Except where otherwise indicated, this work is licensed under a Creative Commons Attribution 4.0 International License
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Rohrmann GF. Baculovirus Molecular Biology [Internet]. 4th edition. Bethesda (MD): National Center for Biotechnology Information (US); 2019.
Cells encode all of the genes necessary for the replication of their own DNA. Viruses that infect these cells can either exploit this apparatus for their DNA replication, or they can independently encode some or all the genes involved in this process. However, it is likely that most viruses that replicate in nuclei use a combination of host and virally encoded proteins. Determining the viral factors involved in DNA replication provides fundamental information on how viral DNA synthesis is accomplished. In addition, because so much is known about the proteins involved in DNA replication in other systems, the identification of proteins that are encoded by the virus can lead by inference to proteins that are likely contributed by the host cell.
Some questions are fundamental to understanding viral DNA replication. These include: i) the identification of sequences, called origins of replication, that specify where DNA replication begins; ii) the identification of gene products involved in DNA replication; iii) understanding the role of these proteins in DNA replication; iv) understanding how the DNA is replicated; and v) understanding how the DNA is incorporated into nucleocapsids. Rapid advances in the first three of these areas were made for baculoviruses by the application of approaches developed for similar studies in other viruses. However, these methods often employ transient assays that involve the transfection of combinations of cloned viral DNA into insect cells, and although they have yielded significant information, they do not result in the production of mature, functional genomes. Below is a summary of the current information in this area.
Genes required for DNA synthesis
The genes involved in baculovirus DNA synthesis were originally identified by a combination of the characterization of temperature sensitive mutants (15) and by transient replication assays (16, 17). Transient replication assays involve the transfection of regions of the viral genome along with a plasmid containing a replication origin and the use of the DpnI digestion assay described above. Starting with enough of the genome to give positive replication, subsequent experiments involve the elimination of sequences until a minimal set of genes is identified. Since the original experiments were conducted on AcMNPV before the complete genome sequence was published, the final set of plasmids was sequenced which allowed identification of the essential genes. Six genes were identified that are essential for transient DNA replication and several others were found to be stimulatory. The essential genes include dnapol, helicase (p143), lef-1 (late expression factor-1), lef-2, lef-3 and ie1 (Table 1). In addition to dnapol and helicase, these genes have the following functions: LEF-1, primase; LEF-2, primase accessory factor; LEF-3, single stranded DNA binding protein (SSB); and IE-1, unknown but it may bind to origins of replication. Homologs of the first four have been identified in all sequenced baculovirus genomes (18). The lack of homologs of lef-3 and ie1 does not necessarily imply their absence in evolutionary distant genomes. It could reflect the incorporation of so many changes that the homology of the primary sequences can no longer be identified. This set of genes is similar to those identified as being required for herpes simplex 1 transient DNA replication (19) (Table 1). In both these viruses it is likely that host enzymes also participate in this process. The similarity of the complement of herpes virus and baculovirus replication and other genes has led to the suggestion that these virus are members of a lineage distinct from other large DNA viruses of eukaryotes (20).
Initiation of DNA replication in well-characterized systems such as E. coli is an orderly process that begins with an origin binding protein interacting with an origin of replication and unwinding this region, thereby allowing access by a helicase that induces more extensive unwinding allowing access by the DNA polymerase. Many of the factors involved in baculovirus DNA replication are similar to those found in other organisms and are described below starting with IE1 that has properties somewhat similar to an origin binding protein (also see Figure 1).
IE1 (Ac147). IE1 has been identified in all NPV genomes sequenced from the Lepidoptera and also appears to be present in GV genomes, but the homology is very low. Homologs have not been identified in the hymenopteran and dipteran virus genomes that have been sequenced (18). As described in Chapter 4, IE1 is a transcriptional activator that binds to hr sequences. The identification of hrs as putative replication origins and of IE1 as an hr binding protein would suggest that IE1 is an origin binding protein and might serve as a marker for enucleating the assembly of a replication complex. This would allow viral DNA to be distinguished from host DNA and would lead to the specific replication of viral sequences. However, the identification of non- hr sequences that also act as replication origins and to which IE1 does not appear to specifically bind, suggests that IE1 might not be an origin binding protein in all cases. It was observed that the N-terminal 23 residues of AcMNPV IE1 are involved in specifically binding DNA replication origins, but was not required for transcriptional activation. Furthermore, a conserved cyclin-dependent kinase phosphorylation motif was observed within this 23-amino sequence that was required for IE1 phosphorylation. Mutagenesis of this motif inactivated the ability of IE1 to mediate DNA replication (21). Because IE1 acts as a transcriptional transactivator of baculovirus early genes, and since all the viral genes involved in DNA replication are early genes, IE1 could act as a transactivator of all these genes. Without transactivation by IE1, the replication genes might not be synthesized to a high enough concentration for transient DNA replication to be detected.
As described above, although IE1 may be an origin binding protein, it should be noted that a host protein may be involved as it was observed that the B. mori origin recognition complex subunit 2 had the highest phosphorylation ratio (5.69) of all the cellular proteins during BmNPV infection (22) suggesting that it may be involved in the infection.
LEF-3 (Ac67), a single-stranded DNA binding protein (SSB). A feature of DNA replication systems is a dependence on the presence of SSBs. SSBs bind tightly to ssDNA and prevent the formation of secondary structure. They also prevent the renaturation of ssDNA and may inhibit non-specific interactions of ssDNA with other proteins. They bind in a cooperative manner and can interact with replication proteins.
AcMNPV LEF-3 was identified as being required for DNA replication and was subsequently characterized as an SSB (23). It is a 385 aa protein with a molecular weight of about 44.5 kDa. LEF-3 homologs are present in all lepidopteran baculovirus genomes, but have not been reported in baculoviruses from Hymenoptera or Diptera (18). A LEF-3 homolog may very likely be present in these viruses, but its primary amino acid sequence may have evolved to such an extent that it is not readily identified. For example, the putative XecnGV LEF-3 and AcMNPV LEF-3 show only 14% aa sequence identity.
In addition to being an SSB, LEF-3 has a number of other roles. LEF-3 interacts with alkaline nuclease and may regulate the function of this enzyme (24, 25); it is capable of both unwinding and annealing DNA depending on its concentration or redox state (26, 27); and it can facilitate the production of recombination intermediates via strand exchange between donor and recipient molecules in vitro (28). LEF-3 binds to other LEF-3 molecules which may facilitate its function. LEF-3 also interacts with helicase and is required for its transport into nuclei (29). The nuclear localization signal was localized to about 5 amino acids and was also found to facilitate nuclear transport in mammalian cells (30). LEF-3 is also required for DNA replication independent of this role (31). This feature was localized to the N-terminal 125 amino acids (32).
Helicase (p143) (Ac95). DNA helicases are critical enzymes involved in DNA replication. They are motor proteins that move directionally and unwind DNA by disrupting the hydrogen bonds between bases in the double helix. This exposes the bases so that complementary DNA strands can be synthesized. Energy provided by the cleavage of ATP by an ATPase domain in helicase is required for strand separation.
A helicase homologue was identified in an AcMNPV ts mutant that was defective in DNA replication (15). Helicase was subsequently found to be required for DNA replication in transient assays and was shown to have the biochemical properties of a DNA helicase with both ATPase and helicase activities, and the capacity to bind ss and dsDNA (33). As described above, helicase interacts with LEF-3 and this could facilitate the interaction of helicase with ssDNA.
A DNA primase, LEF-1 (Ac14) and a primase-associated factor, LEF-2 (Ac6). Once a segment of DNA is unwound and stabilized by its interaction with SSBs, DNA replication can be initiated. However, DNA polymerases are unable to independently initiate the synthesis of a new strand of DNA; they can only elongate from an -OH group on an existing strand. Therefore, a short sequence of oligonucleotides that serves as a primer must be synthesized first. Primers are generated by a category of RNA polymerases called primases. This process results in hybrid molecules containing small regions of RNA linked to DNA. Eventually, the RNA primer is removed and replaced by DNA.
In AcMNPV, LEF-1 and LEF-2 were originally identified as factors required for DNA replication in the transient assay system. Subsequently it was found that LEF-1 interacts with LEF-2 (34). It was originally suspected that LEF-1 was a DNA primase because it contained a conserved primase domain (WVVDAD). When this domain was altered to WVVQAD, transient DNA replication activity was eliminated (34). Purified LEF-1 was found to have primase activity, and products of several hundred nucleotides or more were synthesized when M13 ssDNA was employed as a template. Elution profiles of LEF-1 and LEF-2 from ssDNA cellulose and DEAE resin suggested that LEF-2 may bind to both DNA and LEF-1. Although it is essential for transient DNA replication, the exact role of LEF-2 has not been clearly defined (35).
Based on limited amounts of DNA synthesis by a lef-2 bacmid knockout in transfected cells, it was suggested that Lef-2 is not required for the initiation of DNA replication. This was in contrast to a bacmid with the helicase gene deleted that showed no synthesis (36). However, the data in this report could be interpreted in a different manner. The transfected DNA is likely nicked, and therefore, the ends of the nicks could act as primers, resulting in limited amounts of leading strand DNA synthesis in the absence of an active primase complex. A control with the helicase deleted showed no DNA synthesis, indicating that it is required for all DNA synthesis.
Phylogenetic analysis suggests that LEF-1 and LEF-2 are members of a primase lineage common to archaea and eukarya that is distinct from the primase lineage of a number of other large eukaryotic DNA viruses (37).
Baculovirus primers. In contrast to leading strand synthesis, lagging strand synthesis of double-stranded DNA is not continuous and results in the synthesis of non-contiguous oligomers called Okazaki fragments that fill in the single-stranded region generated as the leading strand is synthesized (Figure 1). These relatively short oligomers are transient because they become ligated to the lagging strand. When short-lived DNA intermediates were characterized in S. frugiperda cells, fragments of 80–200 bp were observed. These are similar in size to the Okazaki fragments observed in other eukaryotic cells. However, when these intermediates were characterized in AcMNPV infected cells, they were much larger (0.2–5.0 kb) with an average of 1–2 kb. This is similar in size to what has been observed in prokaryotes, bacteriophage, and herpes virus and may reflect a lower frequency of priming than the host cell. This would result in longer fragments being synthesized to span the distance between the primed regions (38).
DNA polymerase (Ac65). As described above, DNA polymerases extend DNA from the 3' -OH of an RNA primer in a 5' to 3' direction. This results in the synthesis of a leading strand that can continue indefinitely. However, synthesis of the second or lagging DNA strand must occur in the opposite direction of the leading strand in order to progress in a 5' to 3' direction. As the leading strand is synthesized, additional DNA is exposed on the complementary strand. Consequently, whereas leading strand synthesis can be continuous, lagging strand synthesis occurs in segments called Okazaki fragments.
DNA polymerases often have associated exonuclease activities. A 3' to 5' exonuclease activity allows the removal of newly synthesized DNA in the reverse direction of synthesis. This activity is associated with proofreading and allows the polymerase to correct mistakes as it is synthesizing DNA. This may cause DNA polymerase to repeatedly shuttle between polymerizing and editing modes (39). The other activity involves a 5' to 3' exonuclease and is used for the removal of RNA primers. This would be particularly important in lagging strand synthesis because the extended strands would invariably collide with the RNA primer of the upstream Okazaki fragment as they synthesize DNA in the direction opposite of the replication fork. Some DNA polymerases such as DNA pol I of E. coli have both 3’ to 5’ and 5’ to 3’ exonuclease activities so they can proofread and repair newly synthesized DNA and also remove RNA primers. However, many DNA polymerases lack the 3' to 5' activity.
