Except where otherwise indicated, this work is licensed under a Creative Commons Attribution 4.0 International License
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Rohrmann GF. Baculovirus Molecular Biology [Internet]. 4th edition. Bethesda (MD): National Center for Biotechnology Information (US); 2019.
The cell cycle and apoptosis are interrelated. If a cell detects perturbations in different stages of its replicative cycle such as those caused by viral infection, the cell can be induced to undergo apoptosis. Because baculovirus replication causes major alterations in the cell cycle, it has evolved several mechanisms to interfere with apoptosis.
Baculovirus infection and the cell cycle
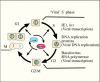
Cell division involves a cycle of defined stages during which DNA precursors and other cellular components are synthesized, chromosomes are duplicated and segregated to opposite poles of a cell during mitosis, followed by division of the cell. These stages in cell growth have been named gap or G phases (G1, G2, and G0), S for the synthetic phase, and M for the mitotic phase (Figure 1). G1 follows mitosis, during which the cell is prepared for DNA replication by the production of synthetic enzymes; S follows G1 and involves the DNA synthesis phase during which chromosomes are duplicated; G2 follows S and involves preparation for cell division, which occurs in M or the mitosis phase. If a cell becomes dormant and is not in a replication cycle it is in stationary or G0 phase. The cell cycle is highly regulated, and the orderly progression through each phase is critical for chromosome replication, separation, and production of daughter cells. The transitions between the different stages of the cycle are governed by the phosphorylation state of a number of proteins called cyclins. They are phosphorylated by cyclin dependent kinases (cdks) (Figure 1). Progress through the cell cycle is also governed by molecular checkpoints that can prevent the continuation of the cell cycle at specific phases. Deregulation of the cell cycle can result in uncontrolled cell proliferation, a hallmark of cancer development. Consequently, regulation of the cell cycle has been intensively investigated.
The cell cycle can be monitored experimentally by measuring the DNA content in each cell in a population by flow cytometry. In this process cells are fixed in ethanol, RNA is removed by treatment with RNase, and the DNA is stained with propidium iodide and quantified for each cell by flow cytometry. Two major populations of cells are normally present, those in G1 phase that contain a set of diploid chromosomes, and those in G2 and M phase (G2/M), in which the chromosomes are doubled and have twice the DNA content of diploid cells. Normally, a diffuse population of cells in S phase is present, and unless the cells have been synchronized, it is not particularly distinct.
Many viruses block the cell cycle (1). This may seem logical, because it would block competition by a dividing cell for the cellular replicative apparatus and components and allow these to be directed towards viral replication. However, blockage of the cell cycle can result in very low levels of enzymes required for transcription and DNA synthesis because they are no longer needed for cell division. For example, in some cells replicative DNA polymerases and other factors associated with DNA replication are amplified threefold or more during S-phase (2). A number of DNA viruses do not encode the substrates required for DNA synthesis. When these viruses infect cells that are in interphase and are not undergoing cell division, there is a reduced availability of compounds necessary for DNA synthesis. Consequently, viruses have devised several methods to induce nonreplicating cells to either enter S phase or to induce a pseudo S phase in which the cellular synthetic machinery is activated, allowing for virus replication independent of cell replication. Some viruses can induce cells to enter S phase by expressing proteins that inactivate a protein called Rb. It is named for the retinoblastoma susceptibility gene that was first implicated in cancer of the retina, a disease that usually occurs before the age of five in about 1 in every 20,000 children. In this tumor, the Rb protein is inactivated and that led to its discovery. Rb governs the G1/S check point by binding to a transcription factor (E2F). If Rb binds to certain viral proteins, it releases the transcription factor (E2F) that can then activate S phase. Normally Rb is regulated by its phosphorylation state—it binds to the S-phase activator when it is not phosphorylated. The inactivation of Rb can lead not only to the entry of the cell into S phase, but to unregulated cell growth and division, which is a characteristic feature of virus-induced tumors. Therefore, the byproduct of the necessity of the virus to induce S phase can be a tumor. This is especially true when the portion of the virus genome encoding the inhibitor of Rb integrates into the cell genome, thereby permanently deregulating the cell cycle. Examples of viruses capable of causing this mutation are tumor viruses, such as human papilloma, SV40, and adenovirus.
