NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Bravata DM, McDonald K, Owens DK, et al. Bioterrorism Preparedness and Response: Use of Information Technologies and Decision Support Systems. Rockville (MD): Agency for Healthcare Research and Quality (US); 2002 Jun. (Evidence Reports/Technology Assessments, No. 59.)
This publication is provided for historical reference only and the information may be out of date.

Bioterrorism Preparedness and Response: Use of Information Technologies and Decision Support Systems.
Show detailsConceptual Model
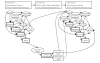
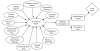
We evaluated the information needs of clinicians and public health officials (to answer Key Question 1) by abstracting information about the decisions they had to make as described in reports of bioterrorism preparedness exercises, outbreaks of naturally occurring infectious diseases, and emergency preparedness plans. We created an influence diagram to represent our conceptual model of the decisions and tasks involved in diagnosis, management, surveillance, and communication by clinicians and public health officials in the event of a bioterrorist attack (Figure 1). The structure of the diagram depicts 3 critical time periods. Time period 1 refers to the interval in which decisions are made by clinicians regarding the events associated with the initial cases, time period 2 refers to the interval in which decisions are made by public health officials regarding the events associated with the initial cases, and time period 3 refers to the interval in which decisions are made by clinicians regarding the events associated with subsequent cases. We recognize that time periods 1 and 2 are likely to occur concurrently but have chosen to represent them as separate events in order to more clearly delineate the decisions made by clinicians and public health officials.
In time period 1, after a bioterrorism event occurs, a population may be exposed to an infectious agent, and those who have been exposed may become infected. The true infection status of any patient is unknown to the clinician. Therefore, the chance node "Infection Status" represents the pre-test probability of disease. After an exposure, a single patient with an unusual clinical syndrome or a cluster of cases may present to a clinician for evaluation. During time period 1, the clinician is faced with 4 decisions: (1) whether or not to perform diagnostic testing; (2) how to manage patients; (3) how to prevent the spread of disease; and (4) whether or not to report to public health officials. Diagnostic testing decisions will be made primarily on the basis of the clinician's estimation of the pre-test probability of disease. The interpretation of test results depends upon the sensitivity and specificity of the test and the pre-test probability of disease. Management decisions include triage, treatment of acutely ill patients, and maintenance of personal safety. Management decisions are influenced by the clinician's interpretation of diagnostic tests. Prevention decisions include prophylaxis and vaccination of exposed individuals; they are similarly affected by the clinician's interpretation of diagnostic tests and by the probability of exposure. Reporting decisions are affected primarily by the clinician's interpretation of diagnostic tests (e.g., if a diagnostic test suggests anthrax, the clinician is likely to report this case to public health officials). Some highly atypical clinical syndromes may also trigger the decision to report. The desired outcome of this decision-making process (denoted by the diamond in Figure 1) could be lives saved or morbidity prevented; it is affected primarily by the patient's true infection status (which is unknown) and by management and prevention decisions.
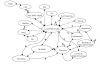
In time period 2, surveillance reports may suggest to public health officials that a potential bioterrorism event has occurred. In Figure 1, we have only shown the surveillance reports submitted by clinicians. However, public health officials could receive surveillance data from a variety of sources, including environmental detection systems, pharmacy sales, veterinarians, zoos, laboratories, first responders (such as ambulance/911 calls), hospital discharges, school/work absenteeism and coroners' reports (Figure 2). Additionally, they could receive alerts of suspected bioterrorist activity from local law enforcement or the intelligence community. If these surveillance reports suggest evidence of a possible bioterrorism event, public health officials are faced with 3 primary decisions: (1) whether to perform outbreak investigation; (2) whether to institute epidemiologic control measures; and (3) whether to issue a surveillance alert to clinicians and first responders. Decisions about performing outbreak investigation will be affected by the methods used to calculate the expected values for each source of surveillance data and to set the thresholds in the surveillance data analyses above which outbreak investigation will be performed. Epidemiologic control measures include actions intended to prevent the spread of disease, such as quarantine, mass vaccination and/or antibiotic distribution, and requesting release of the National Pharmaceutical Stockpile. Decisions about the institution of epidemiologic control measures are based primarily on the results of the outbreak investigation. Decisions about whether to issue a surveillance alert to clinicians, first responders, other public health officials, the intelligence community, the media, and/or interested groups will also be based primarily on these results. The desired outcome of this decision-making process could be lives saved or morbidity prevented; it is affected primarily by the population's infection status and by epidemiologic control measures.
Figure
Figure 2. Conceptual model: Sources of surveillance reports.
In time period 3, clinicians are faced with the identification of subsequent cases. Their estimation of the pre-test probability of disease may be increased secondary to alerts from public health officials, thereby affecting subsequent testing, management, prevention, and reporting decisions.
The formal structure of the influence diagram facilitates the identification of the 4 key decisions faced by clinicians and the 3 key decisions faced by public health officials, the 2 decision makers who are the focus of this Report, in responding to a bioterrorist attack. Additionally, the influence diagram specifies 3 essential features of the decision-making process that could be the targets of IT/DSSs: the relationships between the decisions, the uncertain events that affect the decisions, and the information that is observable by the decision makers at the time they make their decisions. In Figure 1, a double line indicates those decisions that could be affected by DSSs, and the broken lines mark processes in which ITs could play a role.
The information needed by clinicians and public health officials is that which will enable them to make appropriate decisions during a bioterrorism event. We will describe this in greater detail in the next section. Briefly, these information needs include: understanding the clinical presentations of patients exposed to biothreat agents, the best strategies for treating patients thought to have been exposed and for preventing the spread of disease to the unexposed, the characteristics of systems used to detect biothreat agents in the environment and diagnose disease resulting from these agents, the expected values and the thresholds of surveillance data that determine when outbreak investigation should be initiated, and the circumstances under which information should be communicated between interested parties.
Task Decomposition
We used task decomposition, a complementary approach to influence diagramming, to describe in detail the tasks that IT/DSSs would have to perform to facilitate the decision making of clinicians and public health officials. This description of the necessary tasks of IT/DSSs for bioterrorism serves as the foundation of our evaluation system of the currently available IT/DSSs (i.e., Key Question 2). We categorized the information needs of clinicians and public health officials, as described in reports of infectious disease outbreaks, bioterrorism preparedness exercises, conversations with public health officials, and our conceptual model, into 6 top-level tasks and 19 subtasks (Table 2). We then considered the key concepts driving each task and the data requirement for an IT/DSS to assist in that task.
Table
Table 2. Task list and Evaluation criteria.
Clinicians and public health officials are responsible for a complex array of tasks during a bioterrorism event. As a result, they require accurate, current information on a disparate group of topics. This heterogeneity of information needs suggests that a variety of IT/DSSs will be required -- each of which will have to supply a different type of data and perform different computations or manipulations of the data. For example, consider one of the primary tasks of public health officials during a bioterrorism event -- the decision of whether or not to perform an outbreak investigation. This decision will require that the public health official has collected surveillance data, understands the baseline characteristics of these data, and has determined that the current surveillance information is sufficient to warrant the costs associated with the initiation of an outbreak investigation. In Table 2, we have decomposed these processes into 2 main subtasks: the collection and analysis of surveillance data. We first present a selection of the kinds of data that could be collected for bioterrorism surveillance (recognizing that this represents many of the data currently collected but not all the kinds that could be used for this purpose). Additionally, for each source of surveillance data, we describe the primary criteria by which it should be evaluated (e.g., its timeliness, sensitivity, and burden on data collectors). We then describe the 2 primary analytic requirements of a surveillance system: the determination of seasonal and geographic variations in the expected values of the surveillance data, and the evaluation of surveillance data for patterns that fall outside the expected range. For each report of a surveillance system, we recorded whether it described each of these characteristics (i.e., the type of data under surveillance; the methods for establishing both baselines and thresholds; and the factors that affect the system's timeliness, sensitivity, specificity, cost, acceptability, etc.). We describe the evaluation criteria used for each type of IT/DSS in the Results sections that follow.
Search Results
We reviewed a total of 16,888 citations of peer-reviewed articles, 7,685 Web sites of government agencies, and 1,107 Web sites identified through the Copernic® search. Of these, 251 articles, 36 government Web sites, and 54 non-government Web sites met our inclusion criteria. From these, we abstracted descriptions of 217 IT/DSSs of potential use by clinicians and public health officials in the event of a bioterrorist attack. They are comprised of 55 detection systems, 23 diagnostic systems, 18 management and prevention systems, 90 surveillance systems, 26 reporting and communication systems, and 7 systems that integrate surveillance, communication, and command and control functions (Table 3). Most reports only described IT/DSSs; however, 79 studies evaluated 58 systems for at least 1 performance metric (e.g., timeliness, sensitivity, or specificity). Some types of systems have been evaluated more than others. For example, 10 of the 18 management systems have been evaluated in at least 1 study; whereas, none of the 7 integrated surveillance, communication, and command and control systems has been (Table 3).
Table
Table 3. Type of evidence for each category of systems.
Search Results: MEDLINE®
We reviewed 5,173 titles and abstracts from our preliminary searches of MEDLINE® and 11,515 additional titles and abstracts from the final search strategy (Appendix E). Of these, 822 articles were retrieved and cataloged, and 251 met inclusion criteria. These articles report on 162 systems (multiple articles report on the same system).
Search Results: Other Databases of Peer-reviewed Articles and Selected Government Agency Web Sites
Of the 5 databases of peer-reviewed articles, only MEDLINE® and NTIS provided references for articles that described potentially relevant IT/DSSs. None of the articles retrieved from the following databases described potentially relevant IT/DSSs: The Catalog of U.S. Government Publications (search resulted in 51 citations), GrayLIT (search resulted in 55 citations), or Library of Congress (search resulted in 7 citations). Because they were searched using the same search terms, we have included the result of the NTIS search with the result of our search of government agency Web sites (Appendix F). Our review of 7,685 Web sites from our search of the NTIS and government agencies provided us with information on 29 systems (Appendix F).
Search Results: Internet Search
Our review of 1,107 Web sites identified through the Copernic® Internet search added 27 systems to our database from 54 non-government Web sites (Appendix G).
Overview of Included Systems
In the sections that follow, we present the results of the information retrieved on IT/DSSs for detection and diagnosis, management and prevention, surveillance, and reporting and communication. For each of these categories of IT/DSSs, we present general information about the category, including the criteria by which these systems should be evaluated with respect to usefulness for bioterrorism (answer to Key Question 2), information on each system, and summary comments about the potential usefulness of those systems for clinicians and public health officials in the event of an attack (answers to Key Question 3 and 4). We first describe those systems that are commercially available, followed by systems that remain under development or are not currently commercially available (i.e., available only to government agencies or the military). We recognize that there are omissions from the group of systems under development and that numerous systems recently made available in response to the events of September 11, 2001 have not been included. In Appendix H, we present detailed information on the type of evidence found on each system.
We attempted to classify each system according to its principal stated purpose. The considerable overlap in the functionality of some of the included systems made this classification difficult. For example, the National Molecular Subtyping Network for Foodborne Disease Surveillance (PulseNet) is a communication system that facilitates the reporting of laboratory data. Because its primary purpose is communication, we describe it in that section. However, it could be reasonably argued that PulseNet is a component of a laboratory-based surveillance system and therefore should be discussed in the context of other laboratory systems. Similarly, some bioterrorism experts consider detection and diagnostic systems to be a subcategory of surveillance systems since the data generated by them can be integrated into bioterrorism surveillance systems. We present the data on detection and diagnostic systems separately because most of the systems that we describe were not designed for the collection of bioterrorism surveillance data -- although these data could certainly be used for that purpose.
For each of the sections that follow, we present our results in the following order: first, a brief statement of background information on the category of system; second, the criteria by which we attempted to evaluate the systems; third, the information that we found about the systems presented according to the evaluation criteria; and, fourth, summary comments about the systems and the evidence describing them. More detailed information about the included systems is presented in a Table at the end of the section and in the Evidence Tables at the end of this Report. An index of the systems is provided in Appendix I.
Detection and Diagnostic Systems
Our systematic review identified 55 detection systems for the rapid identification of potential biothreat agents in environmental and clinical samples, 6 general diagnostic DSSs used by clinicians to generate a list of possible diagnoses for a given patient, 4 radiologic systems that detect or diagnose interstitial lung disease or pneumonia, 4 telemedicine/teleradiology systems, and 9 other diagnostic systems (Tables 3-12; Evidence Tables 1 and 2).
Detection Systems
The ideal detection system can identify the release of a biothreat agent before a single person becomes infected. Clinicians and public health officials require detection systems than can identify a covert release of a biothreat agent early enough to take action that limits the spread and progression of disease, as well as systems that rapidly evaluate environmental and clinical samples. A comprehensive discussion of biologic detection systems is beyond the scope of this project. We refer interested readers to detailed reports on this topic.10, 36, 37 However, insofar as they provide critical data, which could be used in an IT/DSS to inform diagnostic testing decisions and patient management, we conducted a limited review of the topic. Our discussion of these detection systems is largely informed by the Institute of Medicine's report "Chemical and Biological Terrorism: Research and Development to Improve Civilian Medical Response" 10 and the North American Technology and Industrial Base Organization's report "Biological Detection System Technologies: Technology and Industrial Base Report." 36
Typically, detection systems have 3 parts: a sampler/collector to concentrate the aerosol and preserve samples for further analysis; a trigger component (often a particulate counter or a biomass indicator) that can identify the presence, but not identity, of a possibly harmful biologic material; and an identifier to provide specific identification of the biothreat agent. 36 Additionally, systems often require an information management system to record and send data to a command and control center.10, 36, 37
The relative importance of the characteristics of a detection system varies by its intended use. Identification systems for use by emergency personnel evaluating a suspicious powder in the field require different characteristics than identification systems used by clinicians faced with patients reporting flu-like illnesses and potential biothreat exposures. Depending on the use, the characteristics most desirable in a rapid detection system vary but typically include high specificity (also called the true negative rate or the likelihood that a sample without a biothreat agent has a negative test result), high sensitivity (also called the true positive rate or the likelihood that a sample with a biothreat agent has a positive test result), minimal sample preparation, ability to detect numerous biothreat agents, ability to run numerous samples simultaneously, no expensive or specialized reagents, small/portable size, ability to detect agents in real time (i.e., within minutes), ability to provide an output that is clearly interpretable by the decision maker, and low cost. Often, there is a trade-off between these characteristics such that a more sensitive test may have a higher false positive rate or be less timely than a less sensitive test. For any given detection system, the design of each component -- collector, trigger, and identifier -- will affect its portability, sensitivity, specificity and other detection characteristics.
In the sections that follow, we first describe collection systems, followed by particulate counters and biomass indicators, then identification systems, and, finally, integrated collection and identification systems. 36 For each category, we present general information (e.g., how collection systems generally work, where they are used, and what the important criteria are by which they should be evaluated), a table briefly describing the purpose and relevant test characteristics (such as sensitivity and specificity) of the system, and summary comments describing the usefulness of these systems for bioterrorism preparedness. We provide substantial additional detail on each system in Evidence Table 1.
Collection Systems
Background
Collection systems are used to take samples from either the environment (e.g., air, water, or particulate matter from suspicious surfaces) or from a patient (or an animal) for later identification.10, 36, 37 The principal design considerations for collection systems are that the system must preserve and not harm the collected sample. 36 Many collection systems collect airborne particles onto filters. For those identification systems that require a liquid sample, the collection system must take an aerosol or particulate sample and put it into liquid.10, 36, 37
Evaluation criteria
We evaluated each of the reports of collection systems for the following information (Table 2 -- Detection; Evidence Table 4): the purpose of the system, information regarding the type of sample collected, portability, collection efficiency, limits of size of particulate collected, flow rate, and methods for maintaining the security of the sample.
Findings
We present information on 4 collection systems, none of which was reported in a peer-reviewed evaluation article (Tables 3 and 4; Evidence Table 1; Appendix H). We found descriptions of 4 commercially available stand-alone systems that can be used by public health officials, fire departments, hazardous materials teams, law enforcement, and facility owners to collect environmental samples for ongoing surveillance of high-risk locations (e.g., public buildings and airports) or to monitor clean-rooms: Smart Air Sampler System (SASS) 2000 Plus™ Chem-Bio Air, BioCapture™, SpinCon®, and Portable High-Throughput Liquid Aerosol Air Sampler System (PHTLAAS). We recognize that other collection systems exist and may be currently available; however, we present all of the systems for which we were able to find publicly available reports. Other systems, that include a collection system as part of an integrated collection-detection-identification-communication system, are presented later in this chapter.
Table
Table 4. Collection systems.
The purpose of most of these systems is to collect aerosol environmental samples for use by first responders and for monitoring workplace exposures. We found no collection systems specifically designed for clinicians to obtain a clinical sample from patients (symptomatic or asymptomatic) with suspected exposure to a biothreat agent. Instead, most identification assays can be used on microbiological samples from nasal swabs, sputum, urine, blood, and cerebrospinal fluid collected in standard culture tubes. The descriptions of the 4 collection systems all report that they are portable, and the weight and size dimensions provided seem to justify this claim. The collection efficiency of the devices ranged from 0.5 to 10 microns. (Note that the size of the causative agent of anthrax is 1 to 5 microns, of smallpox is 0.15 to 0.3 microns, of plague is 0.5 to 2 microns, and of tularemia is 0.125 to 0.7 microns. 38 ) Only the SpinCon® reports provided a flow rate, which for this device was 1000 liters per minute (L/min).10, 36, 39-41 None of the reports described methods for maintaining the security of the sample.
The only evaluation information on any of these systems was provided by the manufacturer of the BioCapture™ device, which has been used by fire departments in Seattle, Los Angeles, and New York, among others, and was evaluated at Dugway Proving Ground. 42 The collection efficiency of BioCapture™ was reported to be 50 to 80 percent relative to the All Glass Impinger standard and 60 to 125 percent relative to the Slit Sampler Standard. 42 (We found no additional evaluation information about these standard devices.)
Summary: Collection systems
These portable systems are potentially useful for the collection of environmental samples either as part of ongoing surveillance for a covert release of a biothreat agent or for evaluation of environments suspected of being contaminated. However, there is insufficient evaluative information to determine the utility of these systems for either of these purposes.
Two conditions should be met for collection systems to be maximally useful to first responders, clinicians or public health officials in the event of a bioterrorist attack. First, the collection system must be in use in the affected area. In the event of a covert attack, this is only possible if the collection system is already in place in areas of likely attack (e.g., airports; subways; major sporting, political, or entertainment events). In the event of a known attack, these systems must be portable enough that they can be taken by first responders to the location of suspected release and collect testable environmental samples. The manufacturers of many of these systems claim that they are portable and therefore meet this last condition. Second, the collectors must have sufficiently high flow rate and collection efficiency to be able to collect aerosolized biothreat agents should they exist. According to the available information, only the BioCapture™ device has been subjected to an evaluation of these performance characteristics. (However, we have no additional information on the devices against which it was compared, the All Glass Impinger and Slit Sampler.) Other considerations for which we have no information include: how difficult it is to train first responders in the use of these collection devices, how difficult it is to use these devices to collect samples in a secure manner in the event that they are used as evidence in a criminal investigation of the bioterrorist attack, and how much it would cost to fully implement these systems.
Particulate Counters and Biomass Indicators
Background
A sample collected from one of the collection systems just described could be directly analyzed by an identification system (see following section) to determine the specific identity of biothreat agents contained within the sample. This could be a reasonable analytic strategy when the probability of bioterrorism is particularly high, as in an announced attack. Under other circumstances, it may be preferable to first analyze the sample with a particle counter or biomass indicator to determine if the size, number, or properties of the particles collected suggest the presence of biothreat agents.10, 36 This may be particularly true for environmental samples routinely collected as part of a surveillance system for a covert bioterrorist attack.
Particulate counters and biomass indicators use a variety of methods to determine the presence of potential biothreat agents. We briefly describe 2 methods used by some of the included systems: flow cytometry and biosensor technology. We refer interested readers to reviews on these topics.10, 36, 37, 47-55 Our discussion of these systems was largely informed by these references.
Flow cytometry
Flow cytometers are increasingly common in U.S. hospitals and public health laboratories. 10 Samples introduced into a typical flow cytometer are separated into individual cells.53-55 As cell flow across a laser beam, they scatter light in a characteristic manner and dyes bound to different parts of the cell emit light, or fluoresce.53-55 By measuring the fluorescence and scattered light of the sample, flow cytometers assess a variety of cellular characteristics including: cell size, amount of DNA, presence of specific nucleotide sequences, and cellular proteins.10, 36, 53-55 In an experiment at Los Alamos National Laboratory comparing the ability of flow cytometry and gel electrophoresis to measure bacterial DNA, flow cytometry was about 200,000 times more sensitive than gel electrophoresis (able to detect picogram quantities of DNA) and did so in 10 minutes compared with 24 hours for gel electrophoresis. 55
Biosensor technology
Biosensors use data from living organisms to evaluate environmental samples for potentially toxic substances.37, 47-52 For example, canaries have traditionally been used in coal mines to detect toxic levels of methane gas and, more recently, as nerve agent detectors in the 1995 police raid of the Aum Shinrikyo compound in Japan.37, 47-52 Similarly, fish have been used to monitor water quality. On a smaller scale, data from extracellular recordings of excitable cell types (such as neurons and cardiomyocytes) have been evaluated for their physiologic responses to toxins.37, 47-52 Currently, cytokine production from immune cells used as biosensors for antigens is increasingly a target of research. 56 Technical problems associated with the nutrient media required to keep biosensors alive have prevented cell-based biosensors from becoming widely available; however, these sensors may become a valuable part of the future detection armamentarium -- particularly for the detection of toxins and chemical agents.37, 47-52
Evaluation criteria
We evaluated each of the reports of particulate counters and biomass indicators for the following information (Table 2 -- Detection; Evidence Table 4): the purpose of the system, portability, sensitivity, specificity, the upper and lower limits of the size of particles that can be counted (for the particle counters), the concentration of organisms that can be detected (for the biomass indicators), and methods for maintaining the security of the sample and data about the sample.
Findings
We present information on 14 particulate counters or biomass indicators, 1 of which was reported in a peer-reviewed evaluation (Tables 3 and 5; Evidence Table 1; Appendix H). We found reports of 1 commercially available particle counter (the Met One Aerocet 531 Mass/Particle Counter) and 3 commercially available biomass indicators (the Digital Smell/Electronic Nose, Ameba Biosensor, and Spreeta™) that could be used for biothreat detection but have not been specifically tested for this application. We also found 10 systems that are currently under development or limited to military use, but could be helpful to public health officials for biothreat agent detection. (We report on additional biosensors in the next section on Identification systems. The biosensors listed in the Identification section are designed to specifically identify the presence or absence of a particular organism. In contrast, the biosensors described in this section are designed for the more general purpose of indicating the presence or absence of a biologically active compound.)