All DNA viruses that infect animal cells appear to encode their own DNA polymerases, except for some with small genomes (polyoma, papilloma and parvoviruses). It has been suggested that this benefits the virus because there may be insufficient levels of the cellular DNA polymerase in infected cells since they are not dividing, and therefore, are not in S phase (40). In fact it has been shown that the mRNA and protein levels of several DNA polymerases increase by three-fold or more during S phase (41). However, the benefits of viruses encoding their own polymerases could also be due to the isolation of viral DNA replication to specific foci within nuclei, or simply that it is more efficient to have the DNA polymerase under regulatory control of the virus so that its expression can be coordinated with the infection. A baculovirus specific DNA polymerase was originally characterized from Bombyx mori infected with BmNPV (42). Subsequently, a gene with homology to DNA polymerase was identified in the AcMNPV genome using hybridization of primers to conserved DNA polymerase sequences (43). This gene is most closely related to members of the DNA polymerase B family. Enzymes in this family synthesize both leading and lagging strands of DNA and have a high degree of fidelity that is associated with a strong 3' to 5' exonuclease activity. The enzymes from BmNPV and AcMNPV were shown to have a 3’ to 5’ exonuclease activity (42, 44, 45), and therefore likely have the capacity to proofread newly synthesized DNA and remove DNA sequences if defects such as mismatching are detected. In contrast, a 5' to 3' exonuclease activity was not observed (45). Since this activity is associated with removal of the primer used for the initiation of DNA synthesis, it is not clear how primers are removed during baculovirus DNA replication (see below).
An unexpected observation described in one report employing transient assays for the identification of baculovirus replication genes suggested that DNA polymerase was not essential, since significant levels of DNA replication (12%) were observed in the absence of dnapol (17). This led to the suggestion that it might be stimulatory rather than essential and that the host DNA polymerase may be involved in initiating baculoviral DNA replication. Furthermore, it was observed that in transient replication assays the AcMNPV DNA polymerase could be substituted with the ortholog from OpMNPV (46) or even from an ascovirus (47). This further suggested that the DNA polymerase may be interchangeable in the context of the other AcMNPV replication proteins. However, when the DNA pol gene was deleted from a bacmid containing the AcMNPV genome, no viral DNA replication was observed (48). This suggested that the DpnI resistant DNA generated in the absence of DNA polymerase in the transient assays could be due to repair of the origin containing plasmid caused by the other baculovirus genes and a host cell DNA polymerase.
Additional genes that influence DNA replication
A variety of other genes have been identified that influence DNA replication, but their role in this process has not been characterized. These include the following:
DBP (Ac25), A second SSB. AcMNPV encodes two proteins that possess properties typical of SSBs, LEF-3, and a protein referred to as DNA-binding protein (DBP). Homologs of dbp are found in all sequenced baculovirus genomes except that of the dipteran (CuniNPV), and in some instances, multiple copies of the dbp gene are present. It has properties similar to LEF-3 in that it interacts with itself and is capable of both unwinding and annealing DNA (49). DBP was also able to compete with LEF-3 for binding sites on ssDNA templates and protected ssDNA against hydrolysis by a baculovirus alkaline nuclease (AN)/LEF-3 complex. It is an essential gene, as bacmids lacking Ac25 were non-infectious and appeared to produce defective nucleocapsids; however, unlike LEF-3 it is not required for transient DNA replication. Although not a virion structural protein, fractionation studies indicated that DBP is tightly associated with subnuclear structures, suggesting that it is a component of the virogenic stroma (49). It was found to localize to the virogenic stroma by immuno-electron microscopy and when dbp was deleted from an AcMNPV bacmid, cells transfected with this construct appeared to lack a virogenic stroma and failed to produce normal-appearing nucleocapsids. This suggested that dbp is required for the production of nucleocapsids and the virogenic stroma (50). In addition, although viral DNA synthesis occurred in cells transfected with the dbp knockout, the levels were less than that of the control virus, indicating that DBP may be required for normal levels of DNA synthesis, or for stability of nascent viral DNA. Analysis of the viral DNA replicated by the dbp knockout by pulsed-field gel electrophoresis resulted in DNA of apparent high molecular weight that is retained in the wells of the gel and fragments that are shorter than the full-size viral genome (50). The DNA fragments might represent degradation products of viral genomes or replicative intermediates. Two different activities of DBP might be responsible for these observations. First, DBP may prevent the enzymatic degradation of viral genomes. It has been shown that DBP inhibits hydrolysis of DNA by the proofreading activity of phage T4 DNA polymerase (51) or by the nuclease activity of the AN/L3 complex (49). Therefore, DBP may protect mature viral genomes against nucleases and stabilize them at stages that precede packaging into virions. On the other hand, the unwinding and renaturation activity of DBP may be required for processing of replicative intermediates by annealing and strand invasion reactions involved in DNA recombination that may be essential for the complete replication and processing of baculovirus genomes (9, 52, 53).
LEF-11 (Ac37). Homologs of LEF-11 are present in all baculovirus genomes except the dipteran CuniNPV. No homology to known proteins associated with DNA replication was identified in the LEF-11 sequence. It was found to stimulate late gene transcription, but was not required for transient DNA replication (17). However, an AcMNPV bacmid deleted for lef-11 was unable to synthesize DNA or carry out late gene transcription when transfected into Sf-9 cells (54). In BmNPV, oligomerization of LEF-11 is required for DNA replication (55). Although LEF-11 localizes to nuclei of infected cells, its role in DNA replication is not known (54). BmNPV LEF-11 interacts with and upregulates both a host ATPase and HSPD1 (HSP60) proteins and this appears to facilitate viral DNA replication (56). BmNPV LEF-11 has a novel nuclear localization signal (57, 58)and appears to interact with host importin α-3 and within nuclei it co-localizes with IE-1 and interacts with LEF-3(57).
Ac139 (ME53). Homologs of Ac139 are present in the genomes of all lepidopteran baculoviruses. AcMNPV bacmids deleted for this gene are not viable, fail to replicate their DNA, and do not produce nucleocapsids. However, cells transfected with DNA from the deletion mutant showed early stages of cytopathogenic effects including nuclear enlargement and the formation of granular material in the nucleus (59). No homology to known proteins associated with DNA replication was identified in the Ac139 sequence.
Lef-7 (Ac125). Lef-7 is stimulatory for transient DNA replication (17, 60). When deleted, infection was unaffected in Tn368 cells, but in Sf21 and S. exigua cells DNA replication was 10% of wt (61). When deleted from BmNPV, a reduction in DNA synthesis was also observed (62). Homologs are present in most Group I, a few group II NPVs, and a few GV genomes. LEF-7 was found to be involved in the regulation of the DNA damage response (DDR). It is an F-box protein that interacts with host S-phase kinase-associated protein 1 (SKP1). SKP1 is a component of a complex that interacts with and targets proteins for polyubiquitination. Deletion of lef-7 from the AcMNPV genome resulted in the accumulation of phosphorylated H2AX and activation of the DDR that led to a major reduction in late gene expression and reduced infectious virus production by 100-fold. It was suggested that LEF-7 may interfere with the phosphorylation of H2AX thereby diverting host DDR proteins from cellular chromatin, so that they can be exploited for viral DNA replication (63).
Proliferating cell nuclear antigen (PCNA) (Ac49). Ac 49 has homology to proliferating cell nuclear antigen (PCNA). PCNA homologs have been found in the genomes of a few Group I and Group II lepidopteran NPVs and appear to have been derived from two different lineages (see Chapter 12). Although eukaryotic PCNA lacks an enzymatic function, it plays a role in DNA synthesis, DNA repair, and cell cycle progression. It functions as a sliding circular clamp that mediates protein interactions with DNA and is required for the coordinated synthesis of both leading and lagging strands at the replication fork during DNA replication (64). In AcMNPV it is not an essential gene (65, 66) and did not appear to elevate DNA replication in transient replication assays (16). However, it did appear to be involved in the timing of late gene expression (67). Over expression under the control of the p10 promotor of EGFP fusions of AcMNPV-PCNA and host cell Sf-PCNA has been examined in Sf9 cells. Both stimulated BV production and DNA replication of both host and virus and Ac-PCNA elevated transcription of selected late genes and increased the larval mortality rate (69).
Baculovirus DNA replication genes: What's missing?
Topoisomerase and DNA ligase. Based on the general model of DNA replication, AcMNPV encodes most of the genes involved in this process (Figure 1). However, there are two major factors that have not been identified. These include a topoisomerase and a DNA ligase. As helicases advance and unwind DNA they cause an increase in the number of twists or topoisomers that accumulate in the DNA in advance of the enzyme. This can cause a barrier to the advance of the helicase. This problem is alleviated by topoisomerases that break and rejoin DNA strands thereby allowing the DNA helix to unwind and relieving the barrier to helicase progression. Integrase homologs from other viruses show topoisomerase activity (70). Therefore, it was thought that VLF-1, a homolog of integrase encoded by all baculovirus genomes, may be involved in this process; however, purified VLF-1 showed no topoisomerase activity (71).
Evidence implicating host cell topoisomerase and DNA ligase. Analysis of data of AcMNPV infected T. ni cells from (72), shows that DNA ligase 1 and topoisomerase 1b and -II transcripts increased by a factor of over 1.3 by 6 hr pi, stayed relatively high through 12 hr pi and then declined by about 75% at 18 hr and were down to 3-6% of the 6 hr level at 48 hr pi. This is in contrast to the bulk of the host transcripts that had declined by about 50% by 13 hr pi (72). This suggests that AcMNPV is capable of stimulating host cell topoisomerase and ligase transcription early in infection such that they can be exploited for viral genome replication. It has also been shown in BmNPV infected B. mori cells that the host cell DNA topoisomerase II is phosphorylated with a ratio of 1.43 compared to uninfected cells (22).
Dealing with Okazaki fragments: two DNA ligases and a second helicase. DNA ligase is necessary for the ligation of Okazaki fragments to one another. For viruses lacking a DNA ligase homolog, it is assumed that a host enzyme supplies this function (see above). However, ligase homologs are present in most granulovirus genomes and at least two NPV genomes (LdMNPV and MacoNPV-B). These ligases are most similar to ligase III that is involved in DNA repair. The DNA ligase of LdMNPV has been shown to have enzymatic activity and is able to form a covalent link with (α32P)ATP and to ligate double-stranded synthetic DNA substrates containing a single nick, suggesting that it would be capable of ligating Okazaki fragments. It was not required or stimulatory for DNA replication in transient replication assays, although these assays may not be dependent upon ligase activity (73). A striking feature of the baculovirus ligase homologs is that they are almost always (except MacoNPV-B) accompanied with a helicase homolog that is not found in any of the genomes lacking ligase. This helicase is related to the PIF1 family (73) (note: this is not a per os infectivity factor). Members of this family have a preference for RNA-DNA hybrids and could be involved in the maturation of Okazaki fragments (74). This may involve displacement of the RNA primer producing an RNA flap that would then be cleaved by a flap endonuclease (FEN) (75) or digested by a 5' to 3' exonuclease. Interestingly, the genome of a GV encodes a fusion of helicase 2 and alkaline nuclease (76) suggesting that the helicase may unwind overlapping RNA and DNA hybrids and the alkaline nuclease might remove the RNA although its ability to digest single strand RNA has not been tested. DNA polymerase would then fill in the gap by extending the Okazaki fragment and the ligase could join the fragments. Another enzyme that may be a candidate for a DNA ligase in some viruses is Ac105 (also called HE65). Computer predictions indicate that it contains a domain homologous to eukaryotic DNA ligases. Homologs of Ac105 are present in some Group I and II NPVs and at least two GVs. Therefore, although there are at least two candidate ligases, it is not clear what role they play in baculovirus DNA replication. Ac105 is probably non-essential as insertion/deletion of this gene in BmNPV (Bm89) had no apparent effect on infectivity (77). Vaccinia virus also encodes a DNA ligase most closely related to ligase III. When it is deleted, the virus was still capable of replication and the host cell ligase I was implicated in this process (78).