Another checkpoint governs the transition from G2 to M. It is thought that regulation of this transition is involved in monitoring for DNA damage. If DNA damage is detected, then progression into mitosis is inhibited and the cell may undergo apoptosis. This prevents the establishment of cells with defective DNA genomes. Many viruses appear to block the cell cycle at G2/M, thereby rendering the cells unable to divide (1). This may be caused by the cell checkpoint monitoring systems that detect viral DNA replication and regards it as damaged cellular DNA and thereby stops the transition to metaphase. Concomitant with blocking the transition to M phase, a number of viruses appear to induce what is called a ‘pseudo’ S phase. Baculoviruses likely fall into this category as several reports have indicated that infection causes cells to be blocked at G2/M. One report found that most non-synchronized cells were blocked at G2/M after AcMNPV infection (3). In another report, synchronized Sf9 cells infected in G2/M were arrested in that stage, whereas those infected at G1 were arrested in S phase (4). In yet another study it was reported that cells infected at G2/M, G1, or S phases, were arrested in the S or G2/M phase (5). Investigations on infection with another virus, HaSNPV, in a different cell line, also resulted in a block at G2/M. Normally cyclin B1 is degraded at the onset of metaphase allowing progression through this phase, however in these cells it accumulates and remained at high levels (6). An AcMNPV protein (Ac144) was identified that may contribute to the blockage at G2/M (7) (for more information about Ac144, see Chapter 12).
The various observations regarding baculovirus infection and the cell cycle could be a reflection of the blockage of cellular DNA replication upon infection and the subsequent replication of viral DNA, which would obscure the phase of the host cell cycle. Whereas blocking the cell cycle may be necessary for diverting the cellular replicative machinery for virus replication, it is not understood how the cellular replicative apparatus is exploited by the virus. Clearly, however, for viruses that are dependent on host enzymatic pathways and synthetic machinery, the activity of these systems would either be preserved or elevated during the infection.
Viral synthetic pathways: Induction of a 'pseudo' or ‘viral’ S phase environment
Several factors likely contribute to the ability of baculoviruses to replicate independently of the cell cycle and produce a 'pseudo' S phase-like environment, which might be more appropriately termed ‘viral’ S phase because of the components that the virus contributes to macromolecular synthesis. These include the ability of cells to selectively transcribe viral genes through the transactivator IE1 in combination with hr enhancer sequences, the synthesis of a set of DNA replication proteins so that virus replication is independent of host proteins for DNA replication, and the production of an RNA polymerase so that they are independent of the host RNA polymerase II. These factors are summarized in Figure 1. Another contributing factor would be the shut down of most host RNA polymerase II transcription (8). In addition, there are likely a few other cellular pathways that are exploited by the virus for DNA synthesis. Hints of some of these are likely reflected in enzymes involved in biosynthetic pathways encoded by some, but not all baculoviruses (see Chapter 5). It is likely that there are advantages for the genes encoding these enzymes to be incorporated into the viral genome because then they would be under the regulatory control of the virus. However, in viruses lacking these genes, they likely remain dependent on the host pathways. An example of one such enzyme is ribonucleotide reductase (RR) that is encoded by a minority of baculoviruses and consists of two genes that encode a heterodimer composed of large and small subunits (RR1 and RR2, respectively). RR is involved in the catalysis of ribonucleotides to deoxyribonucleotides as a pathway for providing nucleotides for DNA synthesis. It is found in most Group II NPV, at lease one Group 1 NPV (OpMNPV) and a few GV genomes that have been sequenced. Although it is not clear why some viruses encode this enzyme and others do not, it does suggest that it may be a required component for optimizing the cellular environment for viral replication and indicates that viruses that lack the enzyme are dependent on the host for this pathway. In all of the NPVs and one of the GVs encoding the RR genes, they are accompanied with a companion enzyme, dUTPase. One of the products of RR can be dUTP, which if incorporated into DNA, can be mutagenic. Consequently, the presence of both RR and dUTPase suggests that the latter enzyme mitigates the mutagenic potential of dUTP, which is inactivated by dUTPase.
It has also been noted that many of the most prevalent promoter sequences present in baculovirus genomes are combinations of both early and late promoters (see Chapter 4) (9) such that the genes can be initially expressed by the host RNA polymerase II, but can also be expressed by the viral polymerase. This promoter arrangement would ensure that sufficient levels of the protein are present independent of host cell at both early and late times post-infection.
Apoptosis and baculovirus infections
Apoptosis or programmed cell death is a pathway that is thought to have evolved to allow multicellular organisms to eliminate cells that are no longer required either to facilitate the development of the organism or because they are damaged or malfunctioning. A classic example of this process is the transformation that occurs during insect pupation that causes the metamorphosis of a worm-like insect larva into an adult insect often capable of flight. During this process, larval structures are dismantled, which frees up their components for reuse in the production of new structures. In addition, in some instances apoptosis is induced by malfunctions in the cell cycle. This can include DNA damage or a variety of features of cells that are undergoing virus infection, such as unscheduled DNA replication or RNA synthesis that is independent of the cell cycle. Therefore, apoptosis is also a major method by which organisms can limit and control viral infections.