The purpose of most of the particulate counters is to detect a statistically significant increase in the number of particles in aerosol samples over baseline, and they were generally designed specifically for bioterrorism detection. In contrast, some of the biomass indicators were designed for the general detection of microorganisms, not specifically for bioterrorism. The systems vary widely in terms of size: some, such as the Met One Aerocet 531 Mass/Particle Counter, are designed for use in monitoring clean-rooms and are the size of a large handheld calculator, while others weigh 10 kilograms (kg), which is considerably larger but can still be easily carried by a single person. In contrast, several of the military-designed units are designed for mounting on a ship or motor vehicle. The size limits of particles that can be counted by the included systems varies from 0.3 to 20 micrometers (µm). The Model 3312A Ultraviolet Aerodynamic Particle Sizer (UV-APS)/Fluorescence Aerodynamic Particle Sizer-2 (FLAPS-2) was the only system for which we found information on the concentration of organisms that can be detected (10 agent-containing particles per liter of air). None of the systems reported methods for maintaining the security of the sample or data about the sample.
Of the 3 systems in this section that have been specifically tested for the detection of biothreat agents, only FLAPS-2, about which we have evidence from government reports and the manufacturer, has been tested for both sensitivity and specificity.10, 36, 39, 57, 58 In field tests, FLAPS-2 was able to detect 39 of 40 blind releases of simulant aerosols at a distance of about 1 kilometer with no false alarms over a 3-week period. A description of the Portable Biofluorosensor (PBS), obtained from a government report, suggests that false positives occasionally occur. 43 The meaning of "occasionally" was not explained further. Of the 14 systems, the only evidence available in a peer-reviewed evaluation article was for the Digital Smell/Electronic Nose, a diagnostic system based on the volatile gases given off as metabolites by microorganisms. 59 Holmberg et al. demonstrated that an array of 15 sensors was able to correctly classify 68 of 90 colonies containing 0 or 1 of 5 test organisms and an uninoculated control (22 of 90 were false positives). The commercially available Electronic Nose is marketed for the detection of microorganisms causing bacterial pharyngitis, pneumonia in ventilated patients, urinary tract infections, and bacterial vaginosis. This device has not been evaluated for the detection of biothreat agents.
Of the systems identified, the effort by the Department of Energy (DOE) called the Program for Response Options and Technology Enhancements for Chemical/Biological Terrorism (PROTECT) is particularly geared towards the detection of a covert bioterrorist attack. PROTECT uses a network of particulate collectors and counters set up in public places (e.g., subway stations) to monitor the environment for patterns suggestive of abnormal aerosols.60, 61 This project also facilitates the testing of collection devices and particulate counters. However, there are no published reports of these results.
Summary: Particulate counters and biomass indicators
Many of the particulate counters and biomass indicators are specifically designed to assist in the detection of a covert release of a biothreat agent. The lack of publicly available evaluation data on these systems precludes any conclusions regarding how useful they are likely to be in assisting in the bioterrorism preparations or response planning of clinicians or public health officials.
In particular, the sensitivity and specificity of these systems needs to be carefully evaluated and reported. Because the costs associated with delay in diagnosing a bioterrorism event can be significant in terms of excess morbidity and mortality, these systems must have demonstrated high sensitivity (i.e., low false negative rate). Similarly, because the costs of responding to false alarms and the potential that users may disregard systems with known high false positive rates, these systems must have sufficient specificity to avoid these hazards.
In addition to the criteria by which we evaluated these systems, the following are important considerations for particulate counters and biomass indicators to be useful to clinicians and public health officials. First, they must be located in close proximity to collection systems so that the samples collected may be routinely and promptly tested. Second, they must have the ability to promptly report results to decision makers (often at a remote location) who can either have the sample tested further for specific identification of the biothreat organism or take action to limit additional exposures to the suspected aerosol/contaminated environment. We were not able to assess the capacity of these systems to securely deliver such information to remote decision makers.
Rapid Identification Systems
Background
Traditional methods for the detection and identification of microorganisms, viruses, and/or their products lack the speed and sensitivity to be useful in the field or at the bedside. 49 The systems likely to be of the greatest use to clinicians and public health officials for the identification of biothreat agents are those that provide a result within minutes. Table 6 describes rapid identification systems and is organized according to the type of identification technology: antibody-based methods, nucleic acid-based methods, mass spectrometry, and others. We refer interested readers to the reviews of rapid identification systems that inform the following discussion.10, 36
Table
Table 6. Rapid identification systems.
Antibody-based systems
Antibody-based systems use antibodies developed to recognize specific targets on either antigens or cells of interest to detect potential pathogens.10, 36 An advantage of these systems is that the use of antibodies confers high specificity10, 36 The antigen-antibody binding can be monitored directly or indirectly. For example, sandwich assays use a second antibody, labeled with a fluorescent dye that binds to either the antigen itself or probe antibody to monitor antigen-antibody binding. 10 The detection thresholds of these methods vary between 103 to 104 microbial cells per milliliter (mL). 10 Technical problems with antibody-based sensors include nonspecific binding (which can lead to false positive results), cross reactivity, and degradation of the antibodies over time (which can lead to false negative results). 10 Despite these technical problems, antibody-based systems can be both highly sensitive and specific.10, 36
In response to the recent cases of anthrax in the U.S., considerable interest has been generated in the use of handheld antibody-based detectors by first responders. The CDC recently issued a statement on its Web site stating that the analytical sensitivity of these assays is limited and that a minimum of 10,000 spores is required to generate a positive signal. 75 Given concerns about the sensitivity and specificity of these kits, the CDC has undertaken an independent evaluation of these tests. Conclusions from this study are expected in the near future. 75
Nucleic acid-based systems
The specificity of nucleic acid-based systems (sometimes called polymerase chain reaction- or PCR-based systems) is derived from the selective binding of nucleic acid probes to complementary nucleic acids from the pathogen of interest. 10 Probes are designed to bind specifically to a nucleic acid sequence that is unique to the pathogen or to identify a nucleic acid sequence that is common to several pathogens. 10 The sensitivity of nucleic acid-based systems for bacteria is between 1,000 and 10,000 colony forming units (CFU); 10 however, recent reports suggest that they may be capable of greater sensitivity. 76 Because the reaction occurs within minutes, the time-consuming parts of using nucleic acid systems are the sample preparation and the time required to detect the signal.10, 36 Significant limitations to the use of these methods for bioterrorism include the lack of highly specific probes for all biothreat agents (although the DOE and CDC have entered a collaboration to develop them) and the use of a single probe to test a single sample for the antigen of interest at a given time. Given security concerns, the distribution of highly specific probes will likely remain under strict federal control -- first responders are not likely to have access to these probes for testing samples in the field.
Mass spectrometry
Mass spectrometry is an analytical technique in which materials under analysis are converted into gaseous ions or other characteristic fragments.10, 36 The fragments are separated on the basis of their mass-to-charge ratio.10, 36 The technique can reportedly detect concentrations of as low as 106 cells. 10 When samples are tested in the field, they are likely to contain multiple constituents (contaminants), which must be separated before they can be reliably identified. This separation can be performed by a variety of techniques, including mass spectrometry.10, 36
We note that some technologies are better suited to particular agents. Nucleic acid-based systems, for example, cannot detect toxins (unlike bacteria and viruses, they do not contain nucleic acids). In contrast, mass spectrometry is more effective for the detection of toxins than bacteria.
Evaluation criteria
We evaluated each of the reports of identification systems for the following information (Table 2 -- Detection; Evidence Table 4): the purpose of the system, portability, sensitivity, specificity, the amount of time it takes to run a sample, the number of samples that can be run simultaneously, the number of biothreat agents that can be identified, whether both toxins and organisms can be identified, and methods for maintaining the security of the sample and data about the sample.
Findings
In this section, we report on 27 rapid identification systems, 4 of which were presented in at least 1 peer-reviewed evaluation article (Tables 3 and 6; Evidence Table 1; Appendix H). Table 6 describes 6 antibody-based tests, 7 nucleic acid-based tests, 1 mass spectrometry-based test, 1 flow cytometry-based test, and 12 tests that use other technologies, including biosensors.
In general, the purpose of these systems is to rapidly detect biothreat agents collected from environmental, human, animal, or agricultural samples. The available antibodies limit the antibody-based tests. Assays are commonly reported to be available for Y. pestis, F. tularensis, B. anthracis, V. cholerae, S. enterotoxin B, Brucella species, ricin, and botulinum toxins. Many of these systems are small (portable) enough for use in the field. They all test for a single biothreat agent per assay and run a single assay at a time (except for the Fiber Optic Wave Guide (FOWG)/Rapid Automatic and Portable Fluorometer Assay System (RAPTOR)/Analyte 2000™ Biological Detection system, which can run 4 assays simultaneously). Reports suggest that a result can be obtained from the handheld antibody tests in 5 to 45 minutes. However, several of the reports of these systems suggest that they are prone to false positives (typically attributed to soil contamination). The FOWG/RAPTOR/Analyte 2000 (described in a peer-reviewed article, 77 government report, 36 and by the manufacturer46, 78-81) has the following estimated detection levels (in water): B. anthracis (30-100 CFU/mL), Ricin (less than 10 nanograms per milliliter (ng/mL)), S. enterotoxin (1 ng/mL), F. tularensis (105 CFU/mL), V. cholerae (10 ng/mL), and Y. pestis at levels below 1 parts per billion (ppb) from samples of a few hundred microliters (µL). In their peer-reviewed evaluation of tests for agroterrorism, Von Bredow and colleagues reported that the Luminometer Rapid Detector, designed for portable quantification of the live bacteria on animal carcasses, could detect the presence of 1000 or more organisms (no additional information provided). 82 This article also briefly reported on the Sensitive Membrane Antigen Rapid Test (SMART™) and the Antibody-based Lateral Flow Economical Recognition Ticket (ALERT) assays, for which we also had information from 2 government reports and the manufacturer. The government document reported that during battlefield tests, the SMART™ system had an "alarmingly" high false positive rate. 74 However, the manufacturer reported that the Bengal SMART™ test for V. cholerae O139 has 99 percent sensitivity and 99 percent specificity, the Cholera SMART™ test for V. cholerae O1 can detect as few as 2x103 organisms, and the BengalScreen and CholeraScreen (coagglutination tests for V. cholerae O139 and O1 respectively) each have a sensitivity of 96 percent and a specificity of 94 percent.83, 84
The nucleic acid tests are similar to antibody-based tests in that they are limited by the availability of probes and only test for a single biothreat agent per assay. We have limited information on the availability of highly specific probes (which usually have to be obtained separately from the machinery itself); however, the descriptions of these systems suggest that probes can be obtained at least for B. anthracis and Y. pestis. Unlike antibody-based systems, many more of the nucleic acid-based systems are designed to run multiple assays at a time (as many as 16 in the currently available systems and tens to hundreds of thousands in some of the gene-chip/micro-array technologies that are currently under development) and can do so in 7 to 60 minutes. Many of these systems are small enough to be carried by a single person but, as a group, they tend to be larger than the antibody-based systems. In terms of their sensitivity and specificity, the Advanced Nucleic Acid Analyzer (ANAA) was described in a government report to be able to detect 500 CFUs of Erwinia herbicola. 76 Per the manufacturer, the LightCycler™/Ruggedized Advanced Pathogen Identification Device (RAPID™) was reported to be 99.9 percent specific with the sensitivity set for each assay at half the infective dose (e.g., the infectious dose of Foot and Mouth Disease is 10 virus particles; therefore, RAPID™'s sensitivity is set to detect 5 virus particles.) 85 The SmartCycler® and GeneXpert™, for which we have information from conference proceedings 86 and the manufacturer, 87 is reported to be "specific to 12 B. anthracis strains tested and able to detect 5 genome copies."
Of the other types of rapid identification systems, 2 were presented in peer-reviewed evaluation articles. Biolog is a general identification system for microorganisms, with potential utility for identification of B. anthracis. Using an in-house database (as opposed to the database that is currently commercially-available and does not include B. anthracis), Biolog correctly identified all samples of B. anthracis with "readable profiles" (19 out of 20). However, it falsely identified 5 out of 12 closely related Bacillus strains as B. anthracis (false positives). For both B. anthracis and related strains, roughly 20 percent of the samples gave uniformly false positive reactions, in which all reaction wells were positive. 88 The Fluorescence-based array immunosensor is designed for the simultaneous, antibody-based detection of bioactive analytes in clinical fluids such as whole blood or from a nasal swab in less than 35 minutes. However, in an evaluation of this system, it was unable to detect physiologically relevant S. enterotoxin B levels (less than 125 ng/mL) in experimentally spiked urine, saliva, and blood products, and the detection limit for F1 antigen from Y. pestis was 25 ng/mL. 89
Many of the rapid detection systems provide outputs that can be made available in electronic format. Several of these systems can be used with a laptop computer for storage and communication of test results to remote decision makers. None of the reports of these systems described security measures for handling the samples or the test results.
Summary: Rapid identification systems
The rapid identification of biothreat agents in environmental samples is essential for a swift response to either covert or announced bioterrorist attacks. Without the availability of accurate rapid detection methods, first responders, and clinicians cannot make decisions about triage, management, or prevention of the spread of disease. However, adequate evaluative data about these critical systems have not been published. The available evaluation information suggests that, although the systems are convenient in terms of their portability and speed, they have high false positive rates. Without additional information about the sensitivity and specificity of these tests, their results are uninterpretable: users cannot readily determine the appropriate action to take given a sample producing either a positive or negative result. Reports of independent evaluations of currently available systems against gold standards represent a major gap in the literature.
Additional limitations to the usefulness of these systems for the rapid identification of biothreat agents are: the absence of tests for many of the most worrisome biothreat agents (e.g., smallpox); the lack of available probes for many nucleic acid tests; and the paucity of tests that facilitate the evaluation of a given sample for more than 1 biothreat agent. Similarly, since the identification systems described in this section tend to test for a single organism at a time, and tend to have the ability to detect only a limited number of biothreat agents, a negative result cannot be interpreted as being negative for a bioterrorism event, merely as being negative for the limited number of organisms detected by that assay.
The reports of the antibody-based technologies for the identification of anthrax did not typically specify whether the antibodies recognize the spore or vegetative form of the bacteria. This is an important determinant of the technology's utility. For example, antibodies against the vegetative form would not be useful for monitoring environmental samples (since anthrax is commonly found in the soil and could contaminate environmental samples) but would be useful for screening clinical samples.
Integrated Collection and Identification Systems
Background
In addition to the IT/DSSs that we described in the preceding sections, some systems combine collection and identification functions in a single unit. Often, these systems have a communication component that allows reporting of the results, typically to a remotely located command and control post. Most of these systems have been developed by the military and are likely to have been rigorously evaluated to meet Department of Defense (DOD) standards; however, much of the evaluative data are not publicly available.
Evaluation criteria
We evaluated each of the reports of integrated collection and identification systems according to the same criteria for each of the component systems listed in the 3 preceding sections (Table 2 -- Detection; Evidence Table 4). Specifically, we evaluated each of the reports of these systems for the following: information regarding the purpose of the system, portability, type of sample collected, collection efficiency, limits of size of particulate collected, flow rate, sensitivity, specificity, the upper and lower limits of the size of particles that can be counted (for the particle counters), the concentration of organisms that can be detected (for the biomass indicators), the amount of time it takes to run a sample, the number of samples that can be run simultaneously, the number of biothreat agents it can identify, whether it can identify both toxins and organisms, and methods for maintaining the security of the sample and data about the sample.
Findings
We report on 10 integrated systems that could be of use to public health officials, hospital administrators, or municipal leaders for the collection, detection, identification, and reporting of a biothreat agent; none has been described in a peer-reviewed evaluation article (Tables 3 and 7; Evidence Table 1; Appendix H).
Table
Table 7. Integrated collection and identification systems.
These systems are generally intended to transmit test results electronically to decision makers at some distance from the collection and identification site(s). They have all been designed for military use but may be increasingly available to interested public health officials and national security professionals for ongoing environmental surveillance. These systems are the size of refrigerators or larger and therefore require trucks or similar vehicles for transportation. The Canadian Integrated Biochemical Agent Detection System (CIBADS II)/4WARN system, designed to collect and identify a variety of chemical and biological agents from a commercial sport utility vehicle, is radio-linked to a command and control unit. An evaluation of CIBADSII/4WARN reported in a government document 57 and by the manufacturer 116 suggested that the system was operated at speeds up to 50 miles per hour "without significant degradation of performance."57, 116 The impact of weather patterns on performance was also determined to be low. The exception was immediately after a thunderstorm, when the number of particles in the air rose dramatically and reduced the sensitivity of the system.57, 116 A government report provided evaluation data on the Portal Shield Air Base/Port Biological Detection System, which integrates data from multiple sensors linked to a centralized command post computer. 66 This computer monitors the sensors and evaluates the data to determine if a bioterrorist attack has occurred. In the event a release is detected, the computer alerts the operator. The algorithm looks for a significant increase in at least 2 sensors before it will sound an alarm, giving the system a theoretical false positive rate of 0.25 percent. The report stated that, "after having gone through over 10,000 assays, the Portal Shield system has not had any false positives." 66 The system can reportedly detect 8 agents, although they were not further specified. 66 None of the reports of these integrated detection systems described the methods for maintaining the security of the sample or test results about the sample; very few details were provided about specific collection or identification components.
Summary: Integrated collection and identification systems
Systems that integrate collection and identification (often with communication) functions have potential utility for the detection of a covert release. The large size of several of these integrated systems prohibits their use by first responders and clinicians. However, public health officials and municipal leaders may be interested in using these systems for ongoing surveillance for bioterrorism events in public spaces considered to be likely targets. The military developers of these systems may have performed comprehensive assessments of their important test characteristics; however, no published peer-reviewed evaluative data are currently available to the general public. This lack of evaluative information prohibits drawing conclusions regarding the utility of these systems by groups outside the military.
Summary: Detection systems
The collection, particulate counters and biomass indicators, rapid identification, and integrated collection and identification systems described in the preceding sections have critical roles to play in the detection of a covert release of a biothreat agent. In addition, they are required by first responders and clinicians to test environmental and clinical samples in a known release. However, the paucity of comprehensive evaluative information about these systems prevents conclusions about whether or not one or more of these systems is likely to be useful for these purposes.
The evidence on detection systems was descriptive and predominantly collected from government sources and manufacturers' Web sites. We note that the definitions of what constitutes a "rapid" or "portable" test varied widely. We found no reports that directly compared 2 or more of the commercially available systems in any given category. Additionally, few of these systems have been compared to a gold standard, and their sensitivity and specificity remain poorly characterized. A significant gap in the literature is an analysis performed by an independent research group comparing the most promising technologies to each other and to the gold standard. For most systems, the available information does not describe if reagents are sold with the detector or if they are widely available. We conclude that potential users of these systems must carefully evaluate the data derived from them and consider strategies that include the use of these systems for rapid detection in conjunction with the slower but better-validated methods used in reference laboratories.
General Diagnostic DSSs
Background
General diagnostic DSSs are designed to assist clinicians in generating a list of possible diagnoses for a given patient. For such systems to be useful in the event of a covert bioterrorist attack, they should prompt clinicians to consider biothreat agents as a potential cause of the patient's symptoms. In this way, these systems may increase the clinician's suspicion of bioterrorism, thereby increasing the probability that the clinician performs appropriate diagnostic testing. Most of these systems require that the clinician enter information about the patient's signs and symptoms. Typically, the diagnostic DSS then produces a differential diagnosis or list of possible diagnoses for the patient. These diagnoses are sometimes ranked according to the likelihood of disease. Alternatively, some DSSs provide a calculated probability score for each diagnosis, often based on a clinical prediction rule.
Evaluation criteria
We evaluated each of the reports of general diagnostic DSSs for the following information (Table 2 -- Diagnosis; Evidence Table 4): the purpose of the system, the type of information required by the DSS (e.g., a manually-entered list of signs and symptoms provided by the clinician), the type of information provided by the DSS (e.g., a list of differential diagnoses with or without associated information about the diseases of interest), diagnostic sensitivity and specificity, whether the biothreat agents and their associated illnesses are included in the knowledge base, the method of reasoning used by the inference engine, information regarding the ability to update the probability of biothreat-related illness as the epidemic progresses, and the type of hardware required.
Findings
Our search found 6 currently available general diagnostic DSSs, 3 of which have been clinically evaluated and presented in peer-reviewed reports (Tables 3 and 8; Appendix H).
The purpose of each of these systems is to provide a differential diagnosis based on patient-specific signs and symptoms. Because general diagnostic DSSs typically provide a list of several possible diagnoses, in the event of unrecognized bioterrorism-related illness, even if the system fails to rank the correct diagnosis first, but ranks it among the top few diagnoses, this may prompt a clinician to order a diagnostic test for a biothreat agent.
All of the general DSSs require manual entry of patient information by clinicians. They then use either Bayesian (probabilistic) and/or rules-based methods to compare the patient's information with their knowledge base to generate a differential diagnosis that is typically ranked in descending order of likelihood. Some of the systems provide additional information about the suspected diseases and suggest appropriate tests if clinicians choose to pursue these diagnoses. Most of the general DSSs are available for use on personal computers, although a handheld version of DiagnosisPro® is also available. None of the reports described if it was possible to update the probability of biothreat-related illness as the epidemic progresses. No study of a general diagnostic DSS has specifically evaluated the performance of these systems for the diagnosis of biothreat-related illness.
DXplain™, Iliad, and Quick Medical Reference (QMR) were directly compared in a multi-center trial of 105 diagnostically challenging cases. 126 The DXplain™ knowledge base contained the correct diagnosis for 96 cases (91 percent); the Iliad knowledge base contained the correct diagnosis for 80 cases (76 percent); and the QMR knowledge base contained the correct diagnosis for 77 cases (73 percent). DXplain™ correctly included the ultimate diagnosis in 72 cases (69 percent) with an average rank of 12.4, compared with 64 cases (61 percent) with an average rank of 10.4 for Iliad and 55 cases (52 percent), at an average rank of 6.6 for QMR. (The clinical significance of this difference in rank is not clear. The importance of rank depends on how this information is used. For example, if the clinician only scans the top 5 diagnoses or if the DSS only prints out the top 10 diagnoses, then the rank may well be important. If, however, the clinician reviews the entire list of possible diagnoses specifically seeking the unusual diseases that he or she had not previously considered as a means of enhancing their diagnostic capabilities, then rank is less important.) When considering only the 63 cases for which the correct diagnosis was present in all systems, Dxplain™ identified the correct diagnosis in 50 cases (79 percent) at an average rank of 11.7. Iliad was correct in 48 cases (76 percent) at an average rank of 10.2 and QMR correctly identified the final diagnosis in 45 cases (71 percent) with an average rank of 5.4.
The other evaluations of the general diagnostic DSSs differed with respect to their study designs. Some evaluated physician acceptance of the system. However, high acceptance does not necessarily mean that a clinician would use the system for routine cases (such as a patient presenting with a flu-like illness, a common early presentation of many biothreat-related illnesses). Other study designs addressed the observed phenomenon that different clinicians use a different set of diagnostic terms to describe the same patient.127-129 These differences may result in the DSS producing differing lists of diagnoses. Therefore, some studies compared the terms input into a system by different clinicians, given the same case, and the resulting differential diagnoses.
As of the publication of this Report, the manufacturer of Iliad has stopped selling and providing technical support for that system. 130 We have nonetheless included Iliad in this section because it continues to be available through some retailers, and clinicians continue to use this product.