Rationale for the composition of baculovirus replication genes
As described above, baculoviruses and herpes viruses encode many, but not all, genes that are likely to be involved in DNA replication. To understand the evolution of these systems, it is important to determine the theoretical rationale for the genes that these viruses encode. A main reason for encoding replication genes rather than using host genes, would be to separate the viral genome replication from dependence on the host cell. Since most host genes are likely shut down upon infection (79) or are present in low concentration because the cells are not dividing and therefore are not replicating their DNA, encoding a replication system independent of host cell control may be of compelling importance to the virus. However, this still does not explain the pattern of replication genes that are virally encoded. As shown in Table 1, these two DNA viruses encode a DNA polymerase, an SSB, and a primase. The presence of the SSB may be due to a requirement for its abundant expression, because of all the replication proteins, it may have to be present in the highest concentration (Figure 1). In addition, the presence of virally encoded DNA polymerase/primase may also be due to the need for optimal concentrations higher than the host cell can provide — especially if the host is stalled at the G2/M stage by the viral infection (see Chapter 7). In particular, each nucleotide polymerized would have to be manipulated by the polymerase. Likewise, each nucleotide of the DS DNA would have to be separated by the DNA helicase. In contrast, the lack of the necessity for a viral encoded ligase could be due to it being required only rarely, especially if long Okazaki fragments are produced as may be the case (see above) (38). Under these circumstances, the low levels of ligase present in the host cell may be sufficient. In addition, if the viral DNA is nicked and not covalently closed during initial replication, a topoisomerase activity may not be required to relieve torsional stress during replication of the virus genome. However, it would be necessary for the production of the final covalently closed supercoiled DNA that is packaged into virions. Whether this requires only low levels of a host enzyme remains to be determined.
Location of baculovirus DNA replication; development of the virogenic stroma
As described in Chapter 3, the virogenic stroma is the site of viral genome replication and nucleocapsid assembly. Several investigations have examined the viral gene products involved in the formation of this structure. IE-1 appeared to localize to specific foci before DBP and LEF-3. After DNA replication begins, the foci enlarge and occupy over half the nucleus and DBP, IE-1, and LEF-3 along with newly replicated DNA co-localize to this region. When DNA replication was inhibited with aphidicolin, foci containing all three proteins were present at early times post infection, but were not as uniform as in the absence of the drug. The number of IE-1 foci appears to be restricted to about 15, suggesting that there are a limited number of preexisting sites where DNA replication could be initiated. It was suggested that these might be equivalent to nuclear domain 10 (ND10) sites found in mammalian cells (80). ND10 are sites of concentrations of proteins involved in a variety of cellular processes, and these sites are often associated with virus assembly (81, 82). Similar data on the localization of replication proteins has been described by others, and it was suggested that IE2 may also be associated with these sites (83). Subsequently, it was found that hr sequences were sufficient to cause IE1 to form foci (84) and that the presence of the viral DNA helicase was necessary for LEF-3 to localize to these structures (85). This is consistent with the requirement of LEF-3 for the transport of helicase into nuclei described above (29). In addition, it was suggested that the replication factors DNA helicase, LEF-3, IE1 along with hr sequences are all that is required to produce foci capable of recruiting other replication factors (85). Studies on the relationship of the IE1 foci with virion structural proteins indicated that ODV-E25 (Ac94) and vp91 (Ac83) localize to the periphery of these structures, whereas vp39 was found within the IE1-associated foci (86). It has also been suggested that IE1 interacts with another structural protein BV-ODV-E26 (Ac16) and serves to recruit it to replication sites (87). Using fluorescent-tagged histone H4, the effect of baculovirus infection on this histone was examined. It was found to relocate to the margins of infected nuclei and appeared to be excluded from the viral replication compartment. This marginalization of histone H4 could also be induced by ie1, lef3, p143-helicase and an hr (88).
Additional baculovirus genes: hints of DNA repair
Although, as described above, the 3' to 5' exonuclease activity of the baculovirus DNA polymerase suggests that it has the ability to proofread DNA as it is synthesized, there do not appear to be other DNA repair systems common to all baculoviruses. However, there are enzymes produced by a limited set of baculoviruses that suggest that some viruses may encode additional pathways for repairing their DNA. This suggests that viruses lacking these pathways may be able to co-opt the homologous proteins from the host cell.
Photolyase. A common threat to occluded baculoviruses in the environment is their inactivation by UV light (89). UV light causes crosslinking of adjacent pyrimidine residues. These crosslinked dimers can cause the DNA to bend and this may inhibit the ability of the DNA replication complex to copy beyond the damaged site or cause the incorporation of incorrect nucleotides. This may result in lethal mutations or the inhibition of the interaction of proteins involved in gene regulation. Photolyases are enzymes that bind to the site of such mutations and, after being activated by light, can catalyze the separation of the mutant dimer, thereby correcting the mutation. Homologs of photolyase genes have been found in the genomes of Group II baculovirus that are members of a lineage that infects insects of the subfamily Plusiinae of the family Noctuidae (90) (91) (92). Chrysodeixis chalcites encodes two photolyase genes that are predicted to encode proteins with 45% amino acid sequence identity. When tested, one copy showed photoreactivating activity, whereas the other copy did not (93). Transfection of egfp fusions of photolyase genes into T. ni cells resulted in fluorescence localized to chromosomes and spindles and other structures associated with mitosis. Baculovirus infection of the transfected cells caused fluorescence to localize to the virogenic stroma (94). Evidence suggests that they are associated with mitotic structures (94) and may be involved in circadian clock regulation (95). The incorporation of an algal virus photolyase gene as a means to cause resistance to UV inactivation of AcMNPV has been described. However, although BV survival was increased after exposure to UV light, occluded virion survival was not (96). An active photolyase is also encoded by some poxviruses (97).
dUTPase. Deoxyuridine triphosphate (dUTP) can be mutagenic if incorporated into DNA. The enzyme dUTPase dephosphorylates dUTP to dUMP, which is a substrate for thymidine biosynthesis. Homologs of dUTPase are present in many NPVs and at least one GV genome (98). Baculoviruses may have incorporated this gene to either supplement or substitute for the host gene. The viruses that encode a dUTPase homolog also normally encode both subunits of ribonucleotide reductase (RR). The presence of RR may have selected for the incorporation of dutpase to mitigate the production of the dUTP mutagen by ribonucleotide reductase.
Viral three-prime repair exonuclease (v-trex). A gene with homology to 3' to 5' exonucleases from other systems has been identified in at least two baculovirus genomes. The enzyme from both AgMNPV and CfMNPV demonstrated 3' to 5' exonucleolytic activity (99, 100). These enzymes would appear to carry out proofreading functions similar to those employed by the DNA polymerase. However, it has been suggested that trex proteins may associate with DNA polymerase and increase the fidelity of DNA replication under conditions that cause error-prone nucleotide polymerization (39).
Ac79, a member of the UvrC endonuclease superfamily? It has been suggested that Ac79 is a member of the UvrC superfamily of endonucleases that are involved in DNA repair (101). Homologs of Ac79 are present in all group I and about half group II lepidopteran NPV and also in a few GV genomes. It has homology to orfs found in other insect viruses and a variety of bacteria. It is predicted to be homologous to an endonuclease in a number of these organisms. When Ac79 was deleted from AcMNPV, titers were reduced and plaque sizes were smaller, however differences in DNA replication and protein synthesis and occlusion body production were not observed, but the quantity of infectious virions appeared to be reduced and aberrant capsid-like structures were observed (102). In another study it was found to be associated with AcMNPV ODV (103).
PARP. A homolog of poly (ADP-ribose) polymerase (PARP) is found in one baculovirus genome (AgMNPV – Ag31) (104) (105). PARP is an enzyme found in nuclei that is activated by DNA strand breaks and uses NAD+ as a substrate to synthesize polymers of ADP-ribose on acceptor proteins that are involved in the repair of single-strand breaks in DNA by activating and recruiting DNA repair enzymes. It is also involved in telomere elongation, chromatin structure, and the transcription of a variety of genes involved in immunity, stress resistance, hormone responses, and the possible silencing of retroelements (106). It may also be involved in the regulation of a mitochondrial protein that induces apoptosis (107). PARP is a caspase-3 substrate and its cleavage is used as a measure of apoptosis.
PARG (Ac114). Poly (ADP-ribose) glycohydrolase (PARG) is an enzyme that reverses the products produced by PARP (108). Homologs of Ac114 are found in most Group I NPV genomes. Hhpred analysis (109) indicates that it is a Poly(ADP-ribose) glycohydrolase (PARG) with almost 100% probability. This appears to be a Group I PARG that was not previously identified. The original baculovirus PARG is specific to Group II baculoviruses. The Group I and II PARGs show low levels of relatedness (e.g. AcMNPV and LdMNPV are less than 15% identical) and appear to represent two different lineages of this enzyme. PARG catalyzes the hydrolysis of glycosidic (1’–2’) linkages in poly(ADP-ribose) to produce ADP-ribose (110). Therefore, whereas PARP stimulates a variety of processes (see above), PARG reverses the products of PARP. In AcMNPV it appears to be an ODV (103, 111) and BV associated protein (112) and in HaSNPV, PARG (Ha100) it was ODV associated (113). When deleted in a BmNPV bacmid, the resulting virus appeared similar to wt (77), whereas a deletion mutant of HaSNPV was similar to wt except the lethal time was longer and the LC50 was higher than wt (114).
Nicotinamide riboside kinase 1 (NRK1). Orthologs of NRK1 are found in most group II NPVs and in at least 5 GVs. It plays a role in nicotinamide adenine dinucleotide (NAD+) synthesis. It phosphorylates nicotinamide riboside yielding nicotinate mononucleotide (115). Since PARG reverses the ADP-ribosylation of proteins by PARP and NRK1 is part of the nicotinamide adenine dinucleotide pathway, it is possible that the presence of PARG and NRK1 in many group II baculoviruses is indicative of their ability to manipulate these processes – possibly with NRK1 participating in the salvage of ADP-ribose generated by PARG.
Genes involved in nucleotide biosynthesis
Most baculoviruses do not encode genes involved in nucleotide biosynthesis. However, many Group II and several GVs encode both subunits of ribonucleotide reductase. In addition, these same viruses normally also encode dUTPase (see above).
Ribonucleotide reductase. Ribonucleotide reductase is a heterodimer composed of large and small subunits (RR1 and RR2, respectively). It is involved in the catalysis of ribonucleotides to deoxyribonucleotides as a pathway for providing nucleotides for DNA synthesis. Well-documented RR1 and RR2 genes have been reported in the genomes of at least three GVs, 10 Group II, and a single Group I (OpMNPV) NPVs (98). Two different RR2 genes have been reported for LdMNPV (116). Based on the phylogeny of baculovirus RR1 genes, it is postulated that two different capture events resulted in baculoviruses obtaining this gene (117). For the OpMNPV and LdMNPV RR1 gene lineage, the source was a bacterium, whereas the other lineage (e.g., Spodoptera exigua MNPV (SeMNPV)) appears to have been derived from a eukaryote, most likely an insect. The two RR2 genes from LdMNPV appear to be derived independently, one from each different source rather than via gene duplication. No enzymology has been described for baculovirus RR and it is not known whether they have enzymatic activity, or how they integrate with or substitute for the homologous host enzymes.