Apoptosis is characterized by a specific series of events that are associated with cell death. These include loss of attachment to adjacent cells, cell shrinkage, nuclear fragmentation, chromatin condensation and DNA fragmentation and subsequent disintegration or blebbing of the cell into apoptotic bodies that are eliminated by phagocytosis. For a video of this process see (10). Apoptosis is induced by processes associated with baculovirus DNA replication in Spodoptera frugiperda cells (11) and involves the activation of a series of proteases called caspases. There are two types of caspases involved in apoptosis, initiator and effector caspases. Initiator caspases activate inactive forms of effector caspases (procaspases) by cleaving them. Effector caspases then cleave other cellular proteins resulting in apoptosis. Some effector caspases specifically target cytoskeletal proteins thereby causing the initial morphological changes including detachment and cell shrinkage. The activation of nucleases and the targeting of cell structural elements cause the subsequent fragmentation of the cell. Although it is thought that apoptosis evolved for the removal of cells that are no longer needed by an organism, it was adapted as a cellular defense mechanism for the elimination of cells infected by viruses. This greatly reduces the ability of viruses to establish infections. In AcMNPV infections, yield can be reduced up to 15,000-fold for viruses lacking the anti-apoptotic gene, p35 (12) (13). In some aspects, the monitoring for virus infection is thought to be integrally involved with the monitoring of the cell cycle. When a cell is determined by checkpoint monitoring to have aberrant features such as DNA that is damaged beyond repair, the cell can be directed to undergo apoptosis, and thereby be eliminated. Apoptosis can be induced by DNA damage caused by ionizing radiation and by a variety of toxic chemicals. It is thought that the apparatus that monitors the cell cycle interprets viral DNA replication as aberrant or damaged DNA and induces the cell to undergo apoptosis. In AcMNPV, it was found that by silencing genes required for DNA replication using RNAi, apoptosis could be significantly reduced. In contrast, silencing genes specific to late gene expression did not reduce apoptosis (11) (14). From this it was concluded that either the replication genes themselves, or DNA replication were responsible for the induction of apoptosis.
The baculovirus anti apoptosis gene, p35.
The study of apoptosis is a relatively new field, and baculoviruses have played a major role in understanding this process. Much of the early work on baculoviruses and apoptosis was done in the laboratory of Dr. Lois Miller. For a biography of Dr. Miller, see (15) and for a history of the early discoveries, see (16).
Baculoviruses became widely used as vectors for protein expression in the mid-1980s. Initially, the protocol involved the production of recombinants by homologous recombination at the polyhedrin locus. The polyhedrin gene was the selectable marker, and recombination resulted in occlusion negative virions that could be identified by examination of the plaques that they formed in cultured cells. The first baculovirus apoptotic inhibitor was discovered when a laboratory identified an occlusion negative plaque that was not expressing the intended recombinant protein, but that had aberrant features, including a small plaque size. These investigators brought this to the attention of Lois Miller's laboratory that specialized in baculovirus research. The mutant virus was found to induce apoptosis and consequently had a small plaque size and reduced budded virus production. Investigation of the genome of the mutant virus led to the discovery of a baculovirus gene called p35, much of which was deleted in this aberrant virus. It was found that under normal conditions p35 was capable of blocking the apoptotic pathway in cells infected with the wt virus, but in this mutant it was inactivated (17).
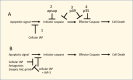
A second fortuitous observation occurred when a laboratory was expressing one of the proteinases involved in apoptosis, caspase 1 (also called ICE) in the baculovirus system and found that it consistently co-purified with a contaminating protein. This contaminant was subsequently shown to be p35 and it was found to be a substrate for an effector caspase, caspase 1, but in the process of its cleavage it irreversibly binds to and inactivates caspase 1. Therefore, when caspase 1 was expressed in the baculovirus system, it bound to p35 that was also being expressed by the baculovirus. This led to understanding how p35 was able to block the apoptotic pathway (18) (Figure 2). Subsequently, p35 was found to block other categories of caspases in a similar manner (19). Although baculoviruses encoding p35 would appear to benefit by its presence, its evolutionary lineage is not clear. Closely related orthologs are only found in a few Group I baculoviruses closely related to AcMNPV. Orthologs of p35 have also been reported in a GV of Choristoneura occidentalis (ChocGV) (20), and in an entomopox virus. The entompox virus p35 ortholog functions similarly to P35 in blocking effector caspases (21). Despite its limited distribution to a few insect viruses, p35 is capable of blocking the apoptotic pathway in diverse organisms from invertebrates to mammals and has been a critical reagent in understanding the molecular interactions involved in the apoptotic pathway. In infected insect cells evidence suggests that the induction of apoptosis is initiated by p53 at the onset of viral DNA replication (22).
P49, a variant of P35. A variant of P35 called P49 was found in a Group II NPV (SpliNPV) (23). In contrast to the inactivation of effector caspases by p35, P49 inhibits both initiator and effector caspases (24) (25), reviewed in (26) (Figure 2). Furthermore, whereas P35 functions as a monomer, P49 acts as a dimer and is capable of binding two caspase molecules and targets a different cleavage motif (27).