Summary: General diagnostic DSSs
The role of general diagnostic DSSs in a bioterrorism response is to enhance the likelihood that clinicians consider the possibility of bioterrorism-related illness. Therefore, these systems could contribute to the detection of a previously unrecognized release of biothreat agents. However, the reports of general diagnostic DSSs have several important limitations that prevent conclusions regarding their ability to serve this role. First, none of the DSSs has been evaluated formally with respect to bioterrorism response. Second, all of these systems require laborious manual entry of patient findings, which may be a substantial barrier to use in clinical settings. Efforts to link general diagnostic DSSs to other hospital information systems, if successful, would reduce the data entry burden substantially. In addition, availability of the system on a handheld computer (as for DiagnosisPro®) might make the system more convenient for clinicians to use. Third, available evaluations do not indicate whether disease caused by biothreat agents are included in the databases for many systems. Thus, we were not able to assess the extent to which biothreat agents are included in any of the general diagnostic DSSs knowledge bases or whether the systems are updated with new information about the clinical presentations of these diseases (except that Iliad has not been updated since 1997). Fourth, general diagnostic systems that use probabilistic information about the likelihood of disease will have inappropriately low pretest probabilities for biothreat agents in the event of a bioterrorism event. To provide a ranking of differential diagnoses, the system relies on estimates of the prevalence or probability of diseases. If a biothreat outbreak was known or strongly suspected, the pretest probability for these agents would change dramatically from the probabilities appropriate during routine clinical use. It would be helpful if the knowledge base could be updated to reflect changes in the likelihood of diseases based on local public health data (i.e., if the system were automatically updated with local incidence and prevalence information) or could be modified in the context of a known bioterrorism event.
Radiologic Systems
Background
Because many biothreat agents cause pulmonary disease, chest X-rays would be a common diagnostic procedure performed on patients presenting after a bioterrorism event. Interstitial disease would be the most likely finding. In the case of inhalation anthrax, a widened mediastinum may be seen; however, this is not always present, even in some advanced cases. 152
Radiology interpretation systems may increase the diagnostic accuracy of radiographic reports. For this Report, we limited our search to those technologies that could be used to automate the interpretation of radiologic images for the diagnosis of biothreat agents. For example, we excluded those systems that detect mammographic lesions or pulmonary nodules. In this section, we discuss 2 types of systems -- those that assist clinicians in the interpretation of radiographic images, and those that use natural language processing methods to abstract information from the reports of radiographic procedures for diagnostic purposes.
Evaluation criteria
We evaluated each of the reports of radiologic DSSs for the following information (Table 2 -- Diagnosis; Evidence Table 4): the purpose of the system, the type of information required by the DSS (e.g., the actual radiological image or the text of a radiologist's report of the image), diagnostic sensitivity and specificity, whether the biothreat agents and their associated illnesses are included in the knowledge base, whether the system uses a standard vocabulary, the method of reasoning used by the inference engine, information regarding the ability to update the probability of biothreat-related illness as the epidemic progresses, the type of hardware required, and the system's security measures.
Findings: Radiologic interpretation systems
Our search found 2 radiologic interpretation systems, 1 of which has been clinically evaluated and described in a peer-reviewed article (Tables 3 and 9; Appendix H).
The first system, described in 3 evaluation articles, scans digitized radiographs for abnormal regions to assist clinicians in the identification of pulmonary infiltrates.153-155 These studies calculated receiver operating characteristic (ROC) curves for each of the systems under evaluation. ROC curves are a plot of the sensitivity of a diagnostic test (typically on the y-axis) against 1 minus its specificity (typically on the x-axis). Because the ideal diagnostic test is 100 percent sensitive and 100 percent specific, the area under an ideal ROC curve would be equal to 1. Minimal improvements in the area under the ROC curve were shown when computer-aided diagnosis was employed for a small set of radiographs. Since the vast majority of infiltrates will not be related to biothreat agents, it remains unclear whether this technology can be translated into improved detection of bioterrorism-related illness.
Researchers at the same institution also found that using an artificial neural network can improve the performance of radiologists in the differential diagnosis of interstitial lung disease. 155 When chest radiographs were viewed in conjunction with network output, the average area under the ROC curve increased from 0.83 to 0.91. The clinical significance of such a change is not clear.
None of the reports described whether the biothreat agents and their associated illnesses are included in the knowledge base, whether the system uses a standard vocabulary, information regarding the ability to update the probability of biothreat-related illness as the epidemic progresses, the type of hardware required, or the system's security measures.
Findings: Natural language processing systems
Natural language processing techniques have been developed to automate identification of disease concepts in free text such as radiology reports. We found reports of 2 such systems, 1 of which has been clinically evaluated and described in a peer-reviewed article (Tables 3 and 10; Appendix H).
Table
Table 10. Natural language processing systems.
The purpose of these programs is to search electronic text for concepts related to pneumonia, and then either alert the clinician or incorporate this information with other data from the electronic medical record into diagnostic or management applications. Neither of the medical language processing systems that we found was specifically designed to diagnose bioterrorism-related illness. None of the reports of these systems described whether the biothreat agents and their associated illnesses were included in their knowledge bases or whether the systems used standard vocabularies, nor did they provide information regarding the ability to update the probability of biothreat-related illness as the epidemic progresses, specify the type of hardware required (minimally, each required an electronic medical record system), or describe the system's security measures.
Two studies have evaluated the ability of medical language processing systems to identify relevant concepts in radiology reports. SymText is a medical language processing system developed by the Latter Day Saints (LDS) Hospital at the University of Utah (an additional description of this system is provided in the Management section of this chapter).156, 157 In one study, researchers compared the ability of SymText to identify pneumonia-related concepts in 298 X-ray reports with those of 2-word search programs, a layperson, and a resident physician. SymText performed better than the word search programs and the layperson but similar to the resident physician. 156 A similar system was evaluated at the Columbia-Presbyterian Medical Center in New York. 157 This study compared differences in the interpretation of 200 radiology reports by groups of 2 physicians, groups of laypersons, and the natural language processor. The differences between the interpretations of the natural language processor and the physicians were similar to the differences among physicians. 157 This suggests that the natural language processor's ability to identify these concepts was similar to that of the physicians.
Researchers at LDS Hospital have integrated SymText into a real-time DSS designed to implement guidelines for community-acquired pneumonia. 158 The radiology department uses speech recognition technology that decreases the time necessary to transcribe radiographic reports. 158 As soon as the radiologist completes his or her dictation, SymText identifies patients who may have pneumonia based on their radiology reports and assesses the severity of their pneumonia. 158 These findings are combined with other clinical and laboratory data to generate management recommendations to clinicians in compliance with clinical practice guidelines. An evaluation of this automated guideline showed that SymText was similar to physicians in identifying patients eligible for the guideline, but worse than the physicians in extracting information about the location and extent of the infiltrates (patient outcomes were not assessed). 158
Summary: Radiologic systems
Our search identified 4 IT systems designed to improve radiographic diagnoses or incorporate data from radiology reports into diagnostic or management DSSs. Their utility in recognizing illnesses caused by bioterrorism is unknown, as none has been formally evaluated for this purpose.
The system from the University of Chicago has established utility for the diagnosis of community-acquired pneumonia. However, because the radiologic findings for most bioterrorism-related illness will be identical to pulmonary diseases of other etiologies and because the presence of a specific radiologic finding associated with bioterrorism-related illness is the exception rather than the rule, it is not clear that these systems could help clinicians, beyond alerting them to the presence of a pulmonary infiltrate, pleural effusion, or widened mediastinum. For radiologic systems to have a significant effect on clinicians' diagnostic decisions in regards to a bioterrorism event, they would have to raise the clinician's index of suspicion that a biothreat agent may be causing the radiologic findings. Incorporating information from these systems with other information from patients' medical records and knowledge bases about the clinical presentations of bioterrorism-related illnesses could be a useful innovation. Specifically, radiologic systems could serve as a component of an integrated management system that incorporates radiologic as well as other clinical information with clinical practice guidelines for the management and reporting of suspected bioterrorism-related illness.
Diagnostic Systems Using Telemedicine
Background
Telemedicine is the use of telecommunications technology for medical diagnostic, monitoring, and therapeutic purposes when distance separates the users. 160 We direct readers interested in this topic to a recent Evidence Report from AHRQ entitled "Telemedicine for the Medicare Population." 160 Briefly, this Report describes 3 types of telemedicine systems: "(1) store-and-forward services that collect clinical data, store them, and then forward them to be interpreted later; (2) self-monitoring/testing services that enable clinicians to monitor physiologic measurements, test results, images, and sounds, usually collected in a patient's residence or care facility; and (3) clinician-interactive services that are real-time distance clinician-patient interactions." 160 They found that telemedicine consults increased steadily throughout the 1990s with most programs designed to serve rural populations, veterans, and the elderly. 160 Additionally, they report that teledermatology is the most-studied clinical specialty in store-and-forward telemedicine; its diagnostic accuracy and patient management decisions are comparable to those of in-person clinical encounters. 160
Evaluation criteria
We evaluated each of the reports of telemedicine systems for the following information (Table 2 -- Diagnosis; Evidence Table 4): the purpose of the system, the settings in which they are used, the sensitivity and specificity of the diagnoses provided by consultants using the system, the type of hardware required, and the system's security measures.
Findings
Our search identified 4 telemedicine/teleradiology systems with potential relevance to bioterrorism; clinical evaluations for 2 of these have been presented in peer-reviewed evaluations (Tables 3 and 11; Appendix H). Since our search strategy was neither designed specifically for telemedicine nor teleradiology, the systems identified may not be representative of the systems that are available. We present these systems in this section because they are related to the radiologic interpretation systems just described, although they share many similarities with the communication systems described later in this Report.
Table
Table 11. Diagnostic systems using telemedicine.
Three of the 4 telemedicine systems were designed by the military to provide telemedicine consultation for military personnel at sites distant from military hospitals. Similarly, the other system, MERMAID, was designed for the European Union to provide telemedicine consultations to members of the merchant marine. None was designed or evaluated specifically for providing telemedicine consultations for disease resulting from bioterrorism. The sensitivity and specificity of the diagnoses provided by consultants using the system, the type of hardware required, and the system's security measures were not described in any report.
The Walter Reed Army Medical Center (WRAMC) Telemedicine Service system was evaluated in a retrospective case review of 171 telemedicine consultations. 161 Of these, 114 consults were reviewed: 39 percent were for dermatology, 16 percent for surgical subspecialties and 15 percent for orthopedics. Telemedicine was felt to affect the diagnosis in 30 percent, the treatment in 32 percent, and the overall patient status in 70 percent of cases.161, 162
Summary: Diagnostic systems using telemedicine
No telemedicine system has been evaluated specifically for bioterrorism. Telemedicine systems are most useful in areas with limited direct access to medical specialists. Since acts of bioterrorism against civilian populations may be less likely to occur in remote areas than in population centers, these systems may be of limited value against bioterrorism. However, since few practicing primary care or emergency physicians have ever seen the rashes associated with smallpox or other bioterrorism-related illness, the use of teledermatology technologies may increase the likelihood of a timely diagnosis by facilitating access to dermatologic experts. In the event of a widespread epidemic reaching geographically isolated areas, existing telemedicine infrastructures could be used by public health officials to relate public health information and alerts to clinicians.
Other Diagnostic Systems
Background
In this section, we present a variety of other types of diagnostic systems. Unlike the general diagnostic DSSs discussed earlier, most of these systems are specifically for the diagnosis of infectious diseases (thereby limiting their use to only those circumstances in which the clinician suspects an infectious etiology).
Evaluation criteria
We evaluated each of the reports of other kinds of diagnostic DSSs for the following information (Table 2 -- Diagnosis; Evidence Table 4): the purpose of the system, the type of information required by the DSS, diagnostic sensitivity and specificity, whether the biothreat agents and their associated illnesses are included in the knowledge base, whether the system uses a standard vocabulary, the method of reasoning used by the inference engine, information regarding the ability to update the probability of biothreat-related illness as the epidemic progresses, the type of hardware required, and the system's security measures.
Findings
In this section, we present a brief description of 9 other diagnostic systems, 7 of which have been described in at least 1 peer-reviewed evaluation (Tables 3 and 12; Evidence Table 2; Appendix H).
Table
Table 12. Other diagnostic DSSs.
The included systems were designed for a variety of purposes: 4 diagnostic DSSs specifically for infectious diseases (The Computer Program for Diagnosing and Teaching Geographic Medicine, GIDEON, a fuzzy logic program to predict the source of bacterial infection from demographic variables, and the Texas Infectious Disease Diagnostic DSS); 2 systems that facilitate the prompt diagnosis of patients with active pulmonary tuberculosis (the first is a neural network-based system from the State University of New York at Buffalo, and the other is based on natural language processing of electronic medical record information from Columbia University); and 3 diagnostic systems with other purposes. We included the tuberculosis diagnostic systems primarily because tuberculosis serves as a model for bioterrorism-related agents that present as pneumonia and require respiratory isolation during the initial treatment period. Those systems that incorporate diagnostic functions with management recommendations are not presented in Table 12 but are described with Management and Prevention systems later in this Report. The methods used by these systems to generate diagnoses include probabilistic and rules-based inference engines and neural networks.
Of all the diagnostic DSSs, we could only verify that GIDEON and The Computer Program for Diagnosing and Teaching Geographic Medicine specifically include most of the worrisome bioterrorism-related organisms in their knowledge bases.168, 169 Both of these systems provide differential diagnoses of infectious diseases based on clinical parameters regarding a patient that are entered into the program. The Computer Program for Diagnosing and Teaching Geographic Medicine also provides general information about infectious diseases, anti-infective agents, and vaccines.
The evaluation of GIDEON compared the diagnostic accuracy of the DSS to that of medical house officers admitting 86 febrile adults to the Boston Medical Center. The house officers listed the correct diagnosis first in their admission note 87 percent (75/86) of the time compared with 33 percent (28/86) for GIDEON. 169 In a study to evaluate the diagnostic accuracy of The Computer Program for Diagnosing and Teaching Geographic Medicine, 6 infectious disease specialists (blinded to the patients' actual diagnoses) were asked to record all positive and negative clinical data for 295 consecutive patients with established diagnoses and 200 hypothetical cases. The computer program correctly identified 75 percent (222 of 295) of actual cases and 64 percent (128 of 200) of hypothetical cases. The clinical diagnosis was included in the computer differential diagnosis list in 94.7 percent of cases. Among the cases included in this evaluation, several were for the causative agents of: anthrax, brucellosis, cholera, cryptosporidiosis, Hantavirus respiratory distress syndrome, Lassa fever, plague, Q fever, Rocky Mountain spotted fever, shigellosis, and tularemia. However, this system was only tested on cases for which the diagnosis was known; therefore, there is no information on how it would perform for cases with unknown outcomes. 168
If a system produces a single diagnosis for a given case as its output, the sensitivity and specificity of the system can be readily determined if the case's actual diagnosis is known. Frequently, systems are designed to provide a list of many possible diagnoses, often ranked according to their probability of being the actual diagnosis. Under these circumstances the clinician will have to determine the lower threshold of probability for which they will make a diagnostic or therapeutic decision (i.e., if a system generates a list of possible diagnoses for a case and suggests that smallpox is on the differential but highly unlikely, he or she may not choose to send a viral culture or notify the local public health official). When diagnostic systems provide a list of possible diagnoses, it may be more appropriate to calculate receiver operator characteristic (ROC) curves to evaluate the performance of the system over a range of probability thresholds. We direct interested readers to an article by Fraser and colleagues measuring the performance of systems that generate differential diagnoses using ROC curves and other methods. 170 Only the neural network for the diagnosis of active pulmonary tuberculosis was evaluated with ROC curves.
For those programs that require a user to input case-specific information, we again found that the differential diagnoses provided by the systems were highly dependent upon the information input about the cases. This was particularly true for DERMIS, where generalists' inputs were less likely than those of specialists to result in a correct diagnosis. This is an unfortunate finding because patients with bioterrorism-related skin lesions are more likely to present to general clinicians than dermatologic specialists and because the early recognition of skin lesions associated with smallpox, Glanders, bubonic plague, and tularemia could significantly reduce bioterrorism-related morbidity and mortality.
None of the reports of these general DSSs discussed barriers to the use of the systems, whether the system uses a standard vocabulary, information regarding the ability to update the probability of biothreat-related illness as the epidemic progresses, the type of hardware required, or the system's security measures.
Summary: Other diagnostic systems
The role of this heterogeneous group of diagnostic DSSs in a bioterrorism response is to enhance the likelihood that clinicians consider the possibility of bioterrorism-related illness. Therefore, these systems could contribute to the detection of a previously unrecognized release of biothreat agents. However, the reports of general diagnostic DSSs have several important limitations that prevent conclusions regarding their ability to serve this role.
As was true for the general diagnostic DSSs, if cases associated with biothreat agents are not included in the system's knowledge base, the diagnosis of bioterrorism-related illness will not be included in a system's differential diagnosis. GIDEON and The Computer Program for Diagnosing and Teaching Geographic Medicine are the only systems for which we were able to obtain a complete list of the diseases included in the knowledge base and could verify that all potential biothreat agents were included. All of the systems presented in Table 12 are limited in that they are not general diagnostic systems but specific for either infectious diseases or another specialized application; thus, if the patient does not present with either a fever or a rash, the clinician may not choose to use these specialized DSSs. Additionally, many of these systems require clinicians to manually enter data -- a laborious step that may be a barrier to the use of these systems and has been demonstrated to increase inter-user variability.
Management and Prevention Systems
Background
In this section, we discuss the systems designed to assist clinicians and public health officials in making management and prevention decisions. Most of the systems included in this section are designed to make recommendations to clinicians, not to public health officials. Typically, they abstract data from an electronic medical record, apply a set of rules, and generate patient-specific management and prevention recommendations. In general, these systems are limited to institutions with electronic medical records and robust medical informatics infrastructures.
Evaluation criteria
We evaluated each of the reports of management systems for the following information (Table 2 -- Management and Prevention; Evidence Table 4): the purpose of the system, the type of information required by the system (e.g., patient information from an electronic medical record), the type of information provided by the system (e.g., antibiotic or quarantine recommendation), information about the manner in which the management recommendations are provided (e.g., whether the recommendations are provided in an unprompted manner to the user), timeliness of management recommendation, the accuracy of the management recommendations, whether the biothreat agents and their associated illnesses are included in the knowledge base, the method of reasoning used by the inference engine, whether the system uses a standard vocabulary, information regarding the ability to update recommendations as the epidemic progresses, type of hardware required, and the system's security measures.
Findings
In this section, we describe 18 systems designed to make management or prevention recommendations; 10 of these have been described in at least 1 clinical evaluation report (Tables 3 and 13; Evidence Table 3; Appendix H). We found no systems specifically designed to provide recommendations to clinicians or public health officials for management of a bioterrorist attack. None of the reports of the management or prevention programs stated that bioterrorism-related diseases were included in their clinical practice guidelines, prediction rules, or knowledge bases. Most of the management systems described in this section provided recommendations at the point of care -- typically, when the clinician entered the electronic medical record of the patient in question. These systems are therefore relatively timely. The reasoning used by these systems varies, including both probabilistic and rules-based methods. Few reports specified whether the system uses a standard vocabulary, whether it would be possible to update management recommendations as the epidemic progresses, the type of hardware required, or the system's security measures (although most are associated with hospital-based electronic medical records that require a user login).
The management DSS that has been the topic of the most numerous descriptive articles and clinical evaluations is the Health Evaluation through Logical Processing (HELP) system at LDS Hospital in Salt Lake City.156, 158, 180-215 The HELP System is a complete computer-based hospital information system designed to support applications including order entry/charge capture, pharmacy, radiology, nursing documentation, and intensive care unit (ICU) monitoring as well as to maintain robust decision support functions. Decision support has been used to provide alerts and reminders, to make patient diagnosis and management recommendations, and to implement clinical protocols. Specifically, it alerts clinicians about infections in normally sterile body sites (e.g., from urine and blood cultures), makes antibiotic recommendations, suggests appropriate timing and duration of prophylactic antibiotics, and identifies adverse drug reactions (as such, the system has considerable similarities to some of the communication systems discussed later in this Report). HELP also generates alerts for infections that are required by law to be reported to state or national public health officials. Earlier in this Report, we briefly described the elements of the HELP program that search radiology reports for pneumonia-related concepts and its decision support module for the diagnosis of community-acquired pneumonia. 156 We also described SymText, a medical language processor developed at the LDS Hospital to analyze free text reports. In this section, we describe 3 additional HELP functions that could serve the information needs of clinicians or public health officials in the event of a bioterrorist attack: its antibiotic management program, its Data Mining Surveillance System (DMSS) for the detection of hospital-acquired infections, and its community-acquired pneumonia diagnosis and management program.
The HELP system has multiple antibiotic protocols to provide comprehensive management recommendations for all antibiotic agents used in the LDS hospital system, including those for prophylactic, empiric, and therapeutic purposes. The HELP investigators convert local physician-derived antibiotic prescribing guidelines regarding drug choice, dosage, and timing into rules, algorithms, and predictive models. These guidelines are combined with 4 knowledge bases to provide timely, patient-specific management recommendations. The knowledge bases include the following information: (1) the probabilities of infectious diseases based on signs, symptoms, risk factors, and diagnostic test results; (2) the expected courses of infectious diseases in terms of morbidity and mortality if left untreated; (3) the expected courses of infectious diseases if treated optimally; and (4) the fraction of patients with each infectious disease expected to respond to each intervention under consideration. 215 During a 7-year study period of its antibiotic management system, HELP improved the timeliness of administration of prophylactic antibiotics. 203 The proportion of patients receiving their first dose within 2 hours before surgical incision increased from 40 percent in 1985 to 99 percent in 1994. During the same period, the rate of antibiotic-associated adverse events decreased from 27 percent to 19 percent, the adjusted antibiotic cost per treated patient decreased from $123 to $52, and antibiotic resistance patterns remained stable. 203 Another study reported reductions in costs of antibiotics, total hospital costs, and length of stay for those patients who always received the antibiotics recommended by HELP compared with the pre-intervention cohort and those who did not always receive the HELP-recommended regimen. 196 Interpretation of this analysis is complicated by the fact that the post-intervention group was divided into those patients who always received recommended antibiotics and those who did not. Patients for whom clinicians chose to override the recommendations of HELP may have been more medically complicated and would, therefore, be expected to have longer, more costly hospitalizations.
For the purpose of surveillance, HELP uses the DMSS, a novel approach for identification of unusual patterns in data.186, 191 The program reviews data from a variety of sources including the microbiology laboratory, nurses' charts, chemistry laboratory, surgical records, and pharmacy to identify association rules over time. For example, a decision rule could be developed to describe the conditional probability of multi-drug resistant Pseudomonas given all Pseudomonas isolates in the ICU. This decision rule can then be used to identify unexpected patterns by statistically comparing the rates of multi-drug resistant Pseudomonas in each time interval with those that preceded it. A daily report detailing any suspicious outbreaks is sent to the hospital epidemiologist.186, 191
HELP has a rapid decision support module designed to identify and manage patients with community-acquired pneumonia on presentation to the emergency department. 182 The HELP system notifies the DSS whenever a new patient registers in the emergency department. The DSS then queries HELP's databases every 5 to 10 minutes for any new clinical information on that patient. It can retrieve up to 42 data elements from which it calculates the probability of community-acquired pneumonia and a severity index score and presents these to clinicians in the emergency department caring for that patient. In an evaluation of this system, the authors compared the diagnostic accuracy of a Bayesian network and an artificial neural network using data from the HELP databases. 182 They set the diagnostic threshold to achieve a sensitivity of 95 percent and calculated the specificity (SP), positive predictive value (PPV), and negative predictive value (NPV) of the Bayesian network and the artificial neural network. For the Bayesian network: SP = 92.3 percent, PPV=15.1 percent, NPV=99.9 percent. For the artificial neural network: SP = 94.0 percent, PPV=18.6 percent, NPV=99.9 percent. These values suggest that the neural network performed somewhat better than the Bayesian network; however, it remains unclear if this difference implies a significant difference in clinical outcomes for pneumonia patients.
