–56. [PubMed: 17763934]
- 92.
- Wormleaton S, Kuzio J, Winstanley D. The complete sequence of the Adoxophyes orana granulovirus genome. Virology. 2003;311:350–65. [PubMed: 12842624]
- 93.
- Lauzon HA, Jamieson PB, Krell PJ, Arif BM. Gene organization and sequencing of the Choristoneura fumiferana defective nucleopolyhedrovirus genome. J Gen Virol. 2005;86:945–61. [PubMed: 15784888]
- 94.
- Duffy SP, Young AM, Morin B, Lucarotti CJ, Koop BF, Levin DB. Sequence analysis and organization of the Neodiprion abietis nucleopolyhedrovirus genome. J Virol. 2006;80:6952–63. [PMC free article: PMC1489044] [PubMed: 16809301]
- 95.
- Garcia-Maruniak A, Maruniak JE, Zanotto PM, Doumbouya AE, Liu JC, Merritt TM, Lanoie JS. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J Virol. 2004;78:7036–51. [PMC free article: PMC421636] [PubMed: 15194780]
- 96.
- Afonso CL, Tulman ER, Lu Z, Balinsky CA, Moser BA, Becnel JJ, Rock DL, Kutish GF. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J Virol. 2001;75:11157–65. [PMC free article: PMC114695] [PubMed: 11602755]
- Introduction to the baculoviruses, their taxonomy, and evolution - Baculovirus M...Introduction to the baculoviruses, their taxonomy, and evolution - Baculovirus Molecular Biology
- DNA replication and genome processing - Baculovirus Molecular BiologyDNA replication and genome processing - Baculovirus Molecular Biology
- RecName: Full=Piwi-like protein 4; Short=mAgo5RecName: Full=Piwi-like protein 4; Short=mAgo5gi|226701333|sp|Q8CGT6.3|PIWL4_MOUSProtein
- AV253931 RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cDNA...AV253931 RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cDNA clone 4921506H11 3', mRNA sequencegi|16387973|gnl|dbEST|9988853|dbj|A 31.2|Nucleotide
Your browsing activity is empty.
Activity recording is turned off.
See more...