Baculovirus infection and the DNA damage response (DDR)
The DNA of individual human cells encounters tens of thousands of lesions each day. If they are not repaired correctly, they can result in mutations incorporated into the cell genome with subsequent threats to the viability of the organism. In additions to errors during replication such as mismatch incorporation or abortive strand breaks introduced by topoisomerase, other factors such as reactive oxygen or nitrogen species, or environmental toxins are major causes of DNA damage. It has been estimated that DNA damage events can total 100,000 per day per cell and that a day in the sun can cause an additional 100,000 lesions per exposed cell per hour. Furthermore, uv induced inflammation can also cause high levels of oxidative damage [reviewed in (118) (119) (120)]. Consequently, a complex system called the DNA damage response (DDR) has evolved to identify and address this damage.
The DNA damage response is a surveillance network maintained in eukaryotic cells that monitors the integrity of their DNA. When DNA damage is detected, the DDR pathway is activated to prevent mutations from being permanently incorporated into DNA due to double or single-strand breaks, or stalled replication forks. A major feature of the DDR is the phosphorylation of a type of histone that acts as signal for the DDR. Histones form protein spools around which DNA is wound, and there are 5 classes, H1, H2A, H2B, H3 and H4. H2AX is a variant of histone H2A and comprises 2-25% of the H2A population. In humans it is 13 amino acids longer than histone H2A.1 and this extension contains a conserved serine that is located 4 amino acid residues from the carboxyl terminus (121). In response to double stranded DNA breaks, this serine is rapidly and massively phosphorylated by members of the phosphatidylinositol-3 kinase-like kinase family (PIKK). The site of phosphorylation may comprise thousands of nucleosomes spanning up to 2 megabases of chromatin surrounding the DNA break and marks them for repair via the DDR. This can lead to the recruitment of an extensive array of repair-related proteins that can immobilize the ends of the break and facilitate their repair [reviewed in (122)].
The outcome of the DDR can be a pause in the cell cycle, repair of the DNA, and the resumption of the cell cycle. However, if the damage it too extensive, it can lead to apoptosis and the death of the cell. The DDR provides a variety of challenges and opportunities for viruses and several have been shown to be dependent upon it for full levels of replication (123). There may be compelling reasons for viral exploitation of this response. Since virus genomes are limited in size and, although larger viral genomes may encode genes involved in DNA replication, they are still dependent on the host cell for a variety of enzymes involved in these processes. As described above, most baculovirus do not encode enzymes required for nucleotide biosynthesis and lack DNA ligases and topoisomerases. Consequently, the activation of the DNA damage response may provide a variety of enzymes that viruses require for their successful replication.
Several studies have investigated the relationships with baculoviruses and the DDR. It was found that the DDR appeared to be activated by factors associated with viral DNA replication, and inhibition of the DDR resulted in up to a 105 fold reduction in BV titers (124) (125). The tumor suppressor gene, p53 is a regulator of apoptosis in many systems. It was found that AcMNPV infection caused the accumulation of P53 and also the phosphorylation of H2AX, the signal for DDR described above. It was observed that despite the accumulation of p53 under conditions of both DNA damage and AcMNPV infection, silencing p53 did not affect the induction of apoptosis suggesting that it might not be absolutely required for activating apoptosis in Sf9 cells (124).
In another study (125), the induction of the DDR in the non-permissive Drosophila cell system was examined. They observed that the DDR was induced by baculovirus DNA replication and H2AX was phosphorylated. They found that activation of the DDR also promoted apoptosis. In contrast, in permissive S. frugiperda cells, they found that the phosphorylation of H2AX was delayed and did not occur until 24 h pi. They suggest that this indicates that the virus might manipulate the activation of the DDR in novel ways to facilitate its replication. Subsequently they found that LEF-7 was involved in the regulation of the DDR. LEF-7 appears to be an F-box protein that interacts with host S-phase kinase-associated protein 1 (SKP1). SKP1 is a component of a complex that interacts with and targets proteins for polyubiquitination. Deletion of lef-7 from the AcMNPV genome resulted in the accumulation of phosphorylated H2AX and activation of the DDR that led to a major reduction in late gene expression and also reduced infectious virus production by 100-fold. They suggested that LEF-7 may interfere with the phosphorylation of H2AX thereby diverting host DDR proteins from cellular chromatin, so that they can be exploited for viral DNA replication (63).
How are baculovirus genomes replicated?
Whereas some genes have been identified that are required for transient DNA synthesis and major advances have been made in understanding the function of most of these genes, it is still not clear how baculovirus genomes are replicated. Evidence suggests that baculovirus replication results in DNA that is larger than unit length genomes. This DNA could be produced by rolling circle replication, DNA recombination, or by a combination of both these processes. The evidence for these two processes is described below.
Rolling circle replication. It was originally suggested that baculovirus genomes replicate by a rolling circle type mechanism because plasmids that were replicated in the transient system showed a ladder of multiples of unit length-sized DNA fragments when they were partially digested by a restriction enzyme with a single site in the plasmid (7). It is not clear how closely the plasmid reflects the viral genome replication because of its much smaller size and minimal complexity. However, evidence has been presented suggesting that AcMNPV might replicate its genome in this manner (8). Furthermore, it was found that plasmid DNA, when co-transfected with AcMNPV DNA, replicated to become high-molecular weight concatemers, some of which were integrated at a variety of locations in the viral genome. This was interpreted to suggest that both rolling circle replication and recombination may be involved in baculovirus DNA replication (5).
Recombination-dependent replication. Baculovirus replication induces a highly recombinogenic state (52, 53, 126, 127), and this contributed to their development as expression vectors because foreign genes could be so readily incorporated into the genomes by homologous recombination. Recombination-dependent replication is a complex yet common mode by which many viruses replicate their DNA. It is unclear why systems have evolved this mode of replication, but evidence for it playing major roles in genome replication is found throughout DNA viruses with large genomes. T4 phage begins DNA replication in an origin-dependent manner and then switches to a recombination-dependent mode (128). Lambda phage also provides an important model for recombination-dependent replication. Lambda DNA replication initially is of the theta type in which DNA is synthesized bi-directionally from a replication origin initially producing a bubble-like structure that progresses to resemble the Greek letter theta. However, later in infection, concatemers are generated by either rolling circle replication or recombination. Lambda encodes a recombination system called the Red system after mutations that were found to be recombination defective. This system includes red α, β and γ that encode an exonuclease, an SSB, and an inhibitor of a host recombination system, respectively. RED α, the exonuclease, digests DNA in a 5' to 3' direction thereby generating 3' overhangs that anneal with complementary strands or invade homologous double strands; RED β, the SSB, facilitates annealing of DNA strands (129). In addition, RED α and β interact forming heterodimers. There is a complex interaction between the host and phage recombination systems. A lambda-type phage, P22, is dependent on recombination and if both the host and phage recombination systems are inactivated, the phage will not replicate. Viability can be restored by the incorporation of the Red system into P22 (130).
It is thought that the replication of herpes virus genomes is recombination-dependent. DNA isolated from herpes simplex virus 1 (HSV-1) infected cells has a nonlinear, apparently branched structure, and much of it will not enter a pulsed-field gel even after digestion with a restriction enzyme that cuts at a single site in the genome. In addition, SV40 DNA normally employs theta replication, but yields complex high MW DNA resembling HSV-1 DNA when replicated in an HSV-1 dependent system (131). HSV-1 encodes a gene (UL12) homologous to the lambda red α exonuclease. The HSV-1 exonuclease also interacts with an SSB (UL29) (132). The two HSV-1 proteins facilitate strand exchange in a manner similar to the lambda Red system (133). When the HSV-1 exonuclease gene is inactivated, the production of infectious virions is severely compromised (134-136).
Similar to herpes virus, baculoviruses produce high molecular weight DNA that fails to enter a pulsed-field gel. When digested with an enzyme that cuts at a single site in the viral genome, much of the DNA is retained in the well. Also similar to herpes viruses, when SV40 is replicated in a baculovirus system, complex, high-molecular weight DNA is produced rather than products of theta replication (137). In addition, all baculoviruses encode a homolog of the lambda red α exonuclease called alkaline nuclease (AN). It forms a stable complex with an SSB, LEF-3, and possesses both a 5'->3' exonuclease and endonuclease activity (24, 25). LEF-3 has been shown to facilitate strand exchange in vitro (28). These activities are consistent with the AN-LEF-3 complex being involved in DNA recombination. An AcMNPV bacmid with the AN gene deleted did not produce infectious virions after transfection into Sf9 cells. Also, although DNA replication levels appeared to be normal, much of the DNA generated appeared to be significantly smaller than observed in the control. These data demonstrated that an is an essential baculovirus gene (138) and suggested that it may be involved in the generation of larger than genome length fragments consistent with its role in a recombination system (9). In one baculovirus, the AN gene was found fused to a helicase gene suggesting that the two genes may function together during baculovirus replication (76).
In summary, replication of baculovirus DNA has properties similar to other systems. These include the generation of what appear to be structurally complex DNA molecules that are larger than genome length, and the expression of two proteins, an exonuclease and an SSB that interact with one another and have the properties of a system that is involved in DNA recombination in phage and other eukaryotic viruses. Furthermore, in viruses from phage to herpes viruses, homologs of the exonuclease are present and in all cases they interact with an SSB.
Implications of recombination-dependent replication: Multiple replication origins, a covalently closed circular genome, and multiple nucleocapsids per envelope
If recombination plays a major role in baculovirus DNA replication, it could explain several features unique to baculoviruses. These could include the possible use of multiple replication origins. Such origins could greatly amplify the amount of viral DNA that can be produced in a given time. The constraints of identifying and initiating replication at a specific site would be avoided, and replication could originate simultaneously at a number of sites. Upon recombination, molecules destined to become genomes would be produced that could begin or end anywhere as long as they were greater than genome length. In addition, the production of covalently closed circular DNA could be mediated by a final recombination event in which two homologous areas near the ends of a greater than unit length linear replication intermediate are joined (Figure 2). Whereas insect cells likely have extensive DNA recombination and repair systems, this final event in baculovirus DNA processing may be crucial for the production of an infectious virus. It is also possible that the propensity for some lepidopteran viruses to produce multiple nucleocapsids in a single envelope may facilitate recombination and repair of those that might be damaged (139). Such damage could occur during their production, after they are released, or during infection in the midgut or within cells.
Processing and packaging of genome-size DNA
Although recombination-based replication appears to solve several problems that are confronted by the replication of baculovirus genomes, recombination in combination with secondary initiation of replication might result in complex branched structures that would likely need to be resolved into covalently closed circular genomes of unit length before they could be packaged. The mechanism leading to the resolution of these structures is not clear. In addition to the extensive effects that alkaline nuclease and LEF-3 may have on genome processing, another protein, VLF-1 may also be involved.
Very late factor 1 (VLF-1) (Ac77). VLF-1 has been implicated in genome processing although its exact role is not clear. It is a member of the lambda integrase (Int) family of proteins. Integrases are a large group of site-specific DNA recombinases that catalyze DNA rearrangements and are found in a variety of organisms including viruses where they are involved in the integration and excision of viral genomes and decatenation of newly replicated chromosomes. A feature of these enzymes is that a conserved tyrosine forms a covalent link with DNA during the cleavage process. VLF-1 homologs are found in all sequenced baculovirus genomes suggesting that it plays a critical role in baculovirus biology. VLF-1 likely has two different functions in baculovirus biology. It is involved in the hyperexpression of very late genes (140) (see Chapter 6) and it also may have another function, independent of transcription, involved in genome processing.