In addition to HELP, we found 17 other management and prevention systems with potential utility during a bioterrorism event: 5 antibiotic recommendation programs, 3 ICU management systems, 1 pneumonia management system, and 8 systems that generate a variety of other management recommendations (Table 13 and Evidence Table 3). ePocrates™, a drug-recommendation program with innovative communication capabilities, is presented in the Reporting and Communication section of this Report.
Table
Table 13. Management and prevention systems.
In general, the antibiotic recommendation programs provide differential diagnoses from infectious disease databases and make patient-specific antibiotic recommendations often taking into consideration cost, pathogen prevalence, and susceptibility patterns. The evaluation evidence for 4 of these systems is mixed: half of them recommended antibiotics with narrower spectra than the clinicians would have otherwise used. This may be the intention of the developers of these systems since it could reduce the general problem of escalating antibiotic resistance. However, if clinicians make antibiotic selection decisions while unaware of the true bioterrorism-related diagnosis and select narrow-spectrum antibiotics, they may not adequately treat the pathogens.
The 2 evaluations of the intensive care management programs suggest inadequate acceptance by users and high false positive rates, limiting their potential utility at this time. However, the results of the system from Queens College that calculates Severity Scores for Community-acquired Pneumonia demonstrated greater diagnostic accuracy. 216 The system generates severity-based pneumonia management recommendations from a clinical prediction rule. The evaluation of this system compared its diagnostic accuracy with an independent expert for 79 patients with community-acquired pneumonia cases. Depending on the information it used to calculate the severity score, the system achieved sensitivities of 87 to 92 percent and specificities of 93 to 98 percent. It was 80 percent accurate in assigning the exact risk class, with the remaining 20 percent differing by only 1 class. 216
Summary: Management and prevention systems
The systems included in this section are designed to make recommendations to clinicians by abstracting clinical information from electronic medical records to make patient-specific recommendations. None of the 18 systems described in this section has been specifically designed or evaluated for utility in providing management or prevention recommendations during a bioterrorism event. We have no information as to whether the knowledge bases and inference engines of these systems include comprehensive information about bioterrorism-related illness. Moreover, none of the evaluations describes effects on patient outcomes other than length of stay in the hospital.
The systems that are not linked to electronic medical records share many of the limitations of the general diagnostic systems -- including, that clinicians may not use the system to seek advice for patients presenting with common viral syndromes (i.e., the bioterrorism-related syndromes). Expert systems that continuously search electronic medical records (including data from the laboratory, radiology reports, and physician notes) for new evidence of an infection and apply clinical practice guidelines to those data have potential utility in bioterrorism management. However, this requires relatively robust hospital IT infrastructures and the incorporation of clinical practice guidelines for biothreat-related illnesses.
Surveillance Systems
Surveillance is the collection, consolidation, and evaluation of morbidity, mortality and other relevant data...and its regular dissemination to all who need to know.
-- A. Langmuir, 1963 236
Surveillance is the routine collection and analysis of relevant data, and their distribution to clinicians, public health officials, and others in the community who use them to take action to prevent further morbidity or mortality. 237 Surveillance data serve the following purposes: to discover the natural incidence of the events under surveillance, to detect abnormal situations that require epidemiologic control measures, to direct preventive actions, to guide resource allocation, and to assess interventions. 237 The most important aspect of a surveillance system is not the nature of the events under observation; rather, it is the ability to detect an outbreak at a stage when intervention may affect the expected course of events. Therefore, the ideal surveillance system for the detection of a covert bioterrorist attack would collect data that are both sensitive and specific for biothreat agents and provide reports of these data to public health officials as soon after the dispersal of the agent as possible.
In the event of a covert aerosol release of a biothreat agent, exposed people will initially present with minor symptoms. Exposed individuals may stay home from work or school, buy over-the-counter medications, or perhaps telephone a triage nurse. During subsequent days their symptoms may worsen, prompting them to seek care from their physicians. Physician visits may in turn result in the use of prescription drugs. If patients become acutely ill, they may call 911, or present directly to an emergency department. A visit to the emergency department may result in laboratory tests or admission to the hospital. The decisions patients make and the data their behaviors generate form the basis of surveillance systems for the early detection of a covert bioterrorist attack.
Data from the intelligence community, environmental detection systems, surveillance reports about food quality, and reports of zoonotic illnesses in livestock, poultry, and wildlife would likely provide the earliest indication of a covert biothreat agent release (Figure 2). Data regarding school and work absenteeism, records of phone calls to triage nurses, and over-the-counter pharmacy sales could provide additional early warnings. Surveillance systems that rely on sentinel clinicians reporting suspicious patients to their local public health officials and pharmacies reporting prescription sales will detect an outbreak somewhat later. Less timely surveillance systems rely on data from emergency departments, 911 calls, laboratories, and hospitals. Because the window period during which antibiotics and antiviral agents are effective against several biothreat agents ends before the patient becomes sick enough to be admitted to the hospital, the outbreak has to be detected early so that effective therapies can be started and excess morbidity and mortality prevented.
In addition to timeliness, these data sources differ in 2 additional ways. First, they differ with respect to their sensitivity and specificity for the detection of biothreat agents. Whereas a large increase in the sales of over-the-counter cold-and-flu preparations may be sensitive for an aerosol biothreat-agent release, it is not likely to be specific. Conversely, viral cultures may be highly specific but not sensitive, particularly if the organism is difficult to collect properly or to grow in culture. Second, these data sources differ with respect to the ease with which they can be collected. For example, some data (such as ICD9 codes) are already routinely collected in electronic format and may be amenable for use by a surveillance system. In contrast, clinicians do not typically report the chief complaint, signs, and symptoms of patients presenting to their clinics; therefore, the routine collection of syndromal surveillance data could significantly add to clinicians' workloads. Additionally, if the data are collected electronically in a systematic manner, they are less likely to be incomplete or to contain errors.
The CDC, the primary agency responsible for the collection of disease surveillance data in the U.S., is working to create an integrated surveillance system -- a "system of systems" that combines many of the existing laboratory and clinical surveillance systems. Through its collaborations with its public health partners (e.g., Association of Public Health Laboratories (APHL), Association of State and Territorial Health Officials, Council of State and Territorial Epidemiologists (CSTE), and National Association of County and City Health Officials), the CDC maintains over 100 separate surveillance and health information systems (not all for infectious diseases). 238 Most of these systems are specific for a particular category of diseases (e.g., sexually transmitted diseases) and are not linked even to related diseases. One example of the integrative efforts of the CDC is the Laboratory Response Network, which now consists of 120 state and local public health laboratories linked to advanced capacity laboratories including clinical, military, veterinary, agricultural, water, and food-testing laboratories. 239 The Laboratory Response Network is described in greater detail in the Laboratory section of this chapter.
To improve data collection and sharing for surveillance purposes at the state and local levels, and to enhance the ability of the public health system to respond to public health threats, the CDC is implementing the National Electronic Disease Surveillance System (NEDSS). 238 This system has not yet been fully deployed or clinically evaluated. When it is completed, NEDSS will include data and information system standards, an Internet-based communications infrastructure, and policy-level agreements on data access, sharing, and security. The NEDSS architecture is designed to enhance the ability to electronically link individual surveillance activities, to improve the integration of information reporting into the provider's workflow through automatic transfer of data from clinical information systems, and to facilitate accuracy and timeliness of surveillance reports. 238 It is intended to integrate and replace several current CDC surveillance systems, including the National Electronic Telecommunications System for Surveillance (NETSS), the HIV/AIDS reporting system, the vaccine preventable disease system, and systems for tuberculosis and infectious diseases. 238
Global infectious disease surveillance is carried out through a loose framework of formal and informal arrangements that the World Health Organization (WHO) characterizes as a "network of networks." 240 The formal partners in this effort include: WHO regional and country offices; national public health authorities; United Nations Children's Fund (UNICEF) and United Nations High Commissioner for Refugees (UNHCR) country offices; WHO collaborating centers/laboratories; epidemiology training networks; and military laboratory networks. 241 The informal contributions come from non-governmental organizations, the media, Internet discussion sites, and the Global Public Health Intelligence Network (GPHIN). 241 Surveillance systems in most nations are limited by a lack of adequately staffed and equipped laboratories; surveillance data already collected in electronic form that can be readily imported into analysis algorithms; trained personnel to collect and analyze additional surveillance data; and information infrastructures to support the communication of surveillance data from remote collection sites to a central analysis site. 241 These limitations are most significant in developing countries. 241 Weaknesses in the detection and response capabilities in poorer countries affect the ability of international efforts to detect and control infectious disease outbreaks. 241
In 1947, an American businessman returned to New York from a sightseeing trip to Mexico during which he had become infected with smallpox. 242 Nine days after arrival, he died, having infected at least 12 others, 2 of whom also died. 242 As a result of concerns that the disease would spread further, more than 6 million people in New York were vaccinated within a month. 242 Given the current state of international travel, importation of food products, and trade in pharmaceuticals and blood products, it is in the best interest of the U.S. to support global surveillance efforts. Infectious diseases from natural sources (such as the as yet unidentified animal reservoirs of Ebola) or from acts of bioterrorism can readily travel from the remotest parts of the world to the U.S.
Since most surveillance systems are identified by the type of data that they collect, we have organized this section accordingly. We found 90 surveillance systems: 7 collecting syndromal data; 6 collecting clinician reports; 11 collecting influenza-related data; 23 collecting laboratory or antimicrobial resistance data; 16 collecting hospital-based infections data; 10 collecting foodborne illness data; 6 collecting zoonotic and animal illness data; and 11 collecting other kinds of surveillance data. We conclude this Surveillance section with a brief description of the standard methods for analysis and presentation of surveillance data.
Evaluation criteria
We evaluated each of the reports of surveillance systems for the following information (Table 2 -- Surveillance; Evidence Table 5): the purpose of the system, the type and method of surveillance data collected by the system; timeliness of data collection, analysis and presentation to the decision maker; methods for determining when an outbreak has occurred; geographic area under surveillance; the type of hardware required; the system's security measures; and information regarding the public health importance of the health event under surveillance, the system's usefulness, simplicity, flexibility, acceptability, sensitivity, specificity, representativeness, and the direct costs needed to operate the system.
Surveillance Systems Collecting Syndromal Reports
Background
The earliest signs and symptoms caused by most biothreat agents are flu-like illness, acute respiratory distress, gastrointestinal symptoms, febrile hemorrhagic syndromes, and febrile illnesses with either dermatologic or neurologic findings. Therefore, patients with these syndromes are the targets of bioterrorism-related syndromal surveillance programs. There is no widely accepted definition for any of these syndromes. As a result, syndromal surveillance systems are widely heterogeneous with respect to the syndromes under surveillance and the definitions of each syndrome.
Findings
We found 7 syndromal surveillance systems, none of which has been described in a peer-reviewed evaluation report (Tables 3, 14 and 15; Figures 3 to 10; Appendix H). We have no specific information about the usefulness of these systems in terms of detecting known infectious disease outbreaks (e.g., no information about whether the syndromal surveillance system detected last season's influenza outbreak). Additionally, we have no specific information on any of these systems' flexibility, acceptability, representativeness, or the direct costs of implementation.

Figure
Figure 3. Examples of data sources for surveillance systems.
The purposes of the systems vary: some systems are designed for ongoing surveillance for infectious disease outbreaks; whereas, others are for short-term surveillance for bioterrorism-related illness before, during, and after major political, economic, or entertainment events. For example, the Border Infectious Disease Surveillance Project (BIDS) collects syndromal surveillance information along the U.S.-Mexican border. BIDS performs ongoing surveillance by routinely collecting and analyzing data at regular intervals (Table 14). In contrast, systems such as the Lightweight Epidemiology Advanced Detection and Emergency Response System (LEADERS), have typically been used to perform event-based surveillance -- that is, they begin collecting data for a brief period before an event thought to be a target for bioterrorists (e.g., a major sporting or political event) and continue collection through a specified time after the event has finished (Tables 14 and 15; Figures 4 and 5). LEADERS has been used for event-based surveillance for the 1999 World Trade Summit, the 2000 Democratic and Republican National Conventions, and the 2001 Presidential Inauguration. (LEADERS can also be used for ongoing surveillance.)
Table
Table 14. Surveillance systems collecting syndromal reports.
The systems also differ with respect to the type of syndromal data they collect. For example, public health officials are evaluating several methods for collecting syndromal surveillance data from clinical personnel. The Syndromal Surveillance Tally Sheet is a paper-based ongoing syndromal surveillance tool used in Santa Clara County, California for the surveillance of 6 syndromes (Tables 14 and 15; Figure 6). 243 Triage nurses in urgent care centers and emergency departments enter data on each patient whom they evaluate and fax the sheet to the local health department at the end of each shift (typically 3 times a day). Health Buddy® is a device that can be used to display syndromal surveillance questions to users in a variety of clinical areas (Tables 14 and 15; Figure 7). 244 It was recently piloted in the Stanford University Medical Center Emergency Department. It is likely that similar tools have been developed and implemented by county health departments throughout the U.S.; however, a comprehensive survey of the syndromal surveillance methods currently in use was outside the scope of our project.

Figure
Figure 6. Syndromal surveillance tally sheet.
The Early Warning Outbreak Recognition System (EWORS),245, 246 developed collaboratively by the Naval Medical Research Unit-2, Department of Defense Global Emerging Infections System (DOD-GEIS), and the Indonesian Ministry of Health, is a global syndromal surveillance system (Tables 14 and 15). EWORS uses a simple computer program designed to enable untrained personnel to collect basic demographic and symptom data that are downloaded daily from remote sites to the Indonesian Ministry of Health.
The Rapid Syndrome Validation Project (RSVP®) is a Web-based collection tool for use by clinicians (currently primarily in use in emergency departments but could be used in any clinical area with a personal computer and Internet connection) (Tables 14 and 15; Figures 8 to 10). Data are entered on only those patients whom clinicians believe fit into 1 or more of 6 syndromal categories. 247 RSVP® facilitates (but does not require) the collection of more detailed information about a particular patient than the other systems (although several of the other systems have flexible interfaces that allow customization and the design of more detailed collection forms). In addition, RSVP® enables public health officials to send alerts and public health information to clinicians.
The Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE) was developed by the DOD-GEIS (Tables 14 and 15). 248 This system downloads ambulatory diagnosis codes grouped into "syndromal clusters." Initially, ESSENCE collected data from the primary care and emergency clinics serving the DOD health care beneficiaries living in and around Washington, DC. The data were downloaded daily onto a server and automatically analyzed with both traditional epidemiologic methods and time-space analyses. Following the events of September 11, 2001, ESSENCE was expanded to include virtually the entire Military Health System. Currently, ESSENCE downloads outpatient data on a daily basis from 121 Army, 110 Navy, 80 Air Force, and 2 Coast Guard installations around the world. Over 2,700 syndrome- and location-specific graphs are prepared each day and automatically analyzed for patterns that suggest a need for further investigation. 248 Beyond these centralized assessments, the graphs are available daily to approved DOD public health professionals on a secure Web site. 248 Two evaluations of ESSENCE are currently ongoing: one to address the issue of data quality, and the other to test the system's sensitivity and specificity over a range of outbreak scenarios. DARPA has awarded a $12-million 4-year grant to the ESSENCE consortium to construct a more powerful system for the Washington, DC area that includes both military and civilian data.
None of these syndromal surveillance systems has been evaluated in a comprehensive manner. However, current evaluations of ESSENCE and LEADERS are ongoing to determine their sensitivity, specificity, and timeliness. In Table 15, we present a comparison of 6 of the syndromal surveillance systems. (The paucity of available data on BIDS precludes its inclusion in this table.) The advantages of the tally sheet and Health Buddy® device are that they are rapidly deployable at relatively low cost, requiring only a fax or phone line. They can be relatively easily integrated into the triage nurses' workflow, thereby increasing the likelihood of compliance with data collection. RSVP® and EWORS are somewhat more costly to install since they require personal computers with Web access (RSVP®) or a modem (EWORS) in the clinical area. For RSVP®, clinicians enter data on only those patients whom they suspect of having one of the syndromes; therefore, the cases collected from this system will have a higher probability of disease than data collected from every patient presenting to a clinical area. Because RSVP® collects more detailed information than the tally sheet or Health Buddy®, it requires more of the clinician's time. ESSENCE or similar systems that perform surveillance on diagnostic codes have the enormous advantage of not adding any additional burden to clinical work flows. Additionally, ESSENCE was relatively low cost to implement and maintain and has demonstrated good scalability. The extensive epidemiologic analyses that ESSENCE performs on a daily basis far exceed what has been described for any of the other systems. However, systems like ESSENCE require that the diagnostic codes be available in an electronic format in a timely manner. Hospital or ambulatory diagnosis codes are not likely to be as sensitive as clinician reports in detecting suspicious cases. LEADERS is the most expensive system -- requiring personal computers with Web access and a subscription fee. It may also be difficult to integrate into the hospital and health departments' IT environments. However, it can incorporate multiple streams of data and is quickly deployable. In terms of data security, LEADERS and RSVP® reported being compliant with the Health Insurance Portability and Accountability Act of 1996 (HIPAA); no other reports specified the security measures of other systems.
Table
Table 15. Comparison of syndromal surveillance systems.
Summary: Syndromal surveillance systems
Syndromal surveillance systems could provide an early indication of cases resulting from a bioterrorism event. However, we have no evidence to determine which of the methods of collecting syndromal data provides the most sensitive, timely, acceptable, and low cost data. Local public health officials are the primary users of these data. Therefore, the ability of the system to present syndromal data in a timely manner, and minimize the necessity for additional analyses, will enhance the system's usefulness and acceptability.
We found no published standard definitions for the most common syndromes under surveillance. Additionally, none of the systems that we found that rely on clinicians to enter syndromal data provided definitions of the syndromes used by the collection tool (i.e., on the data entry screen, there was no definition of "flu-like illness"). A well-accepted list of the key syndromes for surveillance and detailed definitions of these syndromes will facilitate the integration of numerous sources of surveillance data. For example, the definition of "flu-like illness" should include its clinical characteristics so that triage nurses and clinicians can clearly identify patients with the syndrome, the specific ICD9 codes and other administrative data likely to be associated with it, the pharmaceuticals likely to be used to treat it, and the laboratory tests likely to be ordered to diagnose it. Then, each source of syndromal surveillance data can be systematically mapped to each of the syndromes, enabling the integration of all of these data into a single surveillance system.
As public health officials consider which of these systems to implement for ongoing or event-based syndromal surveillance, they are likely to find that a hospital's IT infrastructure may affect the acceptability of a syndromal collection tool. Hospitals and clinics with nascent IT infrastructures and an associated culture of paper may be better suited to the Tally Sheet or Health Buddy®. Within a given county, some hospitals may want to report ICD9 codes and others may prefer reporting triage nurse counts on the Tally Sheet. If different hospitals within a county are reporting syndromal surveillance data from different collection tools, public health officials will need to have the capacity to collect, analyze, and present these data in single system. The Oracle database associated with LEADERS has this capacity but is limited by the lack of standard definitions for the syndromes.
Surveillance Systems Collecting Clinical Reports
Background
Although most industrialized nations mandate the reporting of selected infectious diseases, compliance is typically poor. 252 A study of 176 clinicians in the United Kingdom (U.K.) found that 123 (70 percent) did not know where to obtain notification forms and most were unaware of reporting requirements. 253 For example, 79 clinicians (45 percent) were unaware that cases of pneumococcal meningitis should be reported. 253 Notable exceptions to this trend are the "astute clinicians" who, by reporting suspicious cases or clusters of cases to their local public health officials, have been largely responsible for the timely detection of infectious disease outbreaks (as in the case of West Nile Virus outbreak in New York City in late August 1999 15 and the recent cases of mail-associated anthrax254, 255). Efforts to enhance the awareness of clinicians, educate them about bioterrorism-related illness, and remind them of their reporting obligations are outside the scope of this Report. We refer interested readers to a comprehensive Evidence Report entitled "Training of Clinicians for Public Health Events Relevant to Bioterrorism Preparedness." 256
Increasingly, public health officials have realized that the establishment of networks of practitioners with training in disease reporting could improve the quality of data obtained from voluntary communicable disease notification systems. A sentinel network is a disease surveillance program that involves the collection of health data on a routine basis by clinicians with some training (albeit minimal) in reporting communicable disease. The growth of such sentinel systems has generated demand for information systems capable of automating data collection, analysis, reporting, and communication. In this section, we describe systems for the electronic collection and analysis of clinical reports from individual clinicians and sentinel networks.
Findings
Our search identified 6 IT/DSSs used for the reporting of clinical cases for diseases other than influenza (which we present in a separate section of this Report); 2 of these have been described in peer-reviewed evaluation reports (Tables 3 and 16; Appendix H).
The purpose of each system for collecting data varies. Some monitor the incidence of specific infectious diseases (such as tuberculosis); others collect surveillance data on groups of communicable diseases or on emerging infectious diseases. Accordingly, they also vary with respect to method and timeliness of data collection, type of hardware required, and geographic area under surveillance. In the paragraphs that follow and in Table 16, we present information about these characteristics of the surveillance systems collecting clinical reports. None of the reports of these systems described security measures, information about how an outbreak is determined, or the direct costs needed to operate the system.
Table
Table 16. Surveillance systems collecting clinical reports.