The discovery of the IAP family of antiapoptotic proteins
After the initial discovery of P35, to understand its distribution and diversity, its presence in other baculoviruses was investigated. This involved a granulovirus (CpGV) and an NPV, OpMNPV, that had been shown to have a deletion at the position where p35 was predicted to be located (28). Complementation was used to rescue an AcMNPV mutant deleted for p35. Unexpectedly, in both these viruses, a gene other than p35 was found to compensate for the lack of p35 (29, 30). This gene was called inhibitor of apoptosis (iap). Subsequently, in contrast to the limited distribution of p35, iap orthologs were found in genomes of almost all baculoviruses and the related Nudiviridae and Hytrosaviridae. Orthologs are also found in the Asfarviridae, Iridoviridae, Ascoviridae, Entompoxvirinae, and Malacoherpesviridae (reviewed in (16)). They are also widely distributed in eukaryotes, from yeast to mammals.
In addition to their widespread distribution throughout eukaryotes, where it is often present in multiple copies, six lineages of iap genes (iap 1-6) have been identified in baculoviruses (16). Often an individual baculovirus will have representatives of several iap groups in their genomes, e.g., OpMNPV has representatives of iap -1, 2, -3, and -4 (31). In addition, some viruses have two members of one of the lineages (32). The iap-5 lineage appears to be confined to Betabaculovirus (GV) genomes as is iap-6 which is present in only a few GV genomes. Although, iap-1, -2, -4 are found only in Alphabaculoviruses, iap-3 has a wider distribution and is found in some Alpha-, Beta-, and Gammabaculoviruses (16). Although the genomes of CpGV and OpMNPV were not completely sequenced when their iap genes were initially identified, both of these were later found to be members of the iap-3 lineage. Iap-3 genes are closely related to iap genes of insects. For example, OpMNPV IAP-3 is 57% identical to IAP from B. mori. In addition, iap from S. frugiperda has similar properties to IAP-3 in terms of its structure and function (33). The role of other iap genes in baculovirus biology is not clear (reviewed in (16)). For example, in EppoMNPV, deletion of the iap-1 gene delayed the onset, but did not prevent apoptosis induced by actinomycin. However the iap-2 ortholog from this virus was found to have anti apoptotic activity when expressed from a CMV promoter in S. frugiperda cells (34) (35). It is possible that the different iap genes may function in different cell types (19).
IAP Domains
IAP sequences have a number of distinguishing domains. These include baculovirus IAP repeat (BIR) domains of about 70 amino acids that coordinates a Zn ion. BIR domains are often present in multiple copies, with two copies present in many baculovirus IAPs and up to three copies in some cellular IAPs and is involved in protein-protein interactions. Important interactions include the interaction and inactivation of caspases and the interaction with factors that dissociate IAP-caspase complexes thereby freeing up caspases so that they can participate in apoptosis, reviewed in (36). A RING (‘really interesting new gene’) finger domain of about 40 amino acids is also often present near the C-terminus of the protein and is required for dimerization and for E3 ubiquitin ligase activity. Such enzymes transfer ubiquitin from E2 ubiquitin conjugating enzymes to target proteins.
Cellular IAP genes
Although initially discovered in baculoviruses, homologs of IAP genes were subsequently found to be present in many organisms including yeast, insects, and mammals. It has been found that the BIR domains of cellular IAPs block selected caspases. For example, in human X-linked iap (XIAP), the BIR2 domain binds to and inhibits caspase 3 and 7, whereas BIR3 inhibits caspase-9 (37). Other IAPs have ubiquitin ligase activity associated with their RING domain and thereby target caspases for ubiquitination and subsequent degradation (38). In Drosophila there are several proteins, e.g., HID, Grim and Reaper, that block the activity of Drosophila IAPs and are called IAP antagonists (Figure 2). These antagonists are necessary to allow for normal insect development that is based on the orderly removal of unneeded tissues via apoptosis. Therefore, they facilitate apoptosis by blocking proteins that would inhibit this process.
Baculovirus Iap-3 function. Whereas, XIAP inhibits caspases, the most well-characterized baculovirus IAP, Op-IAP-3 increases the stability of cellular IAPs. Cellular IAP proteins serve as a primary mechanism for blocking apoptosis. Virus infection triggers the rapid depletion of cellular IAP thereby initiating caspase-mediated apoptosis leading to cell death. Spodoptera frugiperda IAP (SfIAP) and related IAPs from other insects contain an instability motif (degron) in their N-terminal leader region that regulates cellular IAP turnover. The baculovirus IAP-3 proteins lack an equivalent degron motif which contributes to their stability. Op-IAP3 can form a heterodimer with SfIAP and prevent its degradation thereby blocking apoptosis. Op-IAP3 also increases the levels and increased the half-life of SfIAP thereby enhancing its anti-apoptotic function. (39, 40)
In summary, evidence suggests that homologs of the IAP proteins are capable of blocking apoptosis by several mechanisms including direct interference with caspases, targeting caspases for degradation via ubiquitin ligase activity, interference with antagonists of other proteins that block apoptosis, and increasing the stability of the host cell IAP. Unlike p35 that has a limited distribution and indeterminate origin, the baculovirus IAP genes have multiple homologs in their insect hosts that would have provided a source for the incorporation of an IAP gene into viral genomes on multiple occasions.