An AcMNPV bacmid with vlf-1 deleted appears to have normal levels of DNA replication but fails to produce infectious virus. Aberrant tube-like capsids are produced by this mutant and appear to lack DNA. Furthermore, viral DNA in this mutant appeared to be completely accessible to DNAse (141). In contrast, other mutants with defective capsid production (e.g., a deletion of alkaline nuclease) showed significant levels of DNase protection (~35%) (Vanarsdall and Rohrmann, unpublished), suggesting that VLF-1 is required for DNA protection. When the bacmid with vlf-1 deleted was rescued with a copy of VLF-1 that carries a mutation of the highly conserved tyrosine (Y355F), it restored the production of nucleocapsids with a normal appearance, but they were not infectious. Furthermore, the mutant appeared to be defective in the movement of nucleocapsids out of the virogenic stroma, suggesting that without active VLF-1, a final virion maturation step was blocked, possibly leaving the capsids tethered to the virogenic stroma by incompletely processed, larger than unit length DNA. Finally, VLF-1 appears to localize to an end region of the nucleocapsid. Collectively, these results indicate that VLF-1 is required for normal capsid assembly and serves an essential function during the final stages of the DNA packaging process (141-143).
In addition to evidence suggesting that VLF-1 is required for both proper capsid and genome structure, VLF-1 is capable of binding to several types of DNA structures that may further suggest that it has a possible role in DNA processing. It shows high affinity binding to cruciform DNA that mimics a structure common to recombination intermediates. No binding was evident to single and double stranded structures, and very low binding was observed to Y-shaped forks (71). These results are consistent with the involvement of VLF-1 in the processing of branched DNA molecules at the late stages of viral genome replication. The ability of VLF-1 to bind homologous region (hr) sequences was also examined and it was found to bind to hrs that form hairpin and H-shaped structures. Although VLF-1 is capable of binding to branched DNA structures, enzymatic activity (endonuclease and topoisomerase) was not detected, suggesting that enzymatic activity of VLF-1, if present, may depend on accessory cellular or viral factors.
DNA packaging and nucleocapsid assembly. Early in the infection, DNA replication may result in the rapid amplification of DNA sequences that are used as templates for the expression of late genes that encode a variety of proteins, including those involved in capsid structure. However, once capsids begin to be assembled and the concentration of viral DNA increases, genome production may be coordinated with the packaging of DNA into preformed capsids. The preassembly of capsids has been suggested by electron microscopy (144) and is a common feature of some large ds DNA viruses. However, genes that affect DNA replication and processing also appear to affect nucleocapsid structure, suggesting that DNA is an integral component of nucleocapsid structure and without it, the capsid itself cannot assemble properly. A good example is the VLF-1 knockout described above that produced empty tube-like capsids. Similar defects are observed with a deletion of the DNA binding protein, DBP (50).
Packaging DNA into a preformed capsid is an energetically unfavorable process and requires an ATP-driven packaging motor. Packaging motor complexes include a channel for the insertion of the DNA into the capsid and a set of enzymes that can compress the genome through the channel into the capsid. In tailed bacteriophage, the channel is composed of a ring-shaped portal structure embedded in the capsid. This forms a point for the nucleation of DNA packaging enzymes. Genome packaging motors of dsDNA viruses are comprised of a pair of components that are not part of the virion structure; the larger component binds to the procapsid, while the smaller component binds to DNA (145, 146) (147). The motor protein associated with genome packaging of lambda phage has an endonuclease activity indicating that it may be involved in both packaging and DNA processing (148).
DNA recognition can be involved in packaging in two ways: a specific sequence may be recognized to initiate DNA packaging, and it may contact the motor complex during packaging. The specific sequence would ensure that only viral DNA would be packaged. Upon packaging within the capsid, DNA is concentrated up to 100-fold, often as a spool of concentric rings, so that it becomes highly condensed to near liquid crystalline density (149). Viruses with concatemeric precursor DNA use either a 'headful' mechanism to measure DNA incorporation or utilize specific recognition sequences that bracket a complete genome. The cleavage at these sequences results in the excision of a complete genome sequence. It has been suggested that the 'headful' mechanism may cause a conformational change in the portal region as the head becomes full, thereby triggering a mechanism to terminate packaging.
Energy for packaging can be derived either from the hydrolysis of ATP or by an electrochemical potential (proton gradient) generated across a membrane. Motor proteins contain active sites that bind ATP and catalyze its cleavage to ADP and Pi, releasing energy that causes a conformation change in the protein, thereby driving the motor. Each cleaved ATP molecule can result in a processive, ratchet type movement driving the DNA into the capsid. In T4, the motor has a packaging rate of 700–2000 bp/sec with a force of >60 pN, one of the most powerful motors documented (150). If similar, it would take less than 5 min to package the AcMNPV genome.
A candidate for a motor protein that might be involved in DNA packaging is Ac66. Homologs of ac66 appear to be present in all baculoviruses. It is related to a variety of motor proteins including myosin heavy chain, a centromere protein, and Smc, a chromosome segregation ATPase that is involved in cell division. Hhpred, the structure prediction program, (109) suggested it was related to formins that are involved in actin polymerization and are associated with the fast-growing end of actin filaments. Ac66 was found to be associated with AcMNPV and HearNPV ODV (103, 113). However, an ac66 knockout bacmid, although not viable, appeared to be normal and produced nucleocapsids with an electron dense core, suggesting that they contained DNA (151). It has been noted that during infection, actin that is normally in the cytoplasm is transported to the nucleus (152) and interacts with structural components of the virion (153). In addition, reagents that block actin polymerization may result in the production of aberrant capsids that appear to lack DNA (154). Collectively, these observations indicate that components of the cytoskeleton could be involved in the insertion of DNA into nucleocapsids.
It has been suggested that a protein called vp1054 (ac54) may be related to a cellular protein called PURα that binds to purine-rich sequences and may be involved in DNA packaging. It was found to be capable of binding single strand DNA or RNA sequences that contained runs of GGN. Therefore it was suggested that it might interact with the orf1629/p/78/83 (ac9) sequence which encodes a series of prolines and therefore is rich in GGN sequences (155).
DNA signals for packaging or processing
Two investigations have identified DNA sequences that may be involved in the regulation of genome packaging or processing. In one investigation, it was observed that an AT-rich conserved non-protein-coding element (CNE) located in the ac152 region of AcMNPV appears to be an essential cis-acting sequence for virion production (156). In the other report, a nucleocapsid assembly element (NAE) was located within Ac83, a virion structural protein (157). These sequences were found only in Alphabaculoviruses and appeared to have a conserved position relative to one another.
Coordinating DNA replication and packaging: A process for avoiding DNA recombination? A common feature of investigations that identify proteins associated with ODV is the presence of some proteins involved in DNA replication (103, 113). This association may reflect remnants of macromolecular complexes that are 'frozen' in association with nucleocapsids by the occlusion process. Therefore, the presence of these replication-associated proteins with the nucleocapsid could reflect a highly coordinated set of reactions, including DNA synthesis, processing, and packaging in close proximity to the nucleocapsids. The insertion of DNA into nucleocapsids as it is synthesized could protect the partially packaged DNA from strand invasion or nuclease attack (Figure 3). In addition, it is possible that DNA replication is partitioned such that DNA destined for genome production is highly coordinated with packaging to prevent extensive recombination.
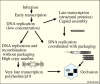
Figure 3.
Hypothetical diagram for roles of two types of baculovirus DNA: genomic DNA that is packaged and DNA that is not packaged and is essential for very late transcription.
Unpackaged DNA is required for very late transcription. Is it the basis for very late gene hyperexpression? The coordination of DNA synthesis with packaging would protect the nascent genomic DNA from random recombination. However, evidence suggests that there is a large component of DNA that is not packaged as genomes. This DNA would be free to recombine and be destined to become templates for gene expression, particularly for very late hyperexpressed genes involved in occlusion body formation. These genes are transcribed after genomic DNA is packaged and inaccessible to transcription (Figure 3) and would be lost after occlusion is completed. Therefore, a major contribution to the ability of baculoviruses to hyperexpress very late genes may be dependent upon the gene copy number required for the massive production of polyhedrin and p10 after genomic DNA is packaged. This is a theory, but in one set of experiments it was observed that late in infection over half the viral DNA in a cell is DNAse sensitive, suggesting that it was not packaged into nucleocapsids (141). Therefore, if this DNA is accessible to the baculovirus RNA polymerase, it could provide a high copy number of very late genes to serve as templates for mRNA synthesis.
A problem with the model of coordination of genome replication with packaging to prevent recombination? A problem with this model of two types of DNA - circular genomic DNA destined for packaging, and the highly branched DNA required for hyperexpression of very late genes model is presented by the ability of the DNA to readily undergo recombination in the laboratory for the production of recombinant virions for gene expression studies. This suggests that DNA destined for viral genome production is not protected from recombination. However, recombinant genomes may be relatively rare under normal replication conditions. In contrast, recombinant production in the laboratory normally involves co-transfection of high concentrations of both viral and target DNA. Under these conditions, recombination might be favored because the DNA is not packaged or in a natural form when it enters nuclei.
More unanswered questions
The lack of genome isomerization. Similar to baculoviruses, herpes simplex virus I (HSV-1) and other herpes viruses are highly recombinogenic (reviewed in (133, 158)). During HSV-1 replication, intra-genomic inversions between two repeated elements within the genome result in a population of four different genome isomers. Since most baculovirus genomes appear to be punctuated with homologous repeated sequences that are distributed throughout their genomes (e.g., AcMNPV has 8 hrs, see above and see Chapter 4, Figure 2) it is surprising that their genomes are replicated with such fidelity. It is clear that inversions do occur as there are examples of inversions bracketed by hrs between different viruses. For example, AcMNPV orfs 1–10 are bordered by hr1 and hr1a and are inverted relative to the homologous sequences in the Orgyia pseudotsugata MNPV genome (159). However, there is no evidence that major populations of isomers are packaged into virions during normal virus replication similar to what occurs in herpes viruses. Restriction enzyme digestion of baculovirus genomic DNA results in a single characteristic pattern for each enzyme, and that pattern conforms to the sequence of the genome. Since one might expect that hr inversions and other forms of recombination between these elements would be common during baculovirus replication, a mechanism must exist to either minimize these events or to eliminate such recombinants from the genome population. As described above, this could involve the partitioning of DNA replication such that DNA destined to become virion genomes is packaged as it is synthesized, whereas other DNA destined for use as templates for transcription is synthesized in a less coordinated manner and is subject to high levels of recombination.
Nucleocapsid length/genome size. Another major unanswered question involves the parameters that determine the length of the capsid and the size of the DNA molecule that is packaged. An examination of capsids associated with defective viral genomes suggested that capsid length may be flexible in response to genome size (160). If capsids are both preassembled and can vary in length, it would suggest that they can be expanded or reduced in response to the size of the genome as part of the packaging process. The facility with which baculoviruses can be engineered to contain additional genetic material could also indicate that a unit size capsid may have some flexibility in the length of DNA that can be accommodated. How the virus senses that a genome is complete and terminates the encapsidation process remains to be determined.
References
- 1.
- Pearson MN, Bjornson RM, Pearson GD, Rohrmann GF. The Autographa californica baculovirus genome: Evidence for multiple replication origins. Science. 1992;257:1382–1384. [PubMed: 1529337]
- 2.
- Kool M, Van Den Berg PM, Tramper J, Goldbach RW, Vlak JM. Location of two putative origins of DNA replication of Autographa californica nuclear polyhedrosis virus. Virology. 1993;192:94–101. [PubMed: 8517035]
- 3.
- Lee HY, Krell PJ. Generation and analysis of defective genomes of Autographa californica nuclear polyhedrosis virus. J Virol. 1992;66:4339–4347. [PMC free article: PMC241240] [PubMed: 1602548]
- 4.
- Pearson MN, Bjornson RM, Ahrens C, Rohrmann GF. Identification and characterization of a putative origin of DNA replication in the genome of a baculovirus pathogenic for Orgyia pseudotsugata. Virology. 1993;197:715–725. [PubMed: 8249294]
- 5.
- Wu Y, Liu G, Carstens EB. Replication, integration, and packaging of plasmid DNA following cotransfection with baculovirus viral DNA. J Virol. 1999;73:5473–80. [PMC free article: PMC112604] [PubMed: 10364295]
- 6.