In 1983, France took a technological lead in electronic disease surveillance with a national telecommunications program that provided videotext home terminals free of charge to French citizens. 252 In 1984, the Institut National de la Santé et de la Recherche Médicale (INSERM), in collaboration with the French Ministry of Health, initiated a program for electronic monitoring of communicable diseases called the French Communicable Disease Network (FCDN).257-259 Today, a volunteer sample of about 1 percent of French general practitioners enters weekly reports on personal computers with either modem or videotext terminals.257-259 These data are available online through a Web site called SentiWeb. A major strength of the FCDN system for surveillance of infectious diseases is the timeliness of the collection, analysis and distribution of data. The reports of influenza-like illness from sentinel general practitioners, combined with information on viral isolates from the French Reference Centers, provide timely information for developers and evaluators of the influenza vaccine. 260 FCDN's early detection system is based on a regression model, which has been demonstrated in a retrospective study to forecast epidemics with a delay of only 1 week. 261 A 1998 evaluation of 500 sentinel practices of the FCDN found that although the system quickly offered estimates of the effectiveness of the influenza vaccine, and all results were available on the Internet within 1 week of data collection, delay could be shortened by updating data from the clinicians daily instead of weekly. 262
The Eurosentinel project depends on an international network of sentinel general practitioners to monitor influenza and other syndromes and diseases in Europe. Volunteer physicians submit weekly reports to a coordinating center in Belgium. Outputs for influenza are within minutes of reporting, while data for other syndromes and diseases are released in a quarterly newsletter. A report describing the experiences of the first 3 years of the project found that discrepancies in disease reporting practices, particularly the use of different denominators, between the sentinel networks of different countries made it difficult to compare the data from each network. 263
In the U.S., each state health department uses a standardized weekly form submitted by e-mail to CDC through the National Electronic Telecommunications System for Surveillance (NETSS).264-267 In the past, CDC returned analyses of this information back to the state health departments via an electronic text message system of the Morbidity and Mortality Weekly Report (MMWR), which these departments received earlier than other subscribers. CDC then established the Public Health Network (PHNET) to provide a tool featuring up-to-date maps and graphs to return information alerts to state health departments. 252 Data on selected notifiable infectious diseases continue to be published weekly in the MMWR and at year-end in the annual Summary of Notifiable Diseases of the United States. An observational study monitoring the delay between the date of disease onset and date of report to CDC found that for states providing the date of disease onset for at least 70 percent of reported cases, the median reporting delay was 23 days for shigellosis, 22 days for salmonellosis, 33 days for hepatitis A, and 20 days for bacterial meningitis. 264 Within 8 weeks of disease onset, more than 75 percent of reports were made for each of these diseases. Reporting delays varied widely between states, particularly for salmonellosis and shigellosis. 264
In addition to the major surveillance efforts just described, we report on 3 other surveillance projects. EUROTB collects data on tuberculosis from multiple European countries and reports these data to the WHO.268-270 EMERGEncy ID NET is a collection of sentinel physicians from 11 U.S. emergency departments who collect data on patients with selected clinical syndromes through multiple-choice forms. The information is entered into a desktop computer and electronically transferred to a central database at the Olive View-UCLA Medical Center for later analysis.271, 272 The Global Emerging Infections Sentinel Network (GeoSentinel)273, 274 collects reports from 25 sentinel clinics to monitor geographic and temporal trends in morbidity among travelers and other mobile populations. Reports are either faxed or electronically submitted to a central database in Georgia. GeoSentinel can also be used to send alerts and surveys to a widespread network of providers. The only additional information available about these systems describe the incidence and prevalence of the diseases under surveillance. It is difficult to determine their potential utility during a bioterrorism event from these data.
Summary: Surveillance systems collecting clinical reports
Because clinicians may be the first to recognize unusual or suspicious illnesses, reports from clinician networks are an essential source of surveillance data for detection of bioterrorism-related diseases. However, the usefulness of the systems described in this section is difficult to assess. First, none of these systems has been evaluated for its ability to detect illness caused by bioterrorism events. Thus, the sensitivity, specificity, and timeliness of the systems have not been documented. Second, the descriptions of many of the systems were published in the early 1990s. It is possible that some have been upgraded to rely on electronic reporting, but current descriptions have not been published.
Systems for detection of bioterrorism require more rapid response times than do systems designed for some other purposes. The timeliness of systems that collect clinical reports depends on the time required to report data, analyze the data, and communicate the results of such analyses to decision makers who can respond appropriately. Systems that collect data weekly will be substantially less useful than systems that can provide more rapid collection and analysis. Of the systems in this section, Eurosentinel provides the timeliest data (but only for influenza, data on other diseases and syndromes has a longer delay). The timeliness of the other systems varies from days to months. Investigation of whether the systems can be modified to increase timeliness should be a high priority.
Surveillance Systems for Influenza
Background
On June 26, 1957, 1,688 delegates from 43 states and foreign countries arrived at a church conference at Grinnell College. 236 Among them were 100 Californian delegates who had made the trip to Iowa in a single chartered railroad coach. 236 Cases of influenza had developed en route and when the 100 Californians interacted with the other delegates, an epidemic exploded. 236 By July 1, more than 200 clinical cases of influenza were documented, the delegates ended their conference early, and returned home -- taking the virus with them. 236
Often perceived as a comparatively low-level threat, the viruses that cause influenza are continually evolving and occasionally undergo sufficient genetic drift that they cause significant morbidity and mortality. 241 For example, the 1918-1919 pandemic killed more than 20 million people around the world. 241 The less severe pandemics in 1957 and 1968 killed a total of 1.5 million people and caused an estimated $32 billion in economic losses worldwide. 241 Established in 1948, the WHO's global network for influenza surveillance currently includes 110 collaborating laboratories in 82 countries, continuously monitoring locally isolated influenza strains.240, 284 These data are used to make recommendations on the 3 virus strains to be included in the next season's influenza vaccine.240, 284 The WHO has also created FluNet, an Internet site for reporting and monitoring clinical cases of influenza.
The U.S. Air Force has actively contributed to the global influenza surveillance effort since 1976 through the efforts of Project Gargle, based at Brooks Air Force Base in San Antonio, Texas.245, 285, 286 In June 1996, President Clinton issued Presidential Decision Directive NSTC-7, which formally expanded the mission of the DOD to support global surveillance, training, research, and response to emerging infectious disease threats.245, 285, 286 The DOD-GEIS was formed in 1997 in response to this Presidential Directive. During the 1999-2000 influenza surveillance season, Project Gargle processed 3,825 throat swabs from 19 sentinel U.S. military bases, 49 nonsentinel bases, and 3 DOD overseas laboratories.245, 285, 286 The Air Force also correlates the immunization status of Air Force personnel with influenza morbidity, providing a marker for vaccine efficacy.245, 285, 286
The lessons of the past 50 years of global surveillance for influenza are applicable to the problem of surveillance for biothreat agents. Effective surveillance for bioterrorism-related diseases requires similar global "networks of networks" integrating clinical and laboratory data collected by governmental, charitable, military, private and professional organizations and reported to local, national, and international public health organizations.
Findings
Our search identified 11 IT/DSSs for influenza surveillance, 3 of which have been described in peer-reviewed evaluation reports (Tables 3 and 17; Appendix H).
The AAH Meditel United Kingdom General Practitioner Reporting System has been used for disease surveillance across the U.K. 287 Each night, data are downloaded from the computers of hundreds of sentinel general practitioner offices to a mainframe computer at AAH Meditel. 287 The system was originally intended to collect prescribing data but has been extended to collect other patient data. A 10-week study compared the AAH Meditel database of clinical reports of influenza cases to the manual influenza surveillance system of the Royal College of General Practitioners' and found the slope of the influenza epidemic curve to be nearly identical for both systems. 287 This suggests that the system was able to detect an outbreak of influenza at least as well as the sentinel General Practitioners against whom it was compared (we have no additional sensitivity or timeliness information on this group). However, the incidence of disease as detected by the General Practitioners was 3 to 4 times higher than that derived from AAH Meditel. 287 The authors suggest that the underreporting of the AAH Meditel system might be due to clinicians inexperienced in surveillance methods, especially in comparison with the trained sentinel Practitioners of the Royal College, and overestimation of the surveillance population. 287
The French Regional Influenza Surveillance Group (GROG) system consists of almost 1500 voluntary general practitioners, pediatricians, pharmacists, and military medical officers who provide a weekly activity summary via telephone.288, 289 A regression-based method for early recognition of influenza epidemics based on time series analysis was used to evaluate surveillance data from various indicators. Sick-leave as prescribed by physicians and recorded by the "Assistance Publique" (Health and Social Security Services), emergency house calls, and numbers of patients with influenza-like illness seen by general practitioners and pediatricians were the most sensitive indicators for the early recognition of influenza epidemics. The detection of influenza epidemics using drug consumption data was 1 week delayed relative to house call and sick-leave indicators.288, 289 The annual epidemic threshold criteria are derived from a combination of these indicators (with virologic data) designed to minimize the false positive rate.288, 289 During the 1992-1993 season, using the threshold criteria developed from the 1984-1992 historical data, GROG detected an influenza B epidemic on week 52 -- 3 weeks after the virus began circulating in the population.288, 289
In 1991, the National Influenza Centre of the Netherlands set up an electronic influenza surveillance project using the existing Medimatica computer network that connected several health care institutions via a central server. The 3000 clinician and 1300 pharmacist subscribers to the Medimatica service receive low cost e-mail service and access to medical database services (including access to patient medical records and current information on a variety of diseases). 290 A new database was added to the Medimatica system providing current information on Dutch influenza outbreaks and served as a means for clinicians and pharmacists to report suspected influenza cases. A prospective evaluation of this influenza reporting system during the 1991-2 influenza season found that no clinicians or pharmacists reported any influenza cases. 290 The investigators suggested that this disappointing result might have been caused by a clinical culture in the Netherlands in which electronic reporting has not yet been embraced. 290
None of the other influenza surveillance systems has been reported in a peer-reviewed evaluation. Many of them collect data on a daily basis with weekly reporting of data to public health officials. The systems vary with respect to their methods of data collection, including telephone, fax, mail, and e-mail. Most incorporate several sources of surveillance data (e.g., virologic data, clinician reports, hospital admissions data, pharmacy data, and work absenteeism data). No reports of these systems discussed information on methods to maintain the security of data or direct costs associated with the program.
Summary: Surveillance systems for influenza
Surveillance systems that collect influenza data are relevant to bioterrorism surveillance in 3 ways. First, sentinel clinicians who report on patients with suspected influenza are experienced at applying a case definition to a clinical population for the collection of public health data. Because many bioterrorism-related illnesses present with a "flu-like illness," this network of trained sentinel clinicians could provide valuable surveillance data. Second, most influenza surveillance systems integrate clinical and laboratory data for the detection of influenza outbreaks. Surveillance for bioterrorism may be aided by similar integration of multiple data sources. Finally, influenza surveillance is a coordinated global effort. Given the current state of international travel, importation of food products, and trade in pharmaceuticals and blood products, infectious diseases from natural sources or from acts of bioterrorism can readily travel to the U.S from remote areas of the world. New programs for the surveillance of bioterrorism-related illness could be derived from the existing IT infrastructures and the historical relationships that have been developed for influenza surveillance. Several of the influenza systems rely on weekly reporting by clinicians -- for bioterrorism surveillance, every effort should be made to reduce this lag.
Surveillance Systems Collecting Laboratory and Antimicrobial Data
Background
Laboratories are critical to the detection of naturally occurring emerging infectious diseases, identification of biothreat agents, and monitoring for unusual patterns of antimicrobial resistance. The laboratory that encounters the first case of a covert bioterrorist attack may be located in a community hospital. 302 Many of the biothreat agents are morphologically similar to the organisms that normal inhabit the human respiratory tract. 302 For example, B. anthracis is morphologically identical to other Bacillus species -- most of which are contaminants of clinical samples. 302 Therefore, if the organism is not speciated, the laboratory technician may consider the isolate a contaminant and discard the sample. 302
APHL and CDC are developing the Laboratory Response Network, a network of civilian public health and private laboratories in the U.S. for routine disease surveillance and detection of biothreat agents. 302 In the Laboratory Response Network, laboratories in local hospitals (Level A) can rule out biothreat agents or refer organisms to Level B laboratories. 302 Level B laboratories can identify certain pathogens, such as anthrax, but must refer other organisms to Level C laboratories for further evaluation. 302 Level C laboratories can definitively identify a broader range of pathogens, but must also refer some samples (such as smallpox and hemorrhagic fever viruses) to Level D laboratories. 302 Only 2 Level D laboratories exist in the U.S., at the CDC and at the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID). 302 An information system is currently being established for the Laboratory Response Network that includes: a password-restricted site (using NEDSS standards and systems) for ordering reagents, distributing procedural information, and standard messaging for results. 302 The Laboratory Response Network has recently received additional federal funds enabling its expansion to 120 laboratories, each equipped with advanced laboratory technologies and trained staff. 303
Findings
Our search identified 12 systems for the surveillance of laboratory data (4 of which were described in peer-reviewed evaluation reports) and 11 systems for the surveillance of antimicrobial data (1 of which was described in a peer-reviewed evaluation report) (Tables 3, 18 and 19; Appendix H).
Table
Table 18. Surveillance systems for laboratory data.
Table
Table 19. Surveillance systems for antimicrobial resistance data.
A clinical evaluation of the Electronic Communicable Disease Reporting System (ECDRS) operated by the Hawaii Department of Health demonstrated that this automated laboratory reporting system improved communicable disease reporting to public health officials relative to conventional reporting methods. 304 At a predetermined time each day, a computer dedicated to reporting purposes in each of the participating laboratories automatically connected to the laboratory information system in routine use at that facility. This connection launched a data extraction program that then transmitted the laboratory data to a public health database. During a 6-month evaluation period, 325 (91 percent) of 357 laboratory reports were reported through ECDRS, compared with 156 (44 percent) reported through conventional methods. 304 For the combined 124 reports received from both reporting mechanisms, electronic reports were received an average of 3.8 (95 percent confidence interval: 2.6-5.0) days earlier than were corresponding reports sent by mail or fax. 304 Of the 21 data fields per report, 12 were significantly more likely to be complete in the electronically reported data; however, the field describing the type of specimen (e.g., from blood or stool) was more likely to be reported by conventional methods. 304
The National Enteric Pathogen Surveillance Scheme (NEPSS)/Salmonella Potential Outbreak Targeting System (SPOT) is designed for the early detection of potential Salmonella outbreaks in Australia. 305 A preliminary retrospective analysis of this system demonstrated a sensitivity of 100 percent (15 out of 15 Salmonella outbreaks), compared with 50 percent for another surveillance method in which epidemiologists "eye-balled" the data. 305 During a separate 3-year retrospective study, NEPSS/SPOT detected 124 out of 134 potential Salmonella outbreaks, thereby demonstrating a sensitivity level greater than 90 percent and a positive predictive value in the range of 53 to 68 percent. 306
The Public Health Laboratory Information System (PHLIS) is used for the investigation and surveillance of outbreaks of specific notifiable diseases in the U.S. 307 Outbreak-specific Data Entry Screens are created and distributed to all reporting sites electronically. Data input and reporting occurs within hours. In addition, there is relational database software on individual personal computers with connections to laboratory and public health computers and databases. An evaluation of PHLIS used different methods to calculate expected values of Salmonella serotype Enteritidis. 307 This retrospective analysis demonstrated that using a 5-week mean as the threshold for what constituted an outbreak was more sensitive than using either the 5-week median or the 15-week mean. Using the 5-week mean as the threshold for determining an outbreak, the system had 3 false negatives, 76 percent sensitivity, 95 percent specificity, and 77 percent false-positive rate. 307 The authors remarked that their evaluation may have been affected by their definition of an outbreak (the system would have missed an outbreak of 3 cases if it occurred in a season with a high background number of reported cases). 307 The authors report that this system, which detected an outbreak of Salmonella serotype Stanley in May 1995, resulted in a more timely investigation of the outbreak than would have occurred with conventional surveillance -- the investigation implicated alfalfa sprouts as the vehicle of infection resulting in "the prompt development of prevention measures." 307
Among the laboratory systems specifically designed for surveillance of antimicrobial resistance, only The Surveillance Network™ (TSN™) Database-USA has been clinically evaluated. Laboratory data obtained from 229 laboratories "chosen for participation on the basis of their geographic and demographic characteristics" and ability to perform antimicrobial susceptibility testing are sent to the TSN™ Database-USA. One study compared in vitro susceptibility testing of isolates from 27 hospital laboratories with surveillance data for 200 laboratories from TSN™ Database-USA for the current status of fluoroquinolone activity against gram-negative species. Despite the fact that the comparison data was from different time periods and for different samples, the 2 surveillance systems agreed in many areas. The authors suggested that the discrepancies between the systems may be due to reporting practices of clinical laboratories, the geographic distribution or number of isolates, or the number of laboratories involved in the studies. 308
In general, the evaluative and descriptive reports of the systems collecting laboratory and antimicrobial resistance data suggest that the electronic systems improve the timeliness and sensitivity of conventional methods. Few reports specifically described how laboratory samples are handled, methods for confirmation of laboratory samples, acceptability, or cost of implementation.
Summary: Surveillance systems collecting laboratory and antimicrobial resistance data
Laboratory testing will be an essential component of any bioterrorism response effort. Systems that facilitate the collection, analysis, and reporting of notifiable pathogens and antimicrobial resistance data could potentially facilitate the rapid detection of a biothreat agent. However, none of these systems has been evaluated for such use.
Limitations of the current U.S. laboratory system for bioterrorism surveillance include lack of staffing and equipment for rapid detection in local laboratories and lack of a robust communication infrastructure among the different levels of laboratories. Efforts are ongoing to correct some of the gaps. Specifically, the Laboratory Response Network, which builds on existing laboratory capacity and is currently under active expansion, was designed so that it can be integrated into surveillance networks (such as NEDSS) and communication networks (such as California's Rapid Health Electronic Alert, Communication, and Training system (RHEACT)).
Surveillance Systems Collecting Hospital-based Infections Data
Background
The primary objective of hospital surveillance is to track hospital-acquired infections, not to identify undiagnosed infections from the community. However, hospital epidemiology systems could play 2 roles in the early detection of a covert bioterrorist attack: the identification of a cluster of recently admitted cases suggestive of a community-based outbreak, and/or the identification of a cluster of cases within the hospital suggestive of patients with an unrecognized communicable disease.
A bioterrorist attack resulting in a large number of people seeking medical attention at the same time would likely be identified by other means. However, a smaller release of a biothreat agent may result in the hospitalization of a few sentinel patients who could be detected by a hospital surveillance system. For example, if a biothreat agent that causes meningitis was released in a covert attack, patients receiving a large inoculum may present to the emergency department of their local hospitals and be admitted to the ICU. Patients receiving a small inoculum may present to their internists or urgent care clinics and be admitted to the medical wards of the same community hospital. Another group of patients may present to a neurologist and be admitted to the neurology service of the hospital. Under these circumstances, no single team of clinicians will have cared for these patients and each is likely to be unaware of the other similar cases within the hospital. The detection of such a cluster would fall to the hospital infection control service.
Alternatively, if cases of smallpox with its characteristic rash present to the emergency department, astute clinicians may recognize the disease and institute appropriate isolation measures. However, if a few patients present with acute onset of fever, chills, myalgia, cough, and chest X-ray revealing patchy infiltrates, they are not likely to be placed in respiratory isolation until Y. pestis is found in their blood cultures. The early identification of patients with contagious infections and their prompt isolation depend on the effectiveness of the hospital epidemiology service.
The hospital epidemiology IT/DSSs that are likely to identify a bioterrorism event are those that function in a timely and sensitive manner. Traditional hospital infection control surveillance relies on the manual review of suspected cases and the retrospective analysis of aggregated surveillance data. This approach tends to be labor intensive, expensive, and slow. Efforts to improve hospital surveillance with IT/DSSs have focused on automated alerts of abnormal microbiology cultures (see also the section of this chapter on Reporting and Communication Systems), identification of high-risk patients, and the detection of infection rates above a statistical background rate.
Findings
We found 16 IT/DSSs designed specifically for hospital surveillance, 10 of which have been described in peer-reviewed evaluation reports (Tables 3 and 20; Appendix H). Several of these systems were principally designed to detect hospital-specific antimicrobial resistance patterns and therefore are similar to systems presented in the preceding section. We have presented them in this section because, unlike the laboratory-based surveillance systems for antimicrobial resistance, whose purpose is largely to provide public health officials with surveillance data, the purpose of these systems is to provide information for use by hospital infection control officers. These data typically are not a part of a public health surveillance system. Several of these systems send alerts to clinicians and hospital infection control personnel (similar to some of the reporting and communication systems discussed below). We present them here because their primary purpose is to detect and reduce hospital infections.
Table
Table 20. Surveillance systems for hospital-based infections data.
Five of the 16 systems have been evaluated for their detection capabilities (others have been evaluated for other outcomes such as user acceptability which will be discussed at the end of this section). As we discussed earlier in this Report, the HELP system at the LDS Hospital in Salt Lake City uses the Data Mining Surveillance System (DMSS), which has the demonstrated ability to identify unusual patterns in surveillance data from sources including the microbiology laboratory, nurses' charts, chemistry laboratory, surgical record, and pharmacy.186, 191
Researchers at the University of Alabama at Birmingham have developed a system that they also call the Data Mining Surveillance System (DMSS).213, 214 They performed an evaluation of the DMSS using all positive inpatient microbiology cultures for a 15-month period. Each month of the study, 475 to 677 records were used in the algorithm with a running time of less than 4 minutes. The system detected 2 outbreaks of a highly resistant strain of Acinetobacter baumannii and changes in the incidence of multi-drug resistant Klebsiella pneumoniae that were not detected by conventional hospital surveillance. It also provided geographic information suggesting that in some units (e.g., the surgical ICU) patterns of antibiotic use may have been associated with changes in antimicrobial resistance, which was unknown before the use of the system. 214
GermWatcher is a system designed to detect both outbreaks of new infections and rising endemic rates of preexisting infections in hospitals.340-343 Each morning, positive laboratory results are automatically transferred from the hospital database to GermWatcher. The system makes recommendations to the infection control officer to keep, discard or watch the cultures, based on the CDC's criteria for potential nosocomial infections. The system was evaluated by comparing these recommendations to those of the hospital control officers. Changes in the detection algorithms between GermWatcher Versions 1 and 2 resulted in improvement from 14 percent to less than 2 percent in rates of disagreement between the system and infection control officers. The final version of the system misclassified 3.5 percent of the 1851 cultures evaluated (2.8 percent false positives and 0.7 percent false negatives). 341 The interpretation of these results is difficult because no additional information was provided regarding the use of the infection control officer as a gold standard (i.e., the sensitivity and specificity of their decision making).
The Danish National Hospital Discharge Registry, a registry of all non-psychiatric patients admitted to Danish hospitals, was developed to determine if hospital discharge data (clinician-provided ICD10 codes) could be used for the detection of bacteremia. 344 Out of 45,000 patients discharged from Aalborg Hospital, Denmark in 1994, a diagnosis of septicemia or sepsis was found in the discharge database 207 times for 186 patients. Of these, 183 episodes (88 percent) were not in the bacteremia database maintained by the regional department of clinical microbiology. Using the clinical microbiology bacteremia database as the gold standard, the sensitivity of septicemia and sepsis registration in the Danish National Hospital Discharge Registry ranged from 4.4 percent to 5.9 percent (18 to 24 cases out of 406) depending on the definition of bacteremia used. The positive predictive value of the registry was 21.7 percent (95 percent confidence interval: 12.8 to 30.5 percent) since only 18 out of 83 episodes of septicemia found in the Danish National Hospital Discharge Registry were confirmed by the clinical microbiology data. 344 These data suggest that the use of ICD10 data as collected in that hospital discharge registry do not significantly add to the information already available in the clinical microbiology data.
The Tucson VA Nosocomial Infection Surveillance System was designed to identify potentially preventable nosocomial infections. 345 During a 6-month study, of the 19 clusters of infections and 3 outbreaks of intravenous catheter-related bacteremias identified by a combination of the system and epidemiological investigations, only 3 of the 22 were identified by standard surveillance methods. Standard surveillance found no additional infections. 345 We conclude that this system significantly improved the detection of nosocomial infections; how this relates to the detection of inpatients with bioterrorism-related illness is unclear.