Apsup, a third baculovirus antiapoptotic gene family
The apoptotic suppressor (apsup) was discovered in the genome (orf109) of the Lymantria dispar MNPV. Ld109 encodes a protein with a predicted mass 39.3 kDa and does not appear to be related to other proteins in the database. It blocks initiator caspases (Figure 2). Homologs were identified in Lymantria xylina MNPV and also in AcMNPV (Ac112/113). Ac112/113 shows about 30% amino acid sequence identity to APSUP, but is truncated and lacks 79 amino acids at its C-terminus and also lacks anti apoptotic activity (26, 35, 41). Apsup blocked apoptosis by inhibiting the proteolytic processing of an initiator caspase (Dronc) of L. dispar. This inhibition may be caused by it physically interacting with Dronc (42).
Baculovirus CIDE domain proteins
The cell death-inducing DFF45-like effector (CIDE) domain is usually present near the N-terminus of a DNAse that is activated by caspase cleavage and is associated with the degradation of DNA during apoptosis and lipid homeostasis (43). CIDE_N domains have been identified (ORF38) in the Mythimna unipuncta GV (MyunGV)) (44) and in a Group I NPV, Choristoneura fumiferana def (CfdefMNPV) (orf142). MyunGV orf38 is related to orfs from 5 other GVs. In contrast, CfdefMNPV orf142 is most closely related to an orf from another Group I NPV (Neophasia sp – the pine butterfly – NespNPV. Both lineages showed structural relatedness to CIDE domains from Mus musculus and Drosophila melanogaster with a probability of almost 100% by Hhpred (45). The predicted CIDE domain proteins from the NPVs and GVs are only distantly related suggesting that this protein may have been incorporated into baculovirus genomes on at least two independent occasions.
AcMNPV orf92 (p33) — a link between apoptosis and the cell cycle?
Orthologs of Ac92 are present in all sequenced baculovirus genomes and it is an essential gene as viable recombinants deleted for this gene have not been isolated (46)(47). Ac92 has been demonstrated to have sulfhydryl oxidase activity, suggesting that it is involved in the formation of disulfide bonds (48). It is also found associated with BV and ODV (46) (47), suggesting that it may be involved in the formation of disulfide bonds in the nuclei of infected cells. The crystal structure of Ac92 was described as a novel dimer composed of two pseudodimers (49). The structure of Ac92 ortholog in BmNPV (Bm75) has also been reported (50). Ac92 also interacts with p53. P53 has been called 'the guardian of the genome' because of a number of roles it plays in protecting cells from damage. Although normally inactive or expressed at low levels, it is induced by factors causing DNA damage, ribonucleotide depletion, and deregulated oncogene expression and can facilitate blocking the division of such cells. This interruption in the cell cycle allows time for the cell to repair the damaged DNA or it can lead to the induction of apoptosis and the destruction of the cell. Similar to the interaction with Rb, some viruses express proteins that inactivate p53 by interfering with its regulation of the cell cycle and preventing it from inducing apoptosis. Therefore, it was of considerable interest when it was observed that Ac92 (p33) forms a stable complex with human p53 (51). When expressed by itself, Ac92 shows diffuse cytoplasmic staining and punctate staining of nuclei. However, when co-expressed with p53, it exclusively localizes to nuclei. Expression of human p53 in Sf cells causes apoptosis which can be blocked by co-expression of baculovirus anti-apoptotic suppressors p35 or OpIAP. However, co-expression of p53 with Ac92 elevated the induction of apoptosis about two-fold. Proteins with sulfhydryl oxidase activity have been implicated in the protection of cells from oxidative stress caused by apoptosis (52) (53). When the role of p53 in baculovirus infection was examined it was found that AcMNPV infection caused the accumulation of P53. It was observed that despite the accumulation of p53 under conditions of both DNA damage and AcMNPV infection, silencing p53 did not affect the induction of apoptosis suggesting that it might not be absolutely required for activating apoptosis in Sf9 cells (54). An ortholog of P53 has been described for S. frugiperda (55) and similar to human p53, Sfp53 was found to interact with Ac92 (56). It interacts with the Sfp53 DNA binding domain and a point mutation in Sfp53 that inactivated DNA binding also inactivated binding of Ac92 to Sfp53. Ac92 was also shown to oxidize Sfp53 in vitro. However, despite the ability of p33 to interact with and oxidize Sfp53 in cultured cells, no effects on Sfp53-mediated apoptosis or virus replication were observed (56). Effects on other cell types or in whole insects was not ruled out by these studies.
References
- 1.
- Davy C, Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology. 2007;368:219–26. [PMC free article: PMC7103309] [PubMed: 17675127]
- 2.