- Habib S, Hasnain SE. Differential activity of two non-hr origins during replication of the baculovirus autographa californica nuclear polyhedrosis virus genome. J Virol. 2000;74:5182–9. [PMC free article: PMC110871] [PubMed: 10799593]
- 7.
- Leisy DJ, Rohrmann GF. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology. 1993;196:722–730. [PubMed: 8372444]
- 8.
- Oppenheimer DI, Volkman LE. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch Virol. 1997;142:2107–2113. [PubMed: 9413520]
- 9.
- Okano K, Vanarsdall AL, Rohrmann GF. A Baculovirus alkaline nuclease knock out construct produces fragmented DNA and aberrant capsids. Virology. 2007;359:46–54. [PMC free article: PMC1852455] [PubMed: 17046043]
- 10.
- Marriott AC, Dimmock NJ. Defective interfering viruses and their potential as antiviral agents. Rev Med Virol. 20:51–62. [PubMed: 20041441]
- 11.
- Lee HY, Krell PJ. Reiterated DNA fragments in defective genomes of Autographa californica nuclear polyhedrosis virus are competent for AcMNPV-dependent DNA replication. Virology. 1994;202:418–429. [PubMed: 8009853]
- 12.
- Kool M, Goldbach RW, Vlak JM. A putative non-hr origin of DNA replication in the HindIII-K fragment of Autographa californica multiple nucleocapsid nuclear polyhedrosis virus. J Gen Virol. 1994;75(Pt 12):3345–52. [PubMed: 7996129]
- 13.
- Pijlman GP, Dortmans JC, Vermeesch AM, Yang K, Martens DE, Goldbach RW, Vlak JM. Pivotal role of the non-hr origin of DNA replication in the genesis of defective interfering baculoviruses. J Virol. 2002;76:5605–11. [PMC free article: PMC137048] [PubMed: 11991989]
- 14.
- Giri L, Feiss MG, Bonning BC, Murhammer DW. Production of baculovirus defective interfering particles during serial passage is delayed by removing transposon target sites in fp25k. J Gen Virol. 2012;93:389–99. [PMC free article: PMC3352349] [PubMed: 21994323]
- 15.
- Lu A, Carstens EB. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology. 1991;181:336–347. [PubMed: 1994581]
- 16.
- Kool M, Ahrens C, Goldbach RW, Rohrmann GF, Vlak JM. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci USA. 1994;91:11212–11216. [PMC free article: PMC45197] [PubMed: 7972036]
- 17.
- Lu A, Miller LK. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. [PMC free article: PMC188666] [PubMed: 7815565]
- 18.
- Okano K, Vanarsdall AL, Mikhailov VS, Rohrmann GF. Conserved molecular systems of the Baculoviridae. Virology. 2006;344:77–87. [PubMed: 16364739]
- 19.
- Wu CA, Nelson NJ, McGeoch DJ, Challberg MD. Identification of Herpes Simplex Virus Type I genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. [PMC free article: PMC250553] [PubMed: 2826806]
- 20.
- Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–84. [PubMed: 16494962]
- 21.
- Taggart DJ, Mitchell JK, Friesen PD. A conserved N-terminal domain mediates required DNA replication activities and phosphorylation of the transcriptional activator IE1 of Autographa californica multicapsid nucleopolyhedrovirus. J Virol. 2012;86:6575–85. [PMC free article: PMC3393540] [PubMed: 22496221]
- 22.
- Shobahah J, Xue S, Hu D, Zhao C, Wei M, Quan Y, Yu W. Quantitative phosphoproteome on the silkworm (Bombyx mori) cells infected with baculovirus. Virol J. 2017;14:117. [PMC free article: PMC5477107] [PubMed: 28629377]
- 23.
- Hang X, Dong W, Guarino LA. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J Virol. 1995;69:3924–3928. [PMC free article: PMC189119] [PubMed: 7745748]
- 24.
- Mikhailov V, Okano K, Rohrmann G. Baculovirus Alkaline Nuclease Possesses a 5’->3’ Exonuclease Activity and Associates with the DNA-Binding Protein LEF-3. J Virol. 2003;77:2436–2444. [PMC free article: PMC141126] [PubMed: 12551981]
- 25.
- Mikhailov V, Okano K, Rohrmann G. Specificity of the Endonuclease Activity of the Baculovirus Alkaline Nuclease for Single-stranded DNA. J Biol Chem. 2004;279:14734–14745. [PubMed: 14736888]
- 26.
- Mikhailov VS, Okano K, Rohrmann GF. The Redox State of the Baculovirus Single-stranded DNA-binding Protein LEF-3 Regulates Its DNA Binding, Unwinding, and Annealing Activities. J Biol Chem. 2005;280:29444–53. [PubMed: 15944160]
- 27.
- Mikhailov V. Helix-destabilizing properties of the baculovirus single-stranded DNA-binding protein (LEF-3). Virology. 2000;270:180–9. [PubMed: 10772990]
- 28.
- Mikhailov VS, Okano K, Rohrmann GF. Structural and functional analysis of the baculovirus single-stranded DNA-binding protein LEF-3. Virology. 2006;346:469–478. [PubMed: 16375940]
- 29.
- Wu Y, Carstens EB. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology. 1998;247:32–40. [PubMed: 9683569]
- 30.
- Au V, Yu M, Carstens EB. Characterization of a baculovirus nuclear localization signal domain in the late expression factor 3 protein. Virology. 2009;385:209–17. [PubMed: 19073335]
- 31.
- Chen Z, Carstens EB. Identification of domains in Autographa californica multiple nucleopolyhedrovirus late expression factor 3 required for nuclear transport of P143. J Virol. 2005;79:10915–22. [PMC free article: PMC1193572] [PubMed: 16103143]
- 32.
- Yu M, Carstens EB. Identification of a domain of the baculovirus Autographa californica multiple nucleopolyhedrovirus single-strand DNA-binding protein LEF-3 essential for viral DNA replication. J Virol. 2010;84:6153–62. [PMC free article: PMC2876648] [PubMed: 20357098]
- 33.
- McDougal VV, Guarino LA. The autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. J Virol. 2000;74:5273–9. [PMC free article: PMC110882] [PubMed: 10799604]
- 34.
- Evans JT, Leisy DJ, Rohrmann GF. Characterization of the interaction between the baculovirus replication factors, LEF-1 and LEF-2. J Virol. 1997;71:3114–3119. [PMC free article: PMC191443] [PubMed: 9060674]
- 35.
- Mikhailov VS, Rohrmann GF. The Baculovirus Replication Factor LEF-1 is a DNA Primase. J Virol. 2002;76:2287–2297. [PMC free article: PMC153802] [PubMed: 11836407]
- 36.
- Wu CP, Huang YJ, Wang JY, Wu YL, Lo HR, Wang JC, Chao YC. Autographa californica multiple nucleopolyhedrovirus LEF-2 is a capsid protein required for amplification but not initiation of viral DNA replication. J Virol. 2010;84:5015–24. [PMC free article: PMC2863805] [PubMed: 20219928]
- 37.
- Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–96. [PMC free article: PMC1176014] [PubMed: 16027112]
- 38.
- Mikhailov VS, Rohrmann GF. Characterization of short-lived intermediates produced during replication of baculovirus DNA. Virus Res. 2009;145:106–11. [PMC free article: PMC2744863] [PubMed: 19560496]
- 39.
- Shevelev IV, Hubscher U. The 3' 5' exonucleases. Nat Rev Mol Cell Biol. 2002;3:364–76. [PubMed: 11988770]
- 40.
- Coen DW. 1996. Viral DNA polymerases. In DePamphilis ML (ed), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
- 41.
- Zeng XR, Hao H, Jiang Y, Lee MY. Regulation of human DNA polymerase delta during the cell cycle. J Biol Chem. 1994;269:24027–33. [PubMed: 7929054]
- 42.
- Mikhailov V, Marlyev K, Ataeva J, Kullyev P, Atrzhev A. Characterization of 3' -> 5' exonuclease associated with DNA polymerase of silkworm nuclear polyhedrosis ivrus. Nucleic Acids Research. 1986;14:3841–3857. [PMC free article: PMC339819] [PubMed: 3012482]
- 43.
- Tomalski MD, Wu J, Miller LK. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology. 1988;167:591–600. [PubMed: 3059678]
- 44.
- McDougal VV, Guarino LA. Autographa californica nuclear polyhedrosis virus DNA polymerase: measurements of processivity and strand displacement. J Virol. 1999;73:4908–18. [PMC free article: PMC112534] [PubMed: 10233952]
- 45.
- Hang X, Guarino LA. Purification of Autographa californica nucleopolyhedrovirus DNA polymerase from infected insect cells. J Gen Virol. 1999;80:2519–26. [PubMed: 10501509]
- 46.
- Ahrens CH, Rohrmann GF. The DNA polymerase and helicase genes of a baculovirus of Orgyia pseudotsugata. J Gen Virol. 1996;77:825–837. [PubMed: 8609478]
- 47.
- Pellock BJ, Lu A, Meagher RB, Weise MJ, Miller LK. Sequence, function, and phylogenetic analysis of an ascovirus DNA polymerase gene. Virology. 1996;216:146–157. [PubMed: 8614981]
- 48.
- Vanarsdall AL, Okano K, Rohrmann GF. Characterization of the replication of a baculovirus mutant lacking the DNA polymerase gene. Virology. 2005;331:175–180. [PubMed: 15582664]
- 49.
- Mikhailov VS, Vanarsdall AL, Rohrmann GF. Isolation and characterization of the DNA-binding protein (DBP) of the Autographa californica multiple nucleopolyhedrovirus. Virology. 2007;370:415–29. [PMC free article: PMC2186299] [PubMed: 17935748]
- 50.
- Vanarsdall AL, Mikhailov VS, Rohrmann GF. Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene. Virology. 2007;364:475–85. [PMC free article: PMC2697660] [PubMed: 17449080]
- 51.
- Mikhailov VS, Mikhailova AL, Iwanaga M, Gomi S, Maeda S. Bombyx mori nucleopolyhedrovirus encodes a DNA -binding protein capable of destabilizing duplex DNA. J Virol. 1998;72:3107–3116. [PMC free article: PMC109761] [PubMed: 9525636]
- 52.
- Hajós JP, Pijnenburg J, Usmany M, Zuidema D, Závodszky P, Vlak JM. High frequency recombination between homologous baculoviruses in cell culture. Arch Virol. 2000;145:159–164. [PubMed: 10664413]
- 53.
- Kamita SG, Maeda S, Hammock BD. High-frequency homologous recombination between baculoviruses involves DNA replication. J Virol. 2003;77:13053–61. [PMC free article: PMC296086] [PubMed: 14645562]
- 54.
- Lin G, Blissard GW. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J Virol. 2002;76:2770–9. [PMC free article: PMC135986] [PubMed: 11861844]
- 55.
- Dong ZQ, Hu N, Zhang J, Chen TT, Cao MY, Li HQ, Lei XJ, Chen P, Lu C, Pan MH. Oligomerization of Baculovirus LEF-11 Is Involved in Viral DNA Replication. PLoS One. 2015;10:e0144930. [PMC free article: PMC4678028] [PubMed: 26660313]
- 56.
- Dong ZQ, Hu N, Dong FF, Chen TT, Jiang YM, Chen P, Lu C, Pan MH. Baculovirus LEF-11 Hijack Host ATPase ATAD3A to Promote Virus Multiplication in Bombyx mori cells. Sci Rep. 2017;7:46187. [PMC free article: PMC5385504] [PubMed: 28393927]
- 57.