The other evaluations of the hospital surveillance systems reported similar improvements in the ability to detect hospital-based infections above what would have been detected by manual methods alone. Several evaluation reports did not report detection outcomes, rather, they primarily presented data regarding the acceptance of the system by hospital infection control officials. For example, the Hospital Infection Standardized Surveillance (HISS)346, 347 system was designed to improve the timeliness and standardization of data collected about nosocomial infections. Data are entered into a handheld computer, and then electronically transferred to a desktop personal computer for storage and analysis using specialized software. In an evaluation of HISS for the reporting of procedure-related infections in public acute care hospitals in New South Wales, Australia, the authors reported that although all 9 infection control officers who used the handheld device "had initial difficulties in adapting to the new technology, personnel at 7 sites consider it a useful tool. The near 100 percent completion rate for the data sets achieved by all 9 sites testify that the handheld computer assisted ICPs (infection control professionals) to collect large data sets." 346
In general, the systems collecting hospital-based infections data with the most compelling evidence for effectiveness are those that are integrated into a robust hospital IT infrastructure. The different systems had different purposes and therefore varied considerably with respect to the information they collected and the kinds of alerts sent to hospital infection control officials. None was specifically designed for integration into a national bioterrorism surveillance program, and it is not clear how to evaluate which of the systems presented would be best suited for that purpose. These incorporate local estimates of pathogenic prevalence and resistance with clinical data to provide hospital infection control personnel with timely, sensitive, and relatively specific analyses. None of the included systems described cost of implementation.
Summary: Surveillance systems collecting hospital-based infections data
The hospital surveillance systems that automate the collection of data from hospital-based laboratories and clinical records (i.e., the LDS and University of Alabama DMSSs, CDR, DHCP in VA hospital, and WING) are likely to be more timely than the manual systems that they replaced.
From the reports of the 5 systems that have been evaluated for their detection capabilities, we conclude that some of these systems could be a valuable tool for hospital infection control officers. However, there is little evidence demonstrating that they have sufficient sensitivity, specificity, or timeliness to detect a community-based bioterrorism event.
The integration of data from hospital-based surveillance systems, already collected in electronic format for use by hospital infection control officers, could be a valuable addition to a surveillance system organized by local public health officials. Similarly, the collection and reporting of hospital-based infections data from networks of hospitals (such as the VA) could contribute to a national bioterrorism surveillance system.
Disease Surveillance of Foodborne Illnesses
Background
On September 17, 1984, the Wasco-Sherman Public Health Department received the first case reports of the 751 victims of the intentional contamination of salad bars in The Dalles, Oregon with Salmonella typhimurium. 7 The lengthy epidemiologic and criminal investigations that followed demonstrated that the nation's largest foodborne outbreak of 1984 was the result of a bioterrorist attack by followers of Bhagwan Shree Rajneesh. 7
As evidenced by this event, terrorists can exploit vulnerabilities to our food supply. Agroterrorists may be motivated to cause human morbidity and mortality through contamination of foods during harvest, processing, or preparation, or they may be interested in creating the economic burden resulting from reduced food supply. 360 Attacks may be made against food and agriculture transportation systems, on water supplies, on farm workers, on food handlers, or on processing facilities. 360 Insect hosts for diseases affecting crop plants or livestock may be imported and released with the intent of creating an epidemic or influencing a nation's ability to export agricultural products abroad. 360 U.S. reliance on imported fresh fruits and vegetables has grown to such an extent that the safety of imported foods has become a source of major public health concern. 360
Most industrialized nations mandate the reporting of foodborne illnesses and have established surveillance systems for foodborne illnesses that collect data from health officials or clinical laboratories. In the U.S., reportable foodborne diseases and organisms include: botulism, brucellosis, cholera, E. coli O157:H7, hemolytic uremic syndrome, post-diarrheal salmonellosis, shigellosis, typhoid fever, Hepatitis A, cryptosporidiosis, cyclosporiasis, and trichinosis. 361
Ours is not a comprehensive review of agroterrorism. We direct interested readers to other reports on the topic.7, 82, 360, 361 Our search produced 2 types of IT/DSSs for surveillance of foodborne illness: those that collect and analyze reports from clinicians and laboratories about the incidence and laboratory characteristics of foodborne pathogens, and those that model microbial growth responses to various food production methods. Our search also found 2 studies on active monitoring systems using implantable sensors in livestock to allow for identification of animals and surveillance of the animals' vital signs.362, 363 Additionally, we found reports on 3 DSSs designed to assist veterinarians and animal handlers with diagnosis and management of common animal diseases (EPIZOO, Associate, and BOVID-3).364, 365 However, these systems are outside the scope of our project and are not described further in this section.
Findings
In this section, we describe 10 IT/DSSs for surveillance of foodborne illnesses, 2 of which have been described in peer-reviewed evaluation reports (Tables 3 and 21; Appendix H). We note that many of the surveillance systems described in other sections (particularly those collecting laboratory data and clinician reports) also collect and report information about Salmonella species and other foodborne pathogens. The 10 surveillance systems for foodborne illness described in this section differ from these general systems in that they are designed specifically for the detection of foodborne pathogens or illness. In this section, we will first describe the systems that collect and analyze reports from clinicians and laboratories about the incidence and laboratory characteristics of foodborne pathogens. Then, we will present information about those systems that model microbial growth responses to various food production methods.
Table
Table 21. Surveillance systems for foodborne illnesses.
In the U.S., the national system for surveillance of food-borne illnesses is dependent on voluntary reporting from clinicians and laboratories. In addition, the Foodborne Diseases Active Surveillance Network (FoodNet), the principal foodborne-disease component of the CDC's Emerging Infections Program (EIP), is an active system that collects data from clinical laboratories and public health officials to estimate the burden and sources of specific foodborne diseases in the U.S.366-372 FoodNet is a collaborative project among the CDC, the 8 EIP state health department sites, the Food Safety and Inspection Service of the United States Department of Agriculture (USDA), and the Food and Drug Administration (FDA). Results from FoodNet are used to identify infection control points, focus future prevention strategies and decision making within food safety regulatory agencies, measure changes in the burden of disease, and evaluate the effects of interventions on rates of infections over time. FoodNet collects data about 9 foodborne diseases (Campylobacter, Cryptosporidium, Cyclospora, E. coli O157, Listeria, Salmonella, Shigella, Vibrio, and Yersinia) in 8 U.S. sites (California, Connecticut, Georgia, Maryland, Minnesota, New York, Oregon, and Tennessee).
Evaluation data about FoodNet are limited to those comparing the detection of the organisms of interest in one year versus another. In Table 21, we describe 5 related systems for the collection and analysis of foodborne diseases and contaminations. Similarly, the reports of these systems provide estimates of disease incidence as identified by the system but do not further describe the systems' sensitivity, specificity, or timeliness.
The CDC has established several networks of laboratories with capabilities to perform DNA fingerprinting of bacterial strains. These methods were instrumental in demonstrating that the strain of Salmonella typhimurium in the Rajneeshpuram was indistinguishable from the organism cultured from the contaminated salad bars in The Dalles. We present 2 of these IT networks: the Salmonella Outbreak Detection Algorithm (SODA), which tracks the 50,000 clinical isolates of Salmonella that are routinely serotyped by state public health laboratories each year, and the National Molecular Subtyping Network for Foodborne Disease Surveillance (PulseNet), a similar IT for the analysis of strains of 4 foodborne pathogens (Table 21).
Other food safety ITs use mathematical models to predict microbial responses to different environmental conditions (e.g., cooking temperature) in order to recommend food preparation methods that minimize microbial contamination. These systems usually specify a hazard (i.e., pathogen), an exposure (i.e., likely amount of consumption), a hazard characterization (i.e., evaluation of the nature of the adverse effects), and a risk characterization (i.e., estimation of adverse effects given the population at risk). 373 These models are used in the production of a variety of foods, including milk, eggs, ground beef, poultry, and cheese products.374-377 Our search found 3 IT/DSSs that incorporate these kinds of microbial prediction models (Table 21). We have included them because they could be used to model intentional contamination of the food supply by bioterrorists during the production process, although none was specifically designed with this purpose in mind. We present them in the same table with the surveillance systems for foodborne illnesses, recognizing that they are not surveillance systems, per se, but that they use surveillance data for their models.
In general, the relatively small number of organisms for which data are collected limits the usefulness of systems that perform surveillance of specific foodborne pathogens. Additionally, the lack of published evaluative information prevents a clear understanding of how these systems would function in the event of a bioterrorist attack on the food supply.
Summary: Disease surveillance of foodborne illnesses
Technologies like SODA and PulseNet have been used extensively in foodborne outbreak investigations with success -- even with investigations of outbreaks resulting from intentional contamination of the food supply. However, none of the foodborne illness surveillance systems that collect disease incidence data was specifically designed for the early detection of bioterrorist attacks on the food supply, nor has any been evaluated for that purpose. We found no evidence regarding the potential sensitivity, specificity, or timeliness of FoodNet, the active surveillance system collecting data to estimate the burden of foodborne illnesses in the U.S. Moreover, even if FoodNet was sufficiently sensitive and timely to be useful for agroterrorism detection, it is limited in that it only collects data from 8 states on 9 foodborne illnesses. The primary means for detecting an agroterrorist attack outside these states or using a different organism would be based on the analysis of voluntary reports from clinicians and laboratories.
Zoonotic and Animal Disease Surveillance Systems
Background
On the morning of March 18, 1996, John Major, then Prime Minister of the U.K., was presented with a memo from the Health and Agriculture Ministers confirming a link between bovine spongiform encephalopathy (BSE) in cattle and Creutzfeldt-Jakob disease (CJD) in humans. 385 The memo described reports of people contracting CJD by consuming beef infected with BSE. 385 Response to BSE in the U.K. has cost the country an estimated £4 billion, caused significant damage to the agricultural industry, and harmed Britain's relations with its European Union partners, who rapidly banned the importation of British beef. 385 Similarly, concerns exist that a bioterrorist attack could involve the dissemination of a zoonotic illness among animal populations with the intention of infecting humans or livestock to cause economic and political chaos.386-388
Findings
We found 6 ITs designed to collect, process, and disseminate information on zoonotic and animal diseases, none of which has been described in a peer-reviewed evaluation (Tables 3 and 22; Appendix H).
Table
Table 22. Zoonotic and animal disease surveillance systems.
The National West Nile Virus Surveillance System (ArboNet) 389 is an electronic-based surveillance and reporting system, developed by the CDC to track West Nile virus activity in humans, horses, other mammals, birds, and mosquitoes. The California Encephalitis Program 390 monitors mosquitoes and flocks of sentinel chickens for mosquito-borne viruses such as West Nile Virus, St. Louis encephalitis, and Western equine encephalomyelitis. ArboNet and the California Encephalitis Program are the only zoonotic surveillance systems in our Report. Reports of these programs describe the type of data they collect and report the weekly incidence of the diseases under surveillance. They provide no additional information about the sensitivity, specificity, representativeness, acceptability, or other criteria by which we evaluated surveillance systems.
The National Animal Health Monitoring System (NAHMS) is an integrated national (U.S.) surveillance system that collects data on animal disease incidence and prevalence, mortality, management practices, and disease costs.391, 392 One of its surveillance programs is the Veterinary Diagnostic Laboratory Reporting System. Data are compiled from several sources including national animal disease control and eradication programs, information on patterns of disease based on laboratory data, selected etiologic agents, global disease distribution and reports of "unusual" laboratory findings. Results are published quarterly in DxMonitor. Another animal health surveillance program within NAHMS is the Sentinel Feedlot Monitoring program, which relies on health data collected from veterinary practitioners, who report inventory and morbidity by cause. A pilot study, involving a large commercial beef feedlot, was performed in 1988. 393 NAHMS also conducts national studies of livestock, which involve processing data obtained from questionnaires, logs, and laboratory analysis of biological specimens. 391 These studies include the swine health survey in 1990 392 and the 1995 National Swine Study. 394 Data from these principally describe animal disease incidence data and related mortality and cost information but not evaluative information about the system itself.
In New Zealand, an epidemiological information system has been developed to assist disease control authorities in the containment and eradication of animal disease outbreaks. The system was initially developed to control a potential incursion of foot and mouth disease, and is called EpiMAN-FMD. By combining disease and viral spread models, farm maps, and epidemiologic parameters, the system provides statistical reports and decision support to help with the management of an outbreak. While the system has not been tested in an actual foot and mouth disease outbreak (or any bioterrorism events), it has been tested in hypothetical scenarios, and the developers hope to adapt EpiMAN-FMD to other veterinary issues.395, 396
None of the reports of the systems in this section described the hardware requirements, the system's security measures, or the costs of operating the system.
Summary: Zoonotic and animal disease surveillance systems
Bioterrorism involving zoonotic and animal diseases represents a substantial threat to our national security and economy. Early detection of such an event requires effective rapid detection systems for use by farm workers, meat inspectors, and veterinarians with real-time reporting capabilities to public health officials. The surveillance systems described above provide some organizational support for national and international anti-agroterrorism efforts. However, none has been evaluated for this purpose. Our search found reports of only 2 zoonotic surveillance systems -- a major gap in bioterrorism surveillance efforts. Most of the reports provided little or no information about the timeliness of these systems; those that did suggest lag times that would be too long for effective bioterrorism surveillance.
Surveillance Systems Collecting Other Kinds of Data
Background
In this section, we present information about surveillance systems that met our inclusion criteria but collected sufficiently different surveillance data that they could not readily be described in the previous sections. These systems could be valuable additions to surveillance networks that integrate surveillance data from clinicians, hospitals, and laboratories. There are likely to be many other systems collecting data with potential utility for bioterrorism surveillance that have not been described in published reports or fall outside the scope of our primary research focus. For example, by September 2002, the American Association of Poison Control Centers will require all of its approximately 60 member centers to electronically report all call data at the time it is collected to the Toxic Exposure Surveillance System (TESS) database. 401 Efforts are currently underway to develop analysis algorithms to systematically search this database for clusters of cases that may represent public health problems.
Findings
We present 11 surveillance systems that collect other kinds of data, 2 of which have been described in peer-reviewed evaluation reports (Tables 3 and 23; Appendix H).
Table
Table 23. Surveillance systems for other data.
These systems differ with respect to the types of surveillance data they collect and therefore differ with respect to sensitivity, timeliness, and cost. For example, EPIFAR collects drug prescription data, which may be more sensitive for a bioterrorism event than a system like Data Web, which performs surveillance on administrative databases of demographic, economic, environmental, and health data collected by a variety of organizations in the U.S. However, Data Web uses data already collected for other purposes and therefore minimizes collection burden.
Two of these 11 systems have been clinically evaluated. EPIFAR is designed to track individual prescription histories in order to provide estimates of disease prevalence in Italy. 402 All drug prescriptions are routinely collected and processed by the Italian National Health Service (NHS). EPIFAR is a computer program used to analyze these data and determine the prevalence of selected diseases. A retrospective study of 2,550 patients who received 1 of 12 drug regimens designed for tuberculosis found that 7 times as many tuberculosis patients were identified by EPIFAR as were officially reported by clinicians. 402 However, 88 of 250 total notified cases (35.2 percent) for the study period were not found in the EPIFAR system. 402 Also, in a survey of physicians prescribing tuberculosis drugs for the included patients, physicians denied any tuberculosis-related problem for nearly 25 percent of the EPIFAR-identified patients. The positive predictive value of the model differed between drug regimes, with a range of 50 to 76.9 percent. 402 To understand whether a system such as EPIFAR is likely to be useful for bioterrorism surveillance, it would be important to have a detailed understanding of the sensitivity, specificity, and timeliness of each of the drugs (or combinations of drugs) under surveillance. For example, if EPIFAR collected surveillance data exclusively for a medication such as isoniazid, which is typically used exclusively for the treatment of mycobacterial diseases, it is likely to have better specificity (albeit perhaps decreased sensitivity) than a combination of medications including those that are used for many diseases. Given the current evidence, it is not clear which drugs (or combination of drugs) should be selected for a bioterrorism surveillance program that attempts to achieve a given degree of sensitivity for all or most of the most worrisome biothreat agents.
Medical Historian electronically asks patients questions about their medical histories to derive a historical database of patient information that can be queried for the purpose of disease surveillance. The system performs a computerized elicitation of a review of symptoms. "Diseases" are defined according to symptom-clusters. The database is then scanned to search for outbreaks of symptom-clusters. In a retrospective evaluation of 288 patients with cough and rhinitis, the computer diagnosis was within the 95 percent confidence interval of expert panel diagnosed-conditions for upper respiratory infection. Sensitivity varied from 27 to 90 percent depending upon the symptom-cluster used. No assessments of specificity were provided. Data on the sensitivity of Medical Historian for 4 other computer-surveyed diseases were not stated, but the authors did note that "the number of encounters identified electronically was always fewer than the number identified by the physician panel." 403 It is difficult to interpret these results in terms of surveillance of bioterrorism-related illness.
The HAWK system for surveillance of reportable diseases in Kansas is a promising system that collects notifiable disease reports from clinicians and a variety of public health agencies in the state using a data warehouse model.404, 405 Authorized public health officials can remotely access the data via the Internet to perform analyses. No evaluative data are available about HAWK, which could serve as a model for similar statewide reporting systems.
None of the reports of the systems in this section described the type of hardware required, the system's security measures, or the costs of operating the system.
Summary: Surveillance systems collecting other kinds of data
None of the specific systems described in this section has been designed or evaluated for surveillance for bioterrorism, and only EPIFAR and Medical History have been clinically evaluated. When we consider the systems in this and all the preceding Surveillance sections, we have described at least 1 surveillance system that collects each of the data sources described in Figure 3. However, there are significantly fewer descriptions of systems collecting the earliest surveillance data -- a significant gap in the literature. For example, given that the 53.1 million school-aged children (aged 5 to 17) represent 18.9 percent of the total U.S. population (281.4 million) and that their absenteeism is collected on a daily basis nationally (albeit not always electronically), an evaluation of the sensitivity, timeliness, and cost of this source of surveillance data seems warranted. 406 The surveillance of work absenteeism rates is complicated by the decline in the types of industries that require employees to clock in and out of work and by the lack of these data in an electronic format. Systems that collect pharmaceutical data, like EPIFAR, hold particular promise for bioterrorism surveillance. Pharmaceutical data, particularly over-the-counter medication sales data, could provide an early, if not specific, indication of an outbreak. Additionally, most pharmaceutical sales are tracked electronically. The evaluation of EPIFAR emphasizes the complexity of selecting specific pharmacy data for bioterrorism surveillance. A careful analysis of the detection characteristics of common prescription and non-prescription medications used for the bioterrorism-related syndromes (e.g., antipyretics, cough suppressants, and antidiarrheals) will be required to determine the utility of these data for bioterrorism surveillance.
Analysis and Presentation of Surveillance Data
It is clearly not the function of surveillance to predict the long-range future, but it is only prudent to anticipate the immediate problems that can be expected on the basis of presently known facts and presently accepted concepts, erroneous though some must be.
-- A. Langmuir, 1963 236
Background
Once surveillance data are collected, analysis -- typically by public health officials -- is required to identify patterns suggestive of bioterrorism-related illness. Our search identified a number of methods for analysis of surveillance data. We note however, that we have not conducted a systematic review of surveillance analysis methods per se but have identified methods that have been or could be applied to the analysis of bioterrorism-related illnesses. In this section, we describe the important features of analytic methods for bioterrorism surveillance data.
The primary analytic question in prospective disease surveillance is whether the currently observed disease process differs from the expected process. Before answering this question, the analyst must determine what elements of the data will be modeled: individual attributes (e.g., gender, age), time, and/or space. Additionally, the analyst must consider how the expected process (i.e., the baseline) will be modeled and what threshold will be used to establish a significant change from baseline (i.e., an aberrant disease process). Traditionally, time has been the major modeling consideration, 416 but recent developments in technology and statistics have greatly facilitated the consideration of spatial information in surveillance analyses. We first examine modeling decisions concerned with generating the expected disease process, and then discuss developments in spatial analysis and their implications for bioterrorism detection.
Findings: Modeling the baseline characteristics of surveillance data
Models of the expected disease process are typically derived from the historical pattern of disease, either over the immediately preceding time interval (e.g., 30-day moving average), or over one or more historically corresponding intervals (e.g., the mean rate for the first week in January over the past 5 years).417, 418 This approach is straightforward, but it ignores the underlying dynamics of disease propagation through the population. Several articles identified in our search describe methods for stochastically modeling the spread of communicable disease epidemics.419-423 These methods have not been widely used for the modeling of disease surveillance, but they may allow more accurate determination of the expected disease rates and deviations from the expected.
More accurate descriptions of the expected and observed disease rates should enable more accurate identification and characterization of data aberrations and disease outbreaks. Because the need for timeliness is particularly acute in bioterrorism surveillance, application of these methods to disease surveillance merits further attention. It should also be possible to apply similar methods to model the disease processes associated with outbreaks of non-communicable diseases of bioterrorism, such as anthrax. Potential drawbacks of this modeling approach include its added complexity and the need to collect or estimate data for a number of parameters, such as the proportion of susceptible individuals, herd immunity, and mixing rate.
Findings: Consideration of space in surveillance analyses
The importance of spatial location for understanding disease processes has been appreciated for many years. Spatial location has a central role in the epidemiological triad of "person, place and time," and maps have long been used to identify important patterns in diseases such as cholera, cancer, leprosy, pneumonia, and smallpox. 252 Consideration of the spatial dimension of surveillance data may enable more timely identification and characterization of important patterns. Recent developments in IT and statistics, most notably the development of user-friendly geographic information system (GIS) technology, can facilitate consideration of space in surveillance data; however, the effective application of these developments to surveillance analysis is not yet well established. 424 In this section, we examine how GIS technology and other spatial analysis tools have been applied to bioterrorism-related diseases.
A GIS is an automated system for collecting, organizing, analyzing and presenting geographically referenced data. Beyond simple mapping, a GIS can perform complex functions such as automated address matching, distance calculations, buffer analyses (i.e., calculation of a buffer zone of variable width around a point, line, or area), spatial queries (i.e., the ability to select observations based on their geographic characteristics), and linking data sources by spatial location. 425 The primary effort to develop global mapping capability for public health data has come from the joint WHO/UNICEF program HealthMap -- a data management, mapping and GIS system for public health. The program was initially created in 1993 to establish a GIS to support management and monitoring of the Guinea Worm Eradication Programme. 426 Since 1995, however, in response to the increasing demand for mapping and GIS technologies from a much wider range of public health administrators, the scope of the work has been broadened to include the promotion and use of GIS technology for other disease control programs. 426
Most applications of GIS that we identified in our review performed retrospective analyses to identify spatial patterns of disease and/or disease vectors. However, several of the syndromal surveillance systems described in this Report (such as ESSENCE and RSVP®) use GIS or spatial analysis in an ongoing and systematic manner to identify disease outbreaks or data aberrations. Some studies used Global Positioning System technology to locate disease cases.427, 428 Analytic methods employed within a GIS environment ranged from simple, descriptive spatial analyses,429-431 to more complex spatial432, 433 and space-time modeling.427, 428 The main objective of the analyses was usually to characterize the relationship between disease vectors and cases. In some instances, this relationship was used to forecast future disease outbreaks based on predicted disease vector ecology.433-436
Summary: Analysis and presentation of surveillance data
The studies presented, with their focus on predicting new outbreaks from previous outbreaks and disease vector distributions, may seem unrelated to bioterrorism surveillance. The lack of previous large-scale bioterrorism attacks, and the fact that most agents of bioterrorism are not vector-borne, prevent this type of predictive modeling for bioterrorism. However, a number of the specific technologies and GIS/spatial analytic methods employed in these studies may be directly applicable to the analysis of bioterrorism surveillance data. These methods and technologies include remote sensing, data integration, spatial interpolation, and space-time statistical analysis. For example, remote sensing data may help to identify potential effects of meteorological conditions for airborne dispersal of a bioterrorism agent. The ability of a GIS to integrate disparate data sources (e.g., pharmacy sales data, ambulance activity, and ICD9 codes) according to their spatial location may enable the identification of important relationships that indicate early disease behavior. Spatial interpolation could be used to predict risk in locations for which similar data are not available. Finally, advanced space-time analytic methods can take advantage of the spatial dimension of data to detect aberrations with greater sensitivity and timeliness. The combination of these analytic methods with rich descriptions of the expected disease process (as described previously) may provide even greater sensitivity for identifying bioterrorism attacks in the midst of normally occurring disease outbreaks. We note that no published report has specifically evaluated whether a surveillance system that uses both temporal and spatial analyses is likely to be more timely or sensitive than a system that performs only temporal analyses.