- Zeng XR, Hao H, Jiang Y, Lee MY. Regulation of human DNA polymerase delta during the cell cycle. J Biol Chem. 1994;269:24027–33. [PubMed: 7929054]
- 3.
- Braunagel SC, Parr R, Belyavskyi M, Summers MD. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology. 1998;244:195–211. [PubMed: 9581791]
- 4.
- Ikeda M, Kobayashi M. Cell-cycle perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology. 1999;258:176–88. [PubMed: 10329579]
- 5.
- Saito T, Dojima T, Toriyama M, Park EY. The effect of cell cycle on GFPuv gene expression in the baculovirus expression system. J Biotechnol. 2002;93:121–9. [PubMed: 11738719]
- 6.
- Zhou R, Yu ZH, Li XQ, Jia F, Wu JH, Chen X. Heliocoverpa armigera single nucleocapsid nucleopolyhedrovirus induces Hz-AM1 cell cycle arrest at the G2 phase with accumulation of cyclin B1. Virus Res. 2004;105:113–20. [PubMed: 15351484]
- 7.
- Belyavskyi M, Braunagel SC, Summers MD. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proc Natl Acad Sci U S A. 1998;95:11205–10. [PMC free article: PMC21620] [PubMed: 9736714]
- 8.
- Nobiron I, O'Reilly DR, Olszewski JA. Autographa californica nucleopolyhedrovirus infection of Spodoptera frugiperda cells: a global analysis of host gene regulation during infection, using a differential display approach. J Gen Virol. 2003;84:3029–39. [PubMed: 14573808]
- 9.
- Xing K, Deng R, Wang J, Feng J, Huang M, Wang X. Analysis and prediction of baculovirus promoter sequences. Virus Res. 2005;113:64–71. [PubMed: 15908030]
- 10.
- Manji GA, Friesen PD. Apoptosis in motion. An apical, P35-insensitive caspase mediates programmed cell death in insect cells. J Biol Chem. 2001;276:16704–10. [PubMed: 11278634]
- 11.
- Schultz KL, Friesen PD. Baculovirus DNA replication-specific expression factors trigger apoptosis and shutoff of host protein synthesis during infection. J Virol. 2009;83:11123–32. [PMC free article: PMC2772793] [PubMed: 19706708]
- 12.
- Clem RJ, Miller LK. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J Virol. 1993;67:3730–8. [PMC free article: PMC237736] [PubMed: 8510202]
- 13.
- Herschberger PA, Dickson JA, Friesen PD. Site-specific mutagenesis of the 35-kilodalton protein gene encoded by Autographa californica nuclear polyhedrosis virus: cell line-specific effects on virus replication. J Virol. 1992;66:5525–5533. [PMC free article: PMC289111] [PubMed: 1501287]
- 14.
- Schultz KL, Wetter JA, Fiore DC, Friesen PD. Transactivator IE1 is required for baculovirus early replication events that trigger apoptosis in permissive and nonpermissive cells. J Virol. 2009;83:262–72. [PMC free article: PMC2612339] [PubMed: 18945761]
- 15.
- Passarelli AL, Westpheling J, Espelie EM, Espelie K. 2014. Lois K. Miller, A Biograhical Memoir. http:
//wwwnasonlineorg /publications/biographical-memoirs /memoir-pdfs /miller-loispdf. - 16.
- Clem RJ. Viral IAPs, then and now. Semin Cell Dev Biol. 2015;39:72–9. [PubMed: 25652775]
- 17.
- Clem RJ, Fechheimer M, Miller LK. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. [PubMed: 1962198]
- 18.
- Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–8. [PubMed: 7569933]
- 19.
- Clem RJ. Baculoviruses and apoptosis: a diversity of genes and responses. Curr Drug Targets. 2007;8:1069–74. [PubMed: 17979666]
- 20.
- Escasa SR, Lauzon HA, Mathur AC, Krell PJ, Arif BM. Sequence analysis of the Choristoneura occidentalis granulovirus genome. J Gen Virol. 2006;87:1917–33. [PubMed: 16760394]
- 21.
- Means JC, Penabaz T, Clem RJ. Identification and functional characterization of AMVp33, a novel homolog of the baculovirus caspase inhibitor p35 found in Amsacta moorei entomopoxvirus. Virology. 2007;358:436–47. [PMC free article: PMC2582192] [PubMed: 17010407]
- 22.
- Makino S, Hamajima R, Saito A, Tomizaki M, Iwamoto A, Kobayashi M, Yamada H, Ikeda M. Bombyx mori homolog of tumor suppressor p53 is involved in apoptosis-mediated antiviral immunity of B. mori cells infected with nucleopolyhedrovirus. Dev Comp Immunol. 2018;84:133–141. [PubMed: 29448034]
- 23.
- Du Q, Lehavi D, Faktor O, Qi Y, Chejanovsky N. Isolation of an apoptosis suppressor gene of the Spodoptera littoralis nucleopolyhedrovirus. J Virol. 1999;73:1278–85. [PMC free article: PMC103951] [PubMed: 9882332]
- 24.