- Zhang J, Dong ZQ, Zhang CD, He Q, Chen XM, Cao MY, Li HQ, Xiao WF, Lu C, Pan MH. Identification of a novel nuclear localization signal of baculovirus late expression factor 11. Virus Res. 2014;184:111–9. [PubMed: 24607592]
- 58.
- Chen T, Dong Z, Hu N, Hu Z, Dong F, Jiang Y, Li J, Chen P, Lu C, Pan M. Baculovirus LEF-11 nuclear localization signal is important for viral DNA replication. Virus Res. 2017;238:133–140. [PubMed: 28648848]
- 59.
- Xi Q, Wang J, R. D, Wang X. Characterization of AcMNPV with a deletion of me53 gene. Virus Genes. 2007:223–32. [PubMed: 17096186]
- 60.
- Morris TD, Todd JW, Fisher B, Miller LK. Identification of lef-7: A baculovirus gene affecting late gene expression. Virology. 1994;200:360–369. [PubMed: 8178427]
- 61.
- Chen CJ, Thiem SM. Differential infectivity of two Autographa californica nucleopolyhedrovirus mutants on three permissive cell lines is the result of lef-7 deletion. Virology. 1997;227:88–95. [PubMed: 9007061]
- 62.
- Gomi S, Zhou CE, Yih W, Majima K, Maeda S. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology. 1997;230:35–47. [PubMed: 9126260]
- 63.
- Mitchell JK, Byers NM, Friesen PD. Baculovirus F-Box Protein LEF-7 Modifies the Host DNA Damage Response To Enhance Virus Multiplication. J Virol. 2013;87:12592–9. [PMC free article: PMC3838121] [PubMed: 24027328]
- 64.
- Johnson A, O'Donnell M. CELLULAR DNA REPLICASES: Components and Dynamics at the Replication Fork. Ann Rev Biochem. 2005;74:283–315. [PubMed: 15952889]
- 65.
- Iwahori S, Ikeda M, Kobayashi M. Generation and characterization of Autographa californica nucleopolyhedrovirus mutants defective in pcna gene homologoue. J Insect Biotechnol Sericology. 2002;71:129–139.
- 66.
- Iwahori S, Ikeda M, Kobayashi M. Association of Sf9 cell proliferating cell nuclear antigen with the DNA replication site of Autographa californica multicapsid nucleopolyhedrovirus. J Gen Virol. 2004;85:2857–62. [PubMed: 15448347]
- 67.
- Crawford AM, Miller LK. Characterization of an early gene accelerating expression of late genes of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1988;62:2773–2781. [PMC free article: PMC253711] [PubMed: 3292791]
- 68.
- Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol. 1999;80:1323–37. [PubMed: 10355780]
- 69.
- Fu Y, Wang R, Liang A. Function analysis of Ac-PCNA and Sf-PCNA during the Autographa californica multiple nucleopolyhedrovirus infection process. Mol Cell Biochem. 2018;443:57–68. [PubMed: 29075988]
- 70.
- Craig NL, Nash HA. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983;35:795–803. [PubMed: 6317202]
- 71.
- Mikhailov V, Rohrmann G. Binding of the baculovirus very late expression factor 1 (VLF-1) to different DNA structures. BMC Mol Biol. 2002;3:14. [PMC free article: PMC130038] [PubMed: 12350233]
- 72.
- Chen YR, Zhong S, Fei Z, Gao S, Zhang S, Li Z, Wang P, Blissard GW. Transcriptome responses of the host Trichoplusia ni to infection by the baculovirus Autographa californica multiple nucleopolyhedrovirus. J Virol. 2014;88:13781–97. [PMC free article: PMC4248991] [PubMed: 25231311]
- 73.
- Pearson MN, Rohrmann GF. Characterization of a baculovirus-encoded ATP-dependent DNA ligase. J Virol. 1998;72:9142–9149. [PMC free article: PMC110332] [PubMed: 9765460]
- 74.
- Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–18. [PMC free article: PMC2034482] [PubMed: 17720711]
- 75.
- Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997:233–40. [PubMed: 9080773]
- 76.
- Ferrelli ML, Salvador R, Biedma ME, Berretta MF, Haase S, Sciocco-Cap A, Ghiringhelli PD, Romanowski V. Genome of Epinotia aporema granulovirus (EpapGV), a polyorganotropic fast killing betabaculovirus with a novel thymidylate kinase gene. BMC Genomics. 2012;13:548. [PMC free article: PMC3496565] [PubMed: 23051685]
- 77.
- Ono C, Kamagata T, Taka H, Sahara K, Asano S, Bando H. Phenotypic grouping of 141 BmNPVs lacking viral gene sequences. Virus Res. 2012;165:197–206. [PubMed: 22421381]
- 78.
- Paran N, De Silva FS, Senkevich TG, Moss B. Cellular DNA ligase I is recruited to cytoplasmic vaccinia virus factories and masks the role of the vaccinia ligase in viral DNA replication. Cell Host Microbe. 2009;6:563–9. [PMC free article: PMC2846536] [PubMed: 20006844]
- 79.
- Nobiron I, O'Reilly DR, Olszewski JA. Autographa californica nucleopolyhedrovirus infection of Spodoptera frugiperda cells: a global analysis of host gene regulation during infection, using a differential display approach. J Gen Virol. 2003;84:3029–39. [PubMed: 14573808]
- 80.
- Okano K, Mikhailov VS, Maeda S. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J Virol. 1999;73:110–119. [PMC free article: PMC103814] [PubMed: 9847313]
- 81.
- Negorev D, Maul GG. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene. 2001;20:7234–42. [PubMed: 11704851]
- 82.
- Wileman T. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu Rev Microbiol. 2007;61:149–67. [PubMed: 17896875]
- 83.
- Mainz D, Quadt I, Knebel-Morsdorf D. Nuclear IE2 structures are related to viral DNA replication sites during baculovirus infection. J Virol. 2002;76:5198–207. [PMC free article: PMC136171] [PubMed: 11967334]
- 84.
- Nagamine T, Kawasaki Y, Iizuka T, Matsumoto S. Focal Distribution of Baculovirus IE1 Triggered by Its Binding to the hr DNA Elements. JVirol. 2005;79:39–46. [PMC free article: PMC538704] [PubMed: 15596799]
- 85.
- Nagamine T, Kawasaki Y, Matsumoto S. Induction of a subnuclear structure by the simultaneous expression of baculovirus proteins, IE1, LEF3, and P143 in the presence of hr. Virology. 2006;352:400–7. [PubMed: 16780915]
- 86.
- Kawasaki Y, Matsumoto S, Nagamine T. Analysis of baculovirus IE1 in living cells: dynamics and spatial relationships to viral structural proteins. J Gen Virol. 2004;85:3575–3583. [PubMed: 15557230]
- 87.
- Kang W, Imai N, Kawasaki Y, Nagamine T, Matsumoto S. IE1 and hr facilitate the localization of Bombyx mori nucleopolyhedrovirus ORF8 to specific nuclear sites. J Gen Virol. 2005;86:3031–8. [PubMed: 16227225]
- 88.
- Nagamine T, Kawasaki Y, Abe A, Matsumoto S. Nuclear marginalization of host cell chromatin associated with expansion of two discrete virus-induced subnuclear compartments during baculovirus infection. J Virol. 2008;82:6409–18. [PMC free article: PMC2447088] [PubMed: 18434402]
- 89.
- Griego VM, Martignoni ME, Claycomb AE. Inactivation of nuclear polyhedrosis virus (Baculovirus subgroup A) by monochromatic UV radiation. Appl Environ Microbiol. 1985;49:709–10. [PMC free article: PMC373576] [PubMed: 3888111]
- 90.
- van Oers MM, Herniou EA, Usmany M, Messelink GJ, Vlak JM. Identification and characterization of a DNA photolyase-containing baculovirus from Chrysodeixis chalcites. Virology. 2004;330:460–470. [PubMed: 15567439]
- 91.
- Willis LG, Siepp R, Stewart TM, Erlandson MA, Theilmann DA. Sequence analysis of the complete genome of Trichoplusia ni single nucleopolyhedrovirus and the identification of a baculoviral photolyase gene. Virology. 2005;338:209–26. [PubMed: 15951000]
- 92.
- Xu F, Vlak JM, van Oers MM. Conservation of DNA photolyase genes in group II nucleopolyhedroviruses infecting plusiine insects. Virus Res. 2008;136:58–64. [PubMed: 18513819]
- 93.
- van Oers MM, Lampen MH, Bajek MI, Vlak JM, Eker AP. Active DNA photolyase encoded by a baculovirus from the insect Chrysodeixis chalcites. DNA Repair (Amst). 2008;7:1309–18. [PubMed: 18547877]
- 94.
- Xu F, Vlak JM, Eker AP, van Oers MM. DNA photolyases of Chrysodeixis chalcites nucleopolyhedrovirus are targeted to the nucleus and interact with chromosomes and mitotic spindle structures. J Gen Virol. 2010;91:907–14. [PubMed: 19955559]
- 95.
- Biernat MA, Eker AP, van Oers MM, Vlak JM, van der Horst GT, Chaves I. A baculovirus photolyase with DNA repair activity and circadian clock regulatory function. J Biol Rhythms. 2012;27:3–11. [PubMed: 22306969]
- 96.
- Petrik DT, Iseli A, Montelone BA, Van Etten JL, Clem RJ. Improving baculovirus resistance to UV inactivation: increased virulence resulting from expression of a DNA repair enzyme. J Invertebr Pathol. 2003;82:50–56. [PubMed: 12581719]
- 97.
- Bennett CJ, Webb M, Willer DO, Evans DH. Genetic and phylogenetic characterization of the type II cyclobutane pyrimidine dimer photolyases encoded by Leporipoxviruses. Virology. 2003;315:10–9. [PubMed: 14592755]
- 98.
- Esteban DJ, Da Silva M, Upton C. New bioinformatics tools for viral genome analyses at Viral Bioinformatics - Canada. Pharmacogenomics. 2005;6:271–80. [PubMed: 16013958]
- 99.
- Slack JM, Shapiro M. Anticarsia gemmatalis multicapsid nucleopolyhedrovirus v-trex gene encodes a functional 3' to 5' exonuclease. J Gen Virol. 2004;85:2863–71. [PubMed: 15448348]
- 100.
- Yang DH, de Jong JG, Makhmoudova A, Arif BM, Krell PJ. Choristoneura fumiferana nucleopolyhedrovirus encodes a functional 3'-5' exonuclease. J Gen Virol. 2004;85:3569–73. [PubMed: 15557229]
- 101.
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–42. [PMC free article: PMC148307] [PubMed: 9973609]
- 102.
- Wu W, Passarelli AL. The Autographa californica M nucleopolyhedrovirus ac79 gene encodes an early gene product with structural similarities to UvrC and intron-encoded endonucleases that is required for efficient budded virus production. J Virol. 2012;86:5614–25. [PMC free article: PMC3347285] [PubMed: 22419804]
- 103.
- Braunagel SC, Russell WK, Rosas-Acosta G, Russell DH, Summers MD. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc Natl Acad Sci U S A. 2003;100:9797–802. [PMC free article: PMC187845] [PubMed: 12904572]
- 104.
- Oliveira JV, Wolff JL, Garcia-Maruniak A, Ribeiro BM, de Castro ME, de Souza ML, Moscardi F, Maruniak JE, Zanotto PM. Genome of the most widely used viral biopesticide: Anticarsia gemmatalis multiple nucleopolyhedrovirus. J Gen Virol. 2006;87:3233–50. [PubMed: 17030857]
- 105.
- de Castro Oliveira JV, de Melo FL, Romano CM, Iamarino A, Rizzi TS, Yeda FP, Hársi CM, Wolff JL, Zanotto PM. Structural and phylogenetic relationship of ORF 31 from the Anticarsia gemmatalis MNPV to poly (ADP-ribose) polymerases (PARP). Virus Genes. 2008;37:177–84. [PubMed: 18648922]
- 106.
- Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 2006;172:363–71. [PMC free article: PMC1456164] [PubMed: 16219773]
- 107.
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–63. [PubMed: 12114629]
- 108.
- Chen X, IJkel W, Tarchini R, Sun X, Sandbrink H, Wang H, Peters S, Zuidema D, Lankhorst R, Vlak J, Hu Z. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J Gen Virol. 2001;82:241–257. [PubMed: 11125177]
- 109.
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430:2237–2243. [PubMed: 29258817]
- 110.
- Miwa M, Tanaka M, Matsushima T, Sugimura T. Purification and properties of glycohydrolase from calf thymus splitting ribose-ribose linkages of poly(adenosine diphosphate ribose). J Biol Chem. 1974;249:3475–82. [PubMed: 4831224]
- 111.
- Liang G, Li G, Chen K, Yao Q, Chen H, Zhou Y. Bombyx mori nucleopolyhedrovirus ORF94, a novel late protein is identified to be a component of ODV structural protein. Curr Microbiol. 2010;61:190–6. [PubMed: 20135318]
- 112.
- Wang R, Deng F, Hou D, Zhao Y, Guo L, Wang H, Hu Z. Proteomics of the Autographa californica Nucleopolyhedrovirus Budded Virions. J Virol. 2010;84:7233–7242. [PMC free article: PMC2898249] [PubMed: 20444894]
- 113.
- Deng F, Wang R, Fang M, Jiang Y, Xu X, Wang H, Chen X, Arif BM, Guo L, Wang H, Hu Z. Proteomics analysis of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus identified two new occlusion-derived virus-associated proteins, HA44 and HA100. J Virol. 2007;81:9377–85. [PMC free article: PMC1951453] [PubMed: 17581982]
- 114.
- Luo S, Zhang Y, Xu X, Westenberg M, Vlak JM, Wang H, Hu Z, Deng F. Helicoverpa armigera nucleopolyhedrovirus occlusion-derived virus-associated protein, HA100, affects oral infectivity in vivo but not virus replication in vitro. J Gen Virol. 2011;92:1324–31. [PubMed: 21325478]
- 115.
- Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. [PubMed: 14704851]
- 116.
- Kuzio J, Pearson MN, Harwood SH, Funk CJ, Evans JT, Slavicek J, Rohrmann GF. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. [PubMed: 9887315]
- 117.
- Hughes AL, Friedman R. Genome-Wide Survey for Genes Horizontally Transferred from Cellular Organisms to Baculoviruses. Mol Biol Evol. 2003;20:979–8. [PubMed: 12716988]
- 118.
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–85. [PubMed: 19812404]
- 119.
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. [PMC free article: PMC2906700] [PubMed: 19847258]
- 120.
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. [PMC free article: PMC2988877] [PubMed: 20965415]
- 121.
- Mannironi C, Bonner WM, Hatch CL. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3' processing signals. Nucleic Acids Res. 1989;17:9113–26. [PMC free article: PMC335118] [PubMed: 2587254]
- 122.
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst). 2004;3:959–67. [PubMed: 15279782]
- 123.
- Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol. 2010;64:61–81. [PubMed: 20690823]
- 124.
- Huang N, Wu W, Yang K, Passarelli AL, Rohrmann GF, Clem RJ. Baculovirus infection induces a DNA damage response that is required for efficient viral replication. J Virol. 2011;85:12547–56. [PMC free article: PMC3209345] [PubMed: 21917957]
- 125.
- Mitchell JK, Friesen PD. Baculoviruses modulate a proapoptotic DNA damage response to promote virus multiplication. J Virol. 2012;86:13542–53. [PMC free article: PMC3503141] [PubMed: 23035220]
- 126.
- Martin DW, Weber PC. DNA replication promotes high-frequency homologous recombination during Autographa californica multiple nuclear polyhedrosis virus infection. Virology. 1997;232:300–309. [PubMed: 9191843]
- 127.
- Crouch EA, Passarelli AL. Genetic requirements for homologous recombination in Autographa californica nucleopolyhedrovirus. J Virol. 2002;76:9323–34. [PMC free article: PMC136457] [PubMed: 12186915]
- 128.
- Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. [PubMed: 9928485]
- 129.
- Li Z, Karakousis G, Chiu SK, Reddy G, Radding CM. The beta protein of phage lambda promotes strand exchange. J Mol Biol. 1998;276:733–44. [PubMed: 9500923]
- 130.
- Poteete A. What makes the bacteriophage lambda Red system useful for genetic engineering: molecular mechanism and biological function. FEMS Microbiol Lett. 2001;201:9–14. [PubMed: 11445160]
- 131.
- Blumel J, Graper S, Matz B. Structure of simian virus 40 DNA replicated by herpes simplex virus type 1. Virology. 2000;276:445–54. [PubMed: 11040135]
- 132.
- Thomas MS, Gao M, Knipe DM, Powell KL. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J Virol. 1992;66:1152–61. [PMC free article: PMC240819] [PubMed: 1309895]
- 133.
- Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life. 2003;55:451–8. [PubMed: 14609200]
- 134.
- Porter I, Stow N. Virus particles produced by the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12 contain abnormal genomes. J Gen Virol. 2004:85. [PubMed: 14993641]
- 135.
- Porter I, Stow N. Replication, recombination and packaging of amplicon DNA in cells infected with the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12. J Gen Virol. 2004;85:3501–10. [PubMed: 15557223]
- 136.
- Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–85. [PMC free article: PMC190043] [PubMed: 8642627]
- 137.
- Martin DW, Weber PC. Replication of simian virus 40 origin-containing DNA during infection with a recombinant Autographa californica multiple nuclear polyhedrosis virus expressing large T antigen. J Virol. 1997;71:501–506. [PMC free article: PMC191078] [PubMed: 8985377]
- 138.
- Okano K, Vanarsdall AL, Rohrmann GF. Characterization of a baculovirus lacking the Alkaline Nuclease Gene. J Virol. 2004;78:10650–10656. [PMC free article: PMC516421] [PubMed: 15367632]
- 139.
- Blissard GW, Rohrmann GF. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology. 1989;170:537–555. [PubMed: 2658304]
- 140.
- McLachlin JR, Miller LK. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J Virol. 1994;68:7746–7756. [PMC free article: PMC237236] [PubMed: 7966564]
- 141.
- Vanarsdall AL, Okano K, Rohrmann GF. Characterization of the role of VLF-1 in baculovirus capsid structure and DNA processing. J Virol. 2006;80:1724–1733. [PMC free article: PMC1367162] [PubMed: 16439529]
- 142.
- Vanarsdall AL, Okano K, Rohrmann GF. Characterization of a baculovirus with a deletion of vlf-1. Virology. 2004;326:191–201. [PubMed: 15262507]
- 143.
- Li Y, Wang J, Deng R, Zhang Q, Yang K, Wang X. vlf-1 deletion brought AcMNPV to defect in nucleocapsid formation. Virus Genes. 2005;31:275–84. [PubMed: 16175333]
- 144.
- Fraser MJ. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J Ultrastruct Mol Struct Res. 1986;95:189–195.
- 145.
- Yang K, Baines JD. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J Virol. 2006;80:5733–9. [PMC free article: PMC1472570] [PubMed: 16731912]
- 146.
- Thoma C, Borst E, Messerle M, Rieger M, Hwang JS, Bogner E. Identification of the interaction domain of the small terminase subunit pUL89 with the large subunit pUL56 of human cytomegalovirus. Biochemistry. 2006;45:8855–63. [PubMed: 16846228]
- 147.
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–73. [PMC free article: PMC2769991] [PubMed: 19794496]
- 148.
- Ortega ME, Gaussier H, Catalano CE. The DNA Maturation Domain of gpA, the DNA Packaging Motor Protein of Bacteriophage Lambda, Contains an ATPase Site Associated with Endonuclease Activity. J Mol Biol. 2007;373:851–65. [PMC free article: PMC2082050] [PubMed: 17870092]
- 149.
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–52. [PubMed: 11607035]
- 150.
- Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–73. [PMC free article: PMC2040459] [PubMed: 17942694]
- 151.
- Ke J, Wang J, Deng R, Wang X. Autographa californica multiple nucleopolyhedrovirus ac66 is required for the efficient egress of nucleocapsids from the nucleus, general synthesis of preoccluded virions and occlusion body formation. Virology. 2008;374:421–31. [PubMed: 18241908]
- 152.
- Volkman LE, Talhouk SN, Oppenheimer DI, Charlton CA. Nuclear F-actin: a functional component of baculovirus-infected lepidopteran cells? J Cell Sci. 1992;103:15–22.
- 153.
- Lanier LM, Volkman LE. Actin binding and nucleation by Autographa california M nucleopolyhedrovirus. Virology. 1998;243:167–77. [PubMed: 9527926]
- 154.
- Volkman LE. Autographa californica MNPV nucleocapsid assembly: inhibition by cytochalasin D. Virology. 1988;163:547–53. [PubMed: 3281373]
- 155.
- Marek M, Romier C, Galibert L, Merten O, van Oers MM. Baculovirus VP1054 is an acquired cellular PURα, a nucleic-acid6n binding protein specific for GGN repeats. J Virol. 2013;87:8465–80. [PMC free article: PMC3719801] [PubMed: 23720732]
- 156.
- Kikhno I. Identification of a Conserved Non-Protein-Coding Genomic Element that Plays an Essential Role in Alphabaculovirus Pathogenesis. PLoS One. 2014;9:e95322. [PMC free article: PMC3989284] [PubMed: 24740153]
- 157.
- Huang Z, Pan M, Zhu S, Zhang H, Wu W, Yuan M, Yang K. The Autographa californica Multiple Nucleopolyhedrovirus ac83 Gene Contains a cis-Acting Element That Is Essential for Nucleocapsid Assembly. J Virol. 2017:91. [PMC free article: PMC5309959] [PubMed: 28031366]
- 158.
- Thiry E, Meurens F, Muylkens B, McVoy M, Gogev S, Thiry J, Vanderplasschen A, Epstein A, Keil G, Schynts F. Recombination in alphaherpesviruses. Rev Med Virol. 2005;15:89–103. [PubMed: 15546129]
- 159.
- Ahrens CH, Russell R, Funk CJ, Evans JT, Harwood SH, Rohrmann GF. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. [PubMed: 9126251]
- 160.
- Kool M, Voncken JW, Van Lier FLJ, Tramper J, Vlak JM. Detection and analysis of Autographa californica nuclear polyhedrosis virus mutants with defective interfering properties. Virology. 1991;183:739–746. [PubMed: 1853572]
- Identification of origins of viral DNA replication
- Genes required for DNA synthesis
- Additional genes that influence DNA replication
- Baculovirus DNA replication genes: What's missing?
- Location of baculovirus DNA replication; development of the virogenic stroma
- Additional baculovirus genes: hints of DNA repair
- Genes involved in nucleotide biosynthesis
- How are baculovirus genomes replicated?
- Implications of recombination-dependent replication: Multiple replication origins, a covalently closed circular genome, and multiple nucleocapsids per envelope
- Processing and packaging of genome-size DNA
- More unanswered questions
- References
- DNA replication and genome processing - Baculovirus Molecular BiologyDNA replication and genome processing - Baculovirus Molecular Biology
- RecName: Full=Piwi-like protein 4; Short=mAgo5RecName: Full=Piwi-like protein 4; Short=mAgo5gi|226701333|sp|Q8CGT6.3|PIWL4_MOUSProtein
- AV253931 RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cDNA...AV253931 RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cDNA clone 4921506H11 3', mRNA sequencegi|16387973|gnl|dbEST|9988853|dbj|A 31.2|Nucleotide
Your browsing activity is empty.
Activity recording is turned off.
See more...