Reporting and Communication Systems
Background
On March 20, 1995, the Aum Shinrikyo sarin attack on the Tokyo subway system resulted in 7 deaths and the medical treatment of an additional 250 people. 10 A group of physicians in Matsumoto, Japan, who had treated victims of a sarin release by the Aum Shinrikyo a year before, heard about the attack, and sent information about typical case presentations to Tokyo hospitals and the Ministry of Health and Welfare. 10 Although this information was reportedly helpful, 10 a more systematic approach to the dissemination of medical and intelligence information could have benefited clinicians, public health officials, and victims. In particular, public health officials could have used intelligence information from law enforcement officers regarding the increased likelihood of such an attack and disseminated medical information about the clinical presentation and therapeutics for nerve agents before such an attack occurred. 10
The purpose of communication in the midst of a bioterrorism event is the timely provision of information to relevant responders and decision makers so that appropriate action is undertaken. As presently configured in the U.S., the communication pathway for public health information (such as treatment, prophylaxis, and laboratory protocols) is intended to move from national and international agencies (principally the CDC and WHO, with intelligence provided by the Central Intelligence Assocation (CIA), Federal Bureau of Investigation (FBI) and law enforcement agencies) to the state and local public health officials responsible for the dissemination of information to local decision makers (Figure 11). During October and November 2001, the dissemination of information about confirmed and suspected cases of anthrax in the U.S. and methods for its detection, laboratory testing, treatment, and prophylaxis tested this pathway. The media reports of anthrax cases created a demand for information that exceeded the capacity of national, state, and local health departments. The information on laboratory protocols, reagents, and training available before October 2001 was not sufficiently detailed to meet the information needs of first responders, clinicians, and others. 437 Just as had been predicted by the Dark Winter Exercise, asynchrony in the provision of information between the media and public health officials contributed to the perception that the public health officials had lost control of the situation. 11
After the anthrax cases of late 2001, the National Association of Counties conducted a telephone survey of county public health directors. Completed surveys were obtained from 300 of the 946 county public health directors. 438 Thirty-five percent of county public health directors indicated that insufficient communication networks were considered obstacles to their health department's ability to respond to a bioterrorism or chemical warfare crisis. 438
The discrepancy in reporting time of public health information between the media and public health officials derives principally from the different missions of these organizations. Whereas it is the media's duty to collect, verify, and report news as it becomes available, public health officials' primary obligation is to ensure the public health, which often requires time-consuming analyses of available data and consideration of appropriate responses before notification of the public. For international news organizations to be able to report on a 24-hour news cycle, they must be supported with sophisticated information systems that facilitate the management of their data. As evidenced by the findings in this section, public health officials typically do not have similarly robust communication infrastructures at their disposal.
Alarmed by news of the anthrax cases and contaminated mail, patients sought information about biothreat agents from credible sources such as local public health departments as well as from their personal physicians. Efforts to respond to calls and faxes from concerned patients and clinicians stretched the limited resources of public health departments. Similarly, emergency departments, urgent care clinics, and clinicians' offices strained to meet the information needs of the "worried well."
Reporting and communication systems that facilitate the secure delivery of information from public health officials to the public and from clinicians to their patients could have helped to dispel the perception of chaos and inundation. Specifically, if clinicians had had an effective means of communicating electronically with their patients, they might have been able to provide reassurance without necessitating office visits and phone calls. Similarly, they could use such a system to notify patients of a suspected bioterrorist attack, describe the characteristic signs and symptoms, and disseminate criteria for seeking medical attention. In a recent survey, 13 percent of doctors responded that they e-mail their patients. 439 This low number may be related to the belief, held by some physicians, that electronic communication carries hazards of its own. Barriers to implementation of electronic communication between physicians and patients include physician concerns that this additional mode of communication will add to already busy practices, 439 that e-mails from patients regarding matters requiring urgent attention that will be missed due to delays in checking e-mail, 439 and that certain e-mail systems will not comply with HIPAA.
On September 11, 2001, as in other times of crisis, the volume of calls from around the world into the affected areas exploded. Local telephone systems overloaded and played a standard message saying there was no phone access to that zone -- a message that, in its lack of specificity about what was happening in the area, may have served to increase the callers' apprehension. 172 Additionally, much of the communication infrastructure in the affected area of New York City was physically interrupted by the loss of electric power, cables, servers, and radio transmitters. One lesson that can be learned from these experiences is that, after a bioterrorist attack -- particularly if it is combined with other acts of terrorism such as a physical, radiologic, or chemical attacks -- local phone, fax, and phone-modems may be unavailable. Clinical information systems that rely on these modes of communication (e.g., laboratory data about patients and surveillance alerts from public health officials to clinicians) may be affected. Under these circumstances, access to the Internet will be limited to wireless connections or cable television. 172 Similarly, e-mail will not be available via the phone lines but only via Web mail.
Security is one of the most critical features of a communication system for bioterrorism. The 3 main types of security concerns for these systems are: (1) maintaining patient confidentiality by ensuring that the information is disclosed only to authorized persons (i.e., this issue is addressed by HIPAA regulations and by systems that use role-based access to information); (2) maintaining the accuracy and completeness of the data (i.e., preventing unintended changes to the original data that would compromise subsequent analyses and conclusions); and (3) maintaining the availability of the system so that it is functional when it is needed (i.e., preventing system overloads so that it will be useful to responders in the event of a bioterrorist attack). Security violations can disturb all 3 of these elements of a system. In the event of a bioterrorist attack, it is possible that there may also be cyberterrorist attacks on information and communication resources. Alternatively, systems may simply overload if demand for access exceeds capacity. Communication systems for bioterrorism must include adequate redundancy to avoid overloads, as well as security measures to prevent and respond to cyberattack.
Evaluation criteria
We evaluated each of the descriptions of reporting and communication systems for the following information (Table 2 -- Reporting and Communication; Evidence Table 4): the purpose of the system, the type of information the system is intended to communicate, the intended provider of the information being communicated, the intended recipient of the information, whether the recipient has to actively seek the information from the provider (e.g., by visiting a Web site) or the information is transmitted by the provider by phone, fax, e-mail, or other means to the recipient (i.e., passive on the part of the recipient), the timeliness of the system, type of hardware required, and the system's security measures.
Findings
We identified 26 ITs that could be used to support the reporting and communication needs of decision makers during a bioterrorism event, 7 of which have been described in a peer-reviewed evaluation report (Tables 3 and 24; Appendix H).
The systems vary with respect to their purposes: 8 for communication among public health officials at local, state and federal levels; 2 for communication among public health officials, clinicians, and the public; 4 for communication between patients and clinicians; 5 for the automated communication of information from electronic medical records of patients to clinicians; 1 for communication from professional clinician organizations to clinicians; 3 for communication between emergency departments and first-line emergency response personnel; and 3 for other kinds of communication.
The technologies that have been subjected to the most evaluations are those that send an alert to a clinician based on a worrisome finding in the patient's medical record (typically a laboratory test result, radiologic finding, or medication error). All 5 of the systems in this category have been clinically evaluated, typically for outcomes related to clinician acceptability, time to respond to alerts, or changes in numbers of medication errors. Although clinicians reported being annoyed by erroneous pages or alerts, in general these systems tended to decrease the interval between a laboratory result becoming available and action being taken by the clinician. If these systems increase the speed with which clinicians receive new information that may affect their management decisions, they may provide a useful tool in the event of a bioterrorist attack.
The Program for Monitoring Emerging Infectious Diseases (ProMED©)440-442 is an independent system that was specifically designed by the Federation of American scientists to provide early warning of possible bioterrorism events caused by infectious agents or toxins, including agroterrorism. Reports of outbreaks of human, animal, and crop diseases from around the world are screened and assessed for quality before being distributed (24 hours a day, 7 days a week) to 24,000 subscribers in over 150 countries. These subscribers include clinicians, public health officials, first responders, veterinarians, bioterrorism and agroterrorism experts, members of the CIA and FBI, school teachers and their pupils, and newswire services and newspapers. 172 ProMED© is both a surveillance and communication system that increasingly serves as a model for the development of similar systems within individual countries. 172 In an internal study, ProMED© provided more timely and more numerous outbreak alerts for emerging diseases and toxins as compared to the WHO, CDC and Cable News Network (CNN). 172 For example, ProMED© reported cholera outbreaks in 11 countries in 1999 between 3 days to 11 weeks earlier than the WHO. 441 Over 97 percent (342 out of 351) of ProMED©'s alerts for a 7-month period were subsequently confirmed by official sources (including 6 official reports that were later retracted). 441
Whereas a system like ProMED© reports the same information about a potential disease outbreak to all subscribers, other types of systems are required to disseminate secure outbreak information to limited members of the public health community. Specifically, the many levels of the U.S. public health system (local, state, and national) need an integrated communication system to which all officials have access. The Health Alert Network (HAN)443, 444 is a secure Web-based information and communication system designed by the CDC to link local and state public health agencies with each other and with other organizations responsible for responding to a bioterrorism attack. Public health officials and first responders can access a variety of useful data, including disease reports, response plans, and management guidelines. In addition, the nationwide system includes early warning broadcast alert and distance-learning functions. Since September 11, 2001, HAN has distributed multiple health alerts via e-mail and fax about bioterrorism preparedness.
The California initiative to develop a Rapid Health Electronic Alert, Communication, and Training system (RHEACT) serves as a model for the expansion of a system like HAN.445, 446 RHEACT is a Web-based system, designed to support NEDSS standards, that is intended to serve as a secure environment for collaborations within the public health community. Its mission includes management of episurveillance and laboratory data, command and control of emergency response, and reporting disease outbreaks both vertically (i.e., from clinicians to local health departments to state health departments to the CDC and vice versa) and horizontally (e.g., among all counties within a state). After signing onto RHEACT, the user has access to numerous software modules, including statistical analysis, word processing, and graphics programs. The developers intend to provide a uniform Web portal that supports a number of commercially available software tools. The security of the system is largely derived from a role-based identification system in which users are registered into the system according to their particular role (e.g., communicable disease officer, laboratory director, clinician, epidemiologist, anthrax expert) in a particular public health jurisdiction. Access to the multiple data sources and reporting functions are restricted according to role and jurisdiction. RHEACT's automated notification system enables the user to type in an alert, select the jurisdiction to which the alert is to be sent, and (if coded as a Low Priority alert) send an alert via e-mail to all selected recipients. If the alert is coded as High Priority, the system will phone all recipients and will continue to try all alternative phone numbers associated with a recipient (e.g., office phone, then home, then cell phone) until the recipient is reached, and then play the recorded message. RHEACT tracks when the recipient has opened the e-mail or when the phone message was delivered. Most local public health departments send out alerts to clinicians via fax and have no way of verifying if the intended recipient received the information. RHEACT's alert system could represent a major advance over these currently used faxing systems. RHEACT's communication features are similar to those of HAN and EpiX (Table 24) and may replace these older systems. 303
Table
Table 24. Reporting and communication systems.
Two additional systems, currently under development, show particular promise for serving the communication needs of clinicians and public health officers in the event of a bioterrorist attack: the Global Disaster Information Network (GDIN) and the Urban Security Initiative. GDIN is an interagency disaster system for information and decision support being developed to reduce the impact of disasters by integrating relevant information from all sources and making it available to decision makers and the public. 447 GDIN was piloted in hurricane simulation exercises in Florida and was reported to be "extremely useful;" however, no additional details about its functions or evaluations are publicly available. 447 The Urban Security Initiative is under development at the Los Alamos National Laboratory. 448 It will create centralized computer systems to help cities respond to emergency situations, including chemical and biological attacks. 448 The objective of the Urban Security Initiative is to develop an Internet-based Web environment that allows multiple organizations to solve collective problems. Its pilot project is a Web-based emergency planning effort in which 20 different agencies are working together for earthquake preparedness in the Los Angeles area. The disaster-preparedness Web environment consists of 3 components: (1) detailed scenario data from earthquake simulations, along with damage estimates from different potential earthquakes; (2) information about the mission and capabilities of each of the 20 involved agencies; and (3) an interactive area where each agency can sort disaster planning issues according to importance, order necessary actions in time sequence, and request resources.
In the event of a bioterrorist attack, it is essential that clinicians, public health officials, and other users of public health information have access to information that is easy to find, current, and correct. We found many physician groups, professional organizations, and news services that send e-mail notifications of news stories, articles, and clinical updates to their members; however, we have no evidence about the quality of the information or how it is used. One example is a Web site developed by researchers in the Center for Disaster Preparedness at the University of Alabama at Birmingham with sponsorship from AHRQ that offers 5 online courses on the clinical features of bioterrorism-related illnesses with free continuing education credits to 315,000 clinicians. 449
The CDC and WHO Web sites satisfy some of the information needs of public health officials and clinicians; however, these organizations have limited resources to devote to immediate, real-time dissemination of information regarding an outbreak resulting from bioterrorism. The CDC regularly holds press conferences and issues press releases and other media alerts on outbreaks. It maintains a Web site, accessible from the CDC home page, which provides updates on various biothreats and ongoing investigations. 450 It incorporates a Media Relations tab, which contains updated press kits and press releases, an archive of press releases, and telebriefing access.
In terms of security measures, the systems vary greatly. Many of the systems described being "HIPAA-complaint" without additionally specifying the measures to maintain patient confidentiality. No system described measures to prevent attack from cyber terrorists or to maintain adequate capacity in the event of a surge in demand.
Summary: Reporting and communication systems
The 26 systems described in this section represent only those for which peer-reviewed analyses, government reports, or Web-based information was available. There are undoubtedly many similar systems for which published data were unavailable. In particular, the development and implementation of systems for use by public health officials is likely to occur without publication of reports since these officials' responsibilities do not typically include preparation and publication of journal articles.
In Figure 12, we mapped each of the included reporting and communication systems to each communication need identified in Figure 11 (and described in our Task list, Table 2). Arrows marked with an asterisk indicate that an IT described in this section transmits information between the noted parties. Arrows marked with an "S" indicate that a surveillance system described in a previous section transmits information between the noted parties. Broken arrows indicate those relationships that are not currently supported by a specific IT or surveillance system included in this Report.
The systems with the most evidence for effectiveness are the alert systems that notify clinicians of abnormal findings in their patient's electronic medical records. Although these systems are limited to institutions with electronic medical records, they could play an important role in improving the timely recognition of disease associated with a biothreat agent.
As was demonstrated in the Dark Winter exercise, formal communication systems between members of the media and public health officials are lacking. 11 The media page of the CDC's Web site is a passive means of communication between public health and the media. ProMED© has subscribers from all of the groups depicted in the communication diagram, including the media, and currently serves as an independent source of bioterrorism and agroterrorism information to all groups with a need to know. ProMED© is a recognized leader in the international effort to rapidly report and disseminate information on a wide range of biothreat agents. However, information from ProMED© to the media does not serve the same purpose as a system specifically designed for public health officials to communicate information about outbreaks to the public. This represents a major gap in the currently available technologies.
We found 8 separate systems that link various members of the public health community. There are ongoing efforts to integrate communication of public health information vertically and horizontally within the U.S. public health system.
In the event of a bioterrorism event, clinicians must be able to rapidly communicate with their patients. Systems exist that enable Web-based communications between these parties in a HIPAA compliant manner. However, their utility in crisis situations may likely remain limited unless their use for routine communications increases. Robust security measures will be a necessary component of any bioterrorism-related communication system.
Integrated Surveillance, Communication, and Command and Control Systems
Background
Some systems are designed primarily for facilitating command and control functions. Typically, these systems are designed to collect a stream of data -- some from detection systems and others from meteorologic forecasts -- and apply a mapping function to perform analyses and make predictions for decision makers in a command and control center. Although they are principally designed for use by incident commanders, we have included these systems because they may have some utility for public health officials.
Evaluation criteria
We evaluated each of the reports of integrated surveillance, communication, and command and control systems for the following information (Table 2 -- Surveillance and Reporting and Communication): the purpose of the system, the type and method of surveillance data collected by the system; timeliness of data collection, analysis and presentation to the decision maker; methods for determining when an outbreak has occurred; geographic area under surveillance; the type of hardware required; the system's security measures; the direct costs needed to operate the system; the intended provider of the information being communicated, the intended recipient of the information, whether the recipient has to actively seek the information from the provider (e.g., by visiting a Web site) or the information is transmitted by phone, fax, e-mail, or other means to the recipient (i.e., passive on the part of the recipient), the timeliness of the system, type of hardware required, and the system's security measures.
Findings
In addition to the systems already described with integrative command and control functionality (e.g., LEADERS, PortalShield, and JWARN), our search found 7 other systems with similar purposes; none has been clinically evaluated (Tables 3 and 25).
Table
Table 25. Integrated surveillance, communication, and command and control systems.
In addition to their utility in managing actual crises, these systems may have utility in preparing for events and training personnel. For example, the Meteorological Information and Dispersion Assessment System Anti-Terrorism (MIDAS-AT)479, 480 could be used by event planners to consider vulnerabilities in event security and train staff accordingly. Most of these systems include mapping functions and several report having analysis capabilities to provide decision makers with current information about the status of the event (e.g., where sensor data are reporting abnormal aerosols and where hospitals have exceeded capacity) and predict the spread of the outbreak.
The information about these systems was primarily derived from Web-based information provided by the manufacturers. These descriptions provided no data about timeliness, security measures, hardware requirements, or most of the criteria by which we had intended to evaluate them.
Summary: Integrated surveillance, communication, and command and control systems
These systems are designed to be used by incident commanders, emergency management personnel, and the military; none has been clinically evaluated. However, data from these systems could be integrated with data from environmental detection and surveillance systems. Related projects, combining multiple sources of disparate data for combined surveillance, communication, and incident command capabilities, are currently in development.
Quality Evaluations
Background
In this section, we present the results of 2 quality evaluations of articles that reported evaluations of systems. First, we applied the quality guidelines from researchers at McMaster University33, 34 to the peer-reviewed evaluations of IT/DSSs for detection, diagnosis, management and prevention, or reporting and communication. Second, we applied the CDC's quality guidelines 35 to peer-reviewed evaluations and descriptions of surveillance systems. If a given quality criterion was not specifically described by the reference or it was not applicable to the material presented in the article, we so noted. We did not attempt to independently evaluate any system; instead, we relied exclusively on the authors' reports.
Findings: Quality evaluation of reports of detection, diagnosis, management, and communication systems
We evaluated each of the included 48 peer-reviewed reports of evaluations of the 35 IT/DSSs for detection, diagnosis, management/prevention or reporting/communication based on 5 criteria: method of allocation to study groups (e.g., random), unit of allocation to study groups (e.g., clinician), baseline group differences (e.g., no baseline differences between study and control groups), type of outcome measures (e.g., objective), and completeness of followup (Evidence Table 4). A study of the DERMIS system 178 is represented twice in Evidence Table 4 because it included 2 separate evaluations of 2 different outcomes. Therefore, the total number of evaluations included in Evidence Table 4 is 49. Twenty-eight of 49 studies evaluated the IT/DSS using objective outcome measures. Fifteen of the 49 studies reported followup rates in excess of 90 percent. However, for 3 of the quality criteria -- unit of allocation, method of allocation, and baseline differences -- fewer than 10 studies fully or partially satisfied the criteria. The overall quality of the studies was difficult to assess because the studies often did not report sufficient information for us to rate the quality criteria (Figure 13).

Figure
Figure 13. Quality evaluation for peer-reviewed evaluations of detection, diagnostic, management, and communication systems.
Findings: Quality evaluation of reports of surveillance systems
We reviewed each of the 31 peer-reviewed evaluations and 30 descriptive reports of 39 systems (note, many systems were reviewed in multiple articles) (Evidence Table 5). A total of 61 articles were reviewed. For each of these articles, we abstracted whether the authors specifically described the following characteristics: usefulness, importance, timeliness of the information, flexibility of the system, acceptability of the system, system sensitivity and specificity, simplicity of system use, and representativeness. Often, the discussion of these characteristics in the report was quite modest, and based on opinion about the system rather than a formal evaluation of the characteristic. No reports addressed all characteristics. At least 40 of 61 reports of evaluations or descriptions of surveillance systems described the timeliness, importance and usefulness of the system (Figure 14). However, less than one third of the reports of evaluations of surveillance systems described the representativeness, simplicity, sensitivity, acceptability or flexibility of the system.

Figure
Figure 14. Quality evaluation for peer-reviewed evaluations of surveillance systems.
Summary: Quality evaluations
The evaluation articles of IT/DSSs for detection, diagnosis, management/prevention, or reporting/communication did not report many of the important characteristics of a clinical evaluation. We emphasize that just because the articles did not report a particular characteristic, does not mean that the study was not appropriately designed. It merely indicates that we are not able to make a judgment about that element of the study design. Similarly, many of the articles on surveillance systems did not report some of the most important characteristics of these systems including their sensitivity and acceptability. Consideration of the quality of the evidence regarding the effectiveness of the systems reported by these articles is thereby limited. The literature on the utility of available IT/DSSs to meet the information needs of clinicians and public health officials would benefit from additional articles reporting in sufficient detail so that the evidence may be objectively considered.
Technical Information About the IT/DSSs
Background
We abstracted technical information about each IT/DSS including what kind of hardware platform it uses, whether it uses standard vocabularies, what kind of reasoning it uses, what security measures it has, and whether access to the system is restricted by user type (Appendix C). Security concerns are important for all systems. The other questions were least relevant to the detection systems and most relevant to the diagnostic, management/prevention, and reporting/communication systems.
Findings
In general, very few of the reports of the IT/DSSs provided complete technical information. For those reports that did describe these technical characteristics, we have included this information in the relevant Table and/or Evidence Table about that system.
Most diagnostic DSSs are operated on desktop personal computers (PCs); however, the radiologic and telemedicine systems require additional hardware, and newer handheld versions of some PC-based systems are increasingly available. The reasoning used by these systems includes both probabilistic (Bayesian) and rules-based methods. Additionally, some of the systems relied on neural networks to generate their outputs. Generally, only the radiologic systems were incorporated into the IT infrastructure of the hospital in which they were operating. None of the reports of these systems specifically described restricting access to the system by user type or other security measures.