- Zoog SJ, Schiller JJ, Wetter JA, Chejanovsky N, Friesen PD. Baculovirus apoptotic suppressor P49 is a substrate inhibitor of initiator caspases resistant to P35 in vivo. Embo J. 2002;21:5130–40. [PMC free article: PMC129042] [PubMed: 12356729]
- 25.
- Jabbour AM, Ekert PG, Coulson EJ, Knight MJ, Ashley DM, Hawkins CJ. The p35 relative, p49, inhibits mammalian and Drosophila caspases including DRONC and protects against apoptosis. Cell Death Differ. 2002;9:1311–20. [PubMed: 12478468]
- 26.
- Ikeda M, Yamada H, Hamajima R, Kobayashi M. Baculovirus genes modulating intracellular innate antiviral immunity of lepidopteran insect cells. Virology. 2013;435:1–13. [PubMed: 23217611]
- 27.
- Guy MP, Friesen PD. Reactive-site cleavage residues confer target specificity to baculovirus P49, a dimeric member of the P35 family of caspase inhibitors. J Virol. 2008;82:7504–14. [PMC free article: PMC2493353] [PubMed: 18508888]
- 28.
- Gombart AF, Blissard GW, Rohrmann GF. Characterization of the genetic organization of the HindIII-M region of the multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata reveals major differences among baculoviruses. J Gen Virol. 1989;70:1815–1828. [PubMed: 2661722]
- 29.
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. Journal of Virology. 1993;67:2168–2174. [PMC free article: PMC240327] [PubMed: 8445726]
- 30.
- Birnbaum MJ, Clem RJ, Miller LK. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. [PMC free article: PMC236730] [PubMed: 8139034]
- 31.
- Ahrens CH, Russell R, Funk CJ, Evans JT, Harwood SH, Rohrmann GF. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. [PubMed: 9126251]
- 32.
- Esteban DJ, Da Silva M, Upton C. New bioinformatics tools for viral genome analyses at Viral Bioinformatics - Canada. Pharmacogenomics. 2005;6:271–80. [PubMed: 16013958]
- 33.
- Cerio RJ, Vandergaast R, Friesen PD. Host Insect Inhibitor-of-Apoptosis SfIAP Functionally Substitutes for Baculovirus IAP but is Differentially Regulated by its N-terminal Leader. J Virol. 2010;84:11448–60. [PMC free article: PMC2953141] [PubMed: 20739517]
- 34.
- Maguire T, Harrison P, Hyink O, Kalmakoff J, Ward VK. The inhibitors of apoptosis of Epiphyas postvittana nucleopolyhedrovirus. J Gen Virol. 2000;81:2803–11. [PubMed: 11038395]
- 35.
- Yamada H, Shibuya M, Kobayashi M, Ikeda M. Baculovirus Lymantria dispar multiple nucleopolyhedrovirus IAP2 and IAP3 do not suppress apoptosis, but trigger apoptosis of insect cells in a transient expression assay. Virus Genes. 2012;45:370–9. [PubMed: 22798056]
- 36.
- Mace PD, Shirley S, Day CL. Assembling the building blocks: structure and function of inhibitor of apoptosis proteins. Cell Death Differ. 2010;17:46–53. [PubMed: 19373243]
- 37.
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–94. [PMC free article: PMC1618369] [PubMed: 17016456]
- 38.
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–97. [PubMed: 15803136]
- 39.
- Byers NM, Vandergaast RL, Friesen PD. Baculovirus Inhibitor-of-Apoptosis Op-IAP3 Blocks Apoptosis by Interaction with and Stabilization of a Host Insect Cellular IAP. J Virol. 2016;90:533–44. [PMC free article: PMC4702543] [PubMed: 26491164]
- 40.
- Vandergaast R, Mitchell JK, Byers NM, Friesen PD. Insect inhibitor-of-apoptosis (IAP) proteins are negatively regulated by signal-induced N-terminal degrons absent within viral IAP proteins. J Virol. 2015;89:4481–93. [PMC free article: PMC4442400] [PubMed: 25653450]
- 41.
- Yamada H, Shibuya M, Kobayashi M, Ikeda M. Identification of a novel apoptosis suppressor gene from the baculovirus Lymantria dispar multicapsid nucleopolyhedrovirus. J Virol. 2011;85:5237–42. [PMC free article: PMC3126171] [PubMed: 21411519]
- 42.
- Yamada H, Kitaguchi K, Hamajima R, Kobayashi M, Ikeda M. Novel apoptosis suppressor Apsup from the baculovirus Lymantria dispar multiple nucleopolyhedrovirus precludes apoptosis by preventing proteolytic processing of initiator caspase Dronc. J Virol. 2013;87:12925–34. [PMC free article: PMC3838142] [PubMed: 24067961]
- 43.
- Choi JY, Qiao Q, Hong SH, Kim CM, Jeong JH, Kim YG, Jung YK, Wu H, Park HH. CIDE domains form functionally important higher-order assemblies for DNA fragmentation. Proc Natl Acad Sci U S A. 2017;114:7361–7366. [PMC free article: PMC5514754] [PubMed: 28652364]
- 44.
- Harrison RL, Rowley DL, Mowery J, Bauchan GR, Theilmann DA, Rohrmann GF, Erlandson MA. The Complete Genome Sequence of a Second Distinct Betabaculovirus from the True Armyworm, Mythimna unipuncta. PLoS One. 2017;12:e0170510. [PMC free article: PMC5245865] [PubMed: 28103323]
- 45.
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430:2237–2243. [PubMed: 29258817]
- 46.
- Wu W, Passarelli AL. Autographa californica M nucleopolyhedrovirus Ac92 (ORF92, P33) is required for budded virus production and multiply-enveloped occlusion-derived virus formation. J Virol. 2010;84:12351–61. [PMC free article: PMC2976406] [PubMed: 20861245]
- 47.
- Nie Y, Fang M, Theilmann DA. Autographa californica multiple nucleopolyhedrovirus core gene ac92 (p33) is required for the efficient budded virus production. Virology. 2011;409:38–45. [PubMed: 20965540]
- 48.
- Long CM, Rohrmann GF, Merrill GF. The conserved baculovirus protein p33 (Ac92) is a flavin adenine dinucleotide-linked sulfhydryl oxidase. Virology. 2009;388:231–5. [PubMed: 19409596]
- 49.
- Hakim M, Mandelbaum A, Fass D. Structure of a baculovirus sulfhydryl oxidase, a highly divergent member of the erv flavoenzyme family. J Virol. 2011;85:9406–13. [PMC free article: PMC3165737] [PubMed: 21752922]
- 50.
- Hou Y, Xia Q, Yuan YA. Crystal structure of Bombyx mori nucleopolyhedrovirus ORF75 reveals a pseudo-dimer of thiol oxidase domains with a putative substrate-binding pocket. J Gen Virol. 2012;93:2142–51. [PubMed: 22764321]
- 51.
- Prikhod'ko GG, Wang Y, Freulich E, Prives C, Miller LK. Baculovirus p33 binds human p53 and enhances p53-mediated apoptosis. J Virol. 1999;73:1227–34. [PMC free article: PMC103944] [PubMed: 9882325]
- 52.
- Morel C, Adami P, Musard JF, Duval D, Radom J, Jouvenot M. Involvement of sulfhydryl oxidase QSOX1 in the protection of cells against oxidative stress-induced apoptosis. Exp Cell Res. 2007;313:3971–82. [PubMed: 17927979]
- 53.
- Thirunavukkarasu C, Wang LF, Harvey SA, Watkins SC, Chaillet JR, Prelich J, Starzl TE, Gandhi CR. Augmenter of liver regeneration: an important intracellular survival factor for hepatocytes. J Hepatol. 2008;48:578–88. [PMC free article: PMC2954779] [PubMed: 18272248]
- 54.
- Huang N, Wu W, Yang K, Passarelli AL, Rohrmann GF, Clem RJ. Baculovirus infection induces a DNA damage response that is required for efficient viral replication. J Virol. 2011;85:12547–56. [PMC free article: PMC3209345] [PubMed: 21917957]
- 55.
- Huang N, Clem RJ, Rohrmann GF. Characterization of cDNAs encoding p53 of Bombyx mori and Spodoptera frugiperda. Insect Biochem Mol Biol. 2011;41:613–9. [PubMed: 21497653]
- 56.
- Wu W, Clem RJ, Rohrmann GF, Passarelli AL. 2013. The baculovirus sulfhydryl oxidase Ac92 (P33) interacts with the Spodoptera frugiperda P53 protein and oxidizes it in vitro. 447:197-207. [PMC free article: PMC3870475] [PubMed: 24210115]
- Baculovirus infection: The cell cycle and apoptosis - Baculovirus Molecular Biol...Baculovirus infection: The cell cycle and apoptosis - Baculovirus Molecular Biology
- Introduction to the baculoviruses, their taxonomy, and evolution - Baculovirus M...Introduction to the baculoviruses, their taxonomy, and evolution - Baculovirus Molecular Biology
- DNA replication and genome processing - Baculovirus Molecular BiologyDNA replication and genome processing - Baculovirus Molecular Biology
- RecName: Full=Piwi-like protein 4; Short=mAgo5RecName: Full=Piwi-like protein 4; Short=mAgo5gi|226701333|sp|Q8CGT6.3|PIWL4_MOUSProtein
- AV253931 RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cDNA...AV253931 RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cDNA clone 4921506H11 3', mRNA sequencegi|16387973|gnl|dbEST|9988853|dbj|A 31.2|Nucleotide
Your browsing activity is empty.
Activity recording is turned off.
See more...