Many of the management and prevention systems were dependent on an electronic medical record. As with the diagnostic systems, the inference engines used a wide variety of reasoning methods. Some systems specifically reported using the Systematized Nomenclature of Medicine (SNOMED®) vocabulary; others simply indicated that "a standard vocabulary was used" but did not specify which one. Since most users of these systems had to log in to the hospital system before any recommendations were generated, this provided some measure of security. None of the reports of these systems specifically described restricting access to the system by user type.
The collection of surveillance data depends on reports sent via mail, e-mail, fax, and the Internet. Very few of the descriptions of surveillance systems addressed security concerns.
The reporting and communications systems vary enormously with respect to their technical characteristics. Most of the clinician alerting systems required an electronic medical record system with a rules-based reasoning system that sent e-mail or pager alerts. Generally, the communication systems within the public health system and between clinicians and their patients are Web-based, require passwords, and have some security mechanisms including encryption of patient information. Several systems reported being HIPAA-compliant, and one system has role-based access for users. None of the reports of these IT/DSSs described measures to prevent system overload in times of surges in demand or to thwart cyber attackers.
Summary: Answers to the Key Questions
Key Question 1: What are the information needs of clinicians and public health officials in the event of a bioterrorist attack?
The information required by clinicians and public health officials while preparing for and responding to bioterrorism events relates to the decisions they have to make and the tasks they have to perform. We have described these decisions in the Conceptual Model (Figure 1) and these tasks in the Task Decomposition (Table 2).
Briefly, clinicians require the necessary information to make diagnostic, management, prevention, and reporting decisions. Diagnostic decisions require information to accurately estimate the pre-test probability of disease for a given patient. This includes information about the probability of a patient's exposure to a biothreat agent, their susceptibility for developing bioterrorism-related illness, and the clinical syndromes associated with bioterrorism-related illnesses. Clinicians' interpretation of test results requires information about the sensitivity and specificity of the test. The probability of disease, given a positive or negative test, cannot be calculated without knowledge of the sensitivity and specificity of the test. Because clinicians' decisions will depend on their assessment of the probability of disease after testing, lack of information about sensitivity and specificity critically limits diagnostic decision making. Management decisions require information about how to appropriately distinguish between those patients who need treatment and those who do not, how best to treat the acutely ill, whom to isolate and how, how to manage scarce resources, and how to maintain personal safety. Prevention decisions require information about prophylaxis and vaccination protocols. Reporting decisions rely on information about what constitutes a reportable case or cluster of cases, and about the kinds of data that public health officials seek.
Similarly, the information that public health officials require to prepare for and respond to a bioterrorism event can be considered in relation to the decisions they have make. The decision to perform an outbreak investigation requires information about the baseline characteristics of the surveillance data and threshold levels that suggest that an outbreak resulting from naturally occurring or bioterrorism-related illness may have occurred. This information includes (for each source of surveillance data): timeliness, sensitivity, and specificity, expected value of rates being monitored, and method for determining the outbreak threshold. Once a bioterrorism event has been identified, public health officials require information that will enable them to perform ongoing surveillance in the midst of the crisis to track the extent and spread of the epidemic. The decisions regarding the institution of epidemiologic control measures that prevent the spread of disease require information about the transmissibility of the suspected biothreat agent(s) and about the criteria for and effectiveness of prophylaxis and quarantine strategies. Decisions to issue a surveillance alert require information about the nature of the suspected bioterrorist attack and the characteristics and expected natural history of the suspected biothreat agent(s). Other communication decisions relate to the specific information that needs to be conveyed to other public health officials, clinicians, the media, and other decision makers.
Key Question 2: Based on the information needs identified for these decision makers, what are the criteria by which IT/DSSs should be evaluated with respect to usefulness during a bioterrorism event?
The evaluation criteria vary depending on the purpose of the IT/DSS. To answer Key Question 2, we present the evaluation criteria for the IT/DSSs in the Task Decomposition (Table 2) and immediately preceding the findings in each of the Results sections.
For detection systems, important evaluation criteria in this Report include the following: the purpose of the system, information regarding the type of sample collected, portability, and methods for maintaining the security of the sample. For the collection systems we also evaluated the collection efficiency, limits of size of particulate collected, and flow rate. For the particle counters, biomass indicators, and identification systems we also evaluated sensitivity, specificity, the upper and lower limits of the size of particles that can be counted (for the particle counters), and the concentration of organisms that can be detected (for the biomass indicators). For the identification systems we also evaluated the amount of time it takes to run a sample, the number of samples that can be run at a time, the number of biothreat agents it can identify, and whether it can identify both toxins and organisms. We evaluated each of the reports of integrated collection and identification systems according to the same criteria for each of the component systems.
For diagnostic DSSs, important evaluation criteria include the following: the purpose of the system, the type of information required by the DSS (e.g., a manually-entered list of signs and symptoms provided by the clinician), the type of information provided by the DSS (e.g., a list of differential diagnoses with or without associated information about the diseases of interest), diagnostic sensitivity and specificity, whether the biothreat agents and their associated illnesses are included in the knowledge base, the method of reasoning used by the inference engine, information regarding the ability to update the probability of biothreat-related illness as the epidemic progresses, and the type of hardware required.
For management systems, important evaluation criteria included the following: the purpose of the system, the type of information required by the system (e.g., patient information from an electronic medical record), the type of information provided by the system (e.g., antibiotic recommendation or quarantine recommendation), information about the manner in which the management recommendations are provided (e.g., whether the recommendations are provided in an unprompted manner to the user), timeliness of management recommendation, the accuracy of the management recommendations, whether the biothreat agents and their associated illnesses are included in the knowledge base, the method of reasoning used by the inference engine, whether the system uses a standard vocabulary, information regarding the ability to update recommendations as the epidemic progresses, type of hardware required, and the system's security measures.
For surveillance systems, important evaluation criteria included the following: the purpose of the system, the type and method of surveillance data collected by the system; timeliness of data collection, analysis and presentation to the decision maker; methods for determining when an outbreak has occurred; geographic area under surveillance; the type of hardware required; the system's security measures; and information regarding the public health importance of the health event under surveillance, the system's usefulness, simplicity, flexibility, acceptability, sensitivity, specificity, representativeness, and the direct costs needed to operate the system.
For reporting and communication systems, important evaluation criteria included the following: the purpose of the system, the type of information the system is intended to communicate, the intended provider of the information being communicated, the intended recipient of the information, whether the recipient has to actively seek the information from the provider (e.g., by visiting a Web site) or the information is transmitted by the provider by phone, fax, e-mail, or other means to the recipient (i.e., passive on the part of the recipient), the timeliness of the system, type of hardware required, and the system's security measures.
Key Question 3: When assessed by these criteria, in what ways could existing IT/DSSs be useful during a bioterrorism event? In what ways are they limited?
Our review identified 217 IT/DSSs, few of which were designed specifically for response to bioterrorism. Rather, most included systems had other intended purposes, but could conceivably be useful to clinicians or public health officials in response to a bioterrorism event. The evidence on which to judge the usefulness of these systems is limited. Many of the systems were not evaluated even for their intended purpose. Of the studies that did evaluate systems for their intended purpose, few adhered to published criteria for high-quality evaluations. Even if a system was found useful for its intended purpose, we cannot infer that the system necessarily would be useful for response to bioterrorism. For example, surveillance systems that may function effectively for their intended purpose (e.g., the detection of naturally occurring outbreaks such as influenza) may not provide information quickly enough to be useful in rapid detection of a bioterrorism event. We now describe the evidence we identified in more detail.
Detection systems
The 55 collection, particulate counters and biomass indicators, and rapid identification systems described in this Report have critical roles to play in the detection of a covert release of a biothreat agent. Additionally, they are required by first responders and clinicians to test environmental and clinical samples after a known release. However, the paucity of comprehensive evaluative information about these systems prevents conclusions about whether or not one or more of these systems is likely to be useful for these purposes.
The evidence on detection systems was descriptive and predominantly collected from government sources and manufacturers' Web sites. We note that the definitions of what constitutes a "rapid" or "portable" test varied widely. We found no reports that directly compared 2 or more of the commercially available systems in any given category. Additionally, few of these systems have been compared to a gold standard and their sensitivity and specificity remain poorly characterized in the publicly available literature. The few reports of evaluations of the antibody-based systems, which are available for less than 10 of the most worrisome biothreat agents, have been characterized by high false positive rates. The nucleic acid-based systems are limited by the availability of sensitive probes but are promising in terms of portability, timeliness, and ability to communicate results to decision makers at remote locations. A significant gap in the literature is an analysis performed by an independent research group comparing the most promising technologies to each other and to a gold standard. For most systems, the available information does not describe if reagents are widely available, how difficult it is to train first responders in the use of these systems, how difficult it is to use these devices to collect and analyze samples in a secure manner in the event that that are used as evidence in a criminal investigation of the bioterrorist attack, and how much it would cost to fully implement these systems.
Diagnostic systems
We identified 23 diagnostic systems with potential utility for enhancing the likelihood that clinicians consider the possibility of bioterrorism-related illness. None of these DSSs has been evaluated formally with respect to bioterrorism response. Three of the general diagnostic DSSs have been evaluated for their intended (non-bioterrorism related) purposes. In these evaluations, the general diagnostic DSSs typically performed better than physicians-in-training but not as well as experienced clinicians, and they performed better on more straightforward cases but less well for difficult cases.
The evidence for the utility of telemedicine systems for bioterrorism is mixed. Telemedicine systems are most useful in areas with limited direct access to medical specialists. Because acts of bioterrorism against civilian populations may be more likely to occur in population centers than in remote areas, the usefulness of these systems may be limited. However, since few practicing primary care or emergency physicians have ever seen the rashes associated with smallpox or other bioterrorism-related illness, the use of teledermatology technologies may increase the likelihood of a timely diagnosis by facilitating access to dermatologic experts. Additionally, in the event of a widespread epidemic reaching geographically isolated areas, public health officials could use existing telemedicine infrastructures to relate public health information and alerts to clinicians.
The radiologic system from the University of Chicago (Table 9) has established utility for the diagnosis of community-acquired pneumonia. However, because the radiologic findings for most bioterrorism-related illness will be similar to pulmonary diseases of other etiologies and because the presence of a specific radiologic finding associated with bioterrorism-related illness is the exception rather than the rule, it is not clear that radiologic systems could help clinicians beyond alerting them to the presence of a pulmonary infiltrate, pleural effusion, or widened mediastinum.
Table
Table 9. Radiologic interpretation systems.
The reports of diagnostic DSSs have several important limitations. First, all of these systems (except the telemedicine and radiologic systems) require clinicians to manually enter data -- a laborious step that may be a barrier to the use of these systems and has been demonstrated to increase inter-user variability. Second, GIDEON and The Computer Program for Diagnosing and Teaching Geographic Medicine (Table 12) are the only systems for which we were able to obtain lists of the diseases included in the knowledge bases and could verify that the most worrisome biothreat agents were included. However, these 2 systems are limited in that they are not general diagnostic systems but specific for infectious diseases. Thus, if the patient does not present with either a fever or other signs or symptoms associated with infectious diseases, even the clinician with access to these specialized systems may not choose to use them. Finally, most diagnostic DSSs use probabilistic information about the likelihood of disease. Because bioterrorism-related illness is relatively rare, in the event of a bioterrorism event they will have inappropriately low pretest probabilities for biothreat agents. None of the diagnostic DSSs reported being able to change the probability of disease based on information about suspected bioterrorism events.
Management and prevention systems
Management and prevention systems are designed to make recommendations to clinicians by abstracting clinical information from electronic medical records to make patient-specific recommendations. None of the 18 systems identified in this review has been specifically designed or evaluated for utility in providing management or prevention recommendations during a bioterrorism event. Moreover, we have no information as to whether the knowledge bases of these systems include comprehensive information about bioterrorism-related illnesses. The systems that are not linked to electronic medical records share many of the limitations of the general diagnostic systems -- including, that clinicians may not use the system to seek advice for patients presenting with common viral syndromes (i.e., the bioterrorism-related syndromes). Expert systems that continuously search electronic medical records (including data from the laboratory, radiology reports, and physician notes) for new evidence of an infection and apply clinical practice guidelines to those data have potential utility in bioterrorism management. However, to establish their utility for improving management or prevention decision making during a bioterrorism response, evaluations of the hospital IT infrastructures and methods for the incorporation of clinical practice guidelines for biothreat-related illnesses are required. Antibiotic recommendation programs are typically designed to provide recommendations for antibiotics with the narrowest possible spectra, thereby reducing the risk of developing resistant organisms. If clinicians make antibiotic selection decisions while unaware of the true bioterrorism-related diagnosis and select narrow-spectrum antibiotics, they may not be effective against the pathogens. Therefore, whether the use of these systems would be helpful or detrimental is not known.
Surveillance systems
None of 90 surveillance systems included in this Report has been evaluated for its utility in detecting a bioterrorism event. Forty of 61 reports of evaluations or descriptions of surveillance systems described the timeliness, importance of the health event under surveillance and usefulness of the system (Figure 14). However, less than one-third of the reports of evaluations of surveillance systems described the representativeness, simplicity, sensitivity, specificity, acceptability or flexibility of the system. The quality of the evidence regarding the effectiveness of the systems reported by these articles is therefore limited.
Syndromal surveillance systems have been designed with the intention of collecting data that could provide an early indication of a bioterrorism event. However, we have no evidence to determine which of the methods of collecting syndromal data (e.g., triage nurses collecting syndromal data on patients presenting to emergency departments, clinicians providing syndromal reports on suspicious patients, or using administrative data such as ICD9 codes or school absenteeism data) provides the most sensitive, timely, acceptable, and low cost data.
Because clinicians may be the first to recognize unusual or suspicious illnesses, reports from clinician networks are an essential source of surveillance data for detection of bioterrorism-related diseases. Of the systems that have been evaluated for the collection of clinician reports, Eurosentinel (Table 17) provides the most timely data (however, this is only true for influenza; data on other diseases and syndromes have a longer delay). The timeliness of the other systems varies from days to months. Systems that collect data on a weekly basis will be substantially less useful for bioterrorism surveillance than systems that can provide more rapid collection and analysis.
Table
Table 17. Surveillance systems for influenza.
Although none of the surveillance systems that collect influenza data has been evaluated specifically for the detection of bioterrorism-related illness, they are potentially useful for bioterrorism surveillance in 3 ways. First, sentinel clinicians who report on patients with suspected influenza are experienced at applying a case definition to a clinical population for the collection of public health data. Because many bioterrorism-related illnesses present with a "flu-like illness," this network of trained sentinel clinicians could provide valuable surveillance data. (We note that the evaluation of these sentinel clinicians is derived from heterogeneous surveillance networks in North America, Europe, and Australia. It is difficult to know whether the cultures of medicine, the training that sentinel clinicians receive, and their commitment to public health surveillance efforts is sufficiently similar that we can assume that the results of an evaluation of a surveillance network in France will be generalizable to clinicians in the U.S.) Second, most influenza surveillance systems integrate clinical and laboratory data for the detection of influenza outbreaks. Surveillance for bioterrorism may be aided by similar integration of multiple data sources. Finally, influenza surveillance -- like bioterrorism surveillance -- requires a coordinated global effort. New programs for the surveillance of bioterrorism-related illness could be derived from the existing IT infrastructures and the historical relationships that have been developed for influenza surveillance. Several of the influenza systems rely on weekly reporting by clinicians -- for bioterrorism surveillance, this time lag is likely to be problematic.
Laboratory testing will be an essential component of any bioterrorism surveillance and response effort. Systems that facilitate the collection, analysis, and reporting of notifiable pathogens and antimicrobial resistance data could potentially facilitate the rapid detection of a biothreat agent. Bioterrorism surveillance in the U.S. is limited by the lack of coordination among the many types of laboratories, reporting and communication systems for laboratory surveillance data, and personnel and equipment for rapid detection of biothreat agents. Efforts are ongoing to correct some of these shortcomings. Specifically, the Laboratory Response Network (Table 18), which builds on existing laboratory capacity and is currently under active expansion, was designed so that it can be integrated into surveillance networks (such as NEDSS) and communication networks (such as RHEACT).
Surveillance systems for hospital-acquired infections were included in this Report because of the possibility that data from these systems, already collected in electronic format for use by hospital infection control officers, might be a useful addition to an integrated surveillance system organized by local public health officials. However, the reports of the surveillance systems for hospital-acquired infections suggest that although these systems could be a valuable tool for hospital infection control officers, there is little evidence to suggest that they have sufficient sensitivity, specificity, or timeliness to detect a community-based bioterrorism event.
Technologies like SODA and PulseNet (Table 21) have been used extensively in foodborne outbreak investigations with success -- even with investigations of outbreaks resulting from intentional contamination of the food supply. We found no evidence regarding the potential sensitivity, specificity and timeliness of FoodNet, the active surveillance system collecting data to estimate the burden of foodborne illnesses in the U.S. Moreover, even if FoodNet was sufficiently sensitive and timely to be useful for agroterrorism detection, it is limited in that it collects data from 8 states on 9 foodborne illnesses. The primary means for detecting an agroterrorist attack outside these states or resulting from different organisms would be based on the analysis of voluntary reports from clinicians and laboratories.
When we consider the 90 surveillance systems described in this Report, there are relatively few systems collecting the earliest surveillance data -- such as school and work absenteeism, calls to telephone care nurses, over-the-counter pharmacy sales, or veterinary or zoonotic illness -- a potentially significant gap in available surveillance systems.
Communication systems
Of the 26 communication systems, those that notify clinicians of abnormal findings in their patient's electronic medical records have the most evidence for effectiveness. These systems are limited to institutions with electronic medical records but potentially could play an important role in decreasing the time to recognition of bioterrorism-related illness. Other communication systems with promise for bioterrorism include ProMED© (Table 24) with its subscribers from all relevant groups including international groups of clinicians, public health officials, veterinarians, agricultural experts, and media professionals. ProMED© has demonstrated the capacity for rapid reporting and dissemination of information on the widest possible range of infectious diseases resulting from both naturally occurring and bioterrorism-related events. We found no single system that effectively links members of the public health community at national, state and local levels. However, there are ongoing efforts (such as The Urban Security Initiative project of the Los Alamos National Laboratory, EpiX, HAN and RHEACT (Table 24)) designed to integrate communication of public health information vertically and horizontally within the U.S. public health system. In the event of a bioterrorist event, clinicians must be able to rapidly communicate with their patients. Systems exist that enable Web-based communications between these parties in a HIPAA-compliant manner. However, their utility in crisis situations will likely remain limited unless their use for routine communications increases. Robust security measures that ensure patient confidentiality and resist cyberattack will be a necessary component of any bioterrorism-related communication system.
Key Question 4: In areas where existing IT/DSSs do not meet the information needs of clinicians or public health officials, what functional and technical considerations are important in the design of future IT/DSSs to support response to bioterrorism events?
There are at least 3 explanations for why the evidence about an existing IT/DSS may fail to demonstrate its utility to meet the information needs of clinicians and public health officials. First, it may be that the evidence actually demonstrates that the system fails to support the information needs of clinicians and public health officials. Second, it may be that the evaluation data are of sufficiently poor quality that they cannot support the conclusion that the system may serve the information needs of clinicians and public health officials. Third, the system may have demonstrated efficacy in a clinical trial; however, when used in a real-world environment or clinical setting, the system lacked effectiveness. We did not identify evaluations or studies that directly assess the functional and technical requirements that are important for future IT/DSSs. In this section, we provide our interpretation of factors that should be considered for the design of future IT/DSSs.
For detection systems to be maximally useful to first responders and clinicians the collection system must be in use in the affected area. In the event of a covert attack, this is only possible if the collection system is already in place in areas of likely attack (e.g., airports; subways; major sporting, political, or entertainment events) as in the PROTECT project (Table 5). In the event of a known attack, these systems must be portable and rapid enough that they can be used by first responders and clinicians in a variety of field and clinical situations. Clinicians and first responders require detection methods for all -- not just some -- of the most worrisome biothreat agents, as well as systems that can simultaneously test a sample for multiple biothreat agents and run multiple samples. Because the costs associated with delay in diagnosing a bioterrorist event can be significant in terms of excess morbidity and mortality, these systems must have demonstrated high sensitivity (i.e., low false negative rate) and be timely. Similarly, because of the costs of responding to false alarms and the potential that users may disregard systems with known high false positive rates, these systems must have high specificity. Because the individuals collecting and analyzing the environmental and clinical samples are often at considerable distance from public health decision makers, it is important for detection systems to have the capacity for secure transmission of data to these decision makers.
Table
Table 5. Particulate counters and biomass indicators.
For diagnostic systems, efforts to link general diagnostic DSSs to other hospital information systems would reduce the data entry burden substantially. Making the current systems available on handheld devices (such as DiagnosisPro® (Table 8)) might make these systems more convenient for clinicians to use, but no studies have addressed this question directly. Perhaps most importantly, the knowledge bases of these systems must be updated to include current information about bioterrorism-related illness and should be sufficiently flexible to reflect dynamic probabilities of bioterrorism-related events.
Table
Table 8. General diagnostic DSSs.
Incorporating information from radiologic systems with other information from patients' medical records and knowledge bases about the clinical presentations of bioterrorism-related illnesses could be a useful innovation. Specifically, radiologic systems could serve as a component of an integrated management system that incorporates radiologic as well as other clinical information with clinical practice guidelines for the management and reporting of suspected bioterrorism-related illness.
A well-accepted list of the key syndromes for surveillance and detailed definitions of these syndromes could facilitate the integration of numerous sources of surveillance data. For example, an improved definition of "flu-like illness" could include its clinical characteristics so that triage nurses and clinicians can clearly identify patients with the syndrome, the specific ICD9 codes and other administrative data likely to be associated with it, the pharmaceuticals likely to be used to treat it, and the laboratory tests likely to be ordered to diagnose it. Then, each source of syndromal surveillance data can be systematically mapped to each of the syndromes, facilitating ongoing efforts to integrate multiple sources of surveillance data into a single system. The factors that affect the timeliness of a clinician surveillance report are: the interval between when the clinician first decides to report a given patient and actually sends the report, the interval between transmission and receipt of the surveillance report, the interval between receipt of the report and completion of data analysis, and the interval between analysis and communication of the results to decision makers who can respond appropriately. No system included in this Report has been evaluated to determine how long each of these intervals take, which intervals are rate-limiting, and what steps can be taken to increase their efficiency.
Surveillance efforts for bioterrorism will benefit from a detailed analysis of the sensitivity, specificity, and timeliness of each source of surveillance data; from improved spatial and temporal analysis methods; from evaluations of methods for the integration of multiple sources of surveillance data; from global investments in laboratory and communications infrastructures; from systems that collect sources of data reflecting disease earlier in the course of illness (e.g., school and work absenteeism and over-the-counter pharmacy sales); and from systems that facilitate ongoing outbreak investigation during the midst of a response to bioterrorism.
All IT/DSSs require security measures to protect patient confidentiality and thwart efforts of cyber attackers.
- Conceptual Model
- Task Decomposition
- Search Results
- Overview of Included Systems
- Detection and Diagnostic Systems
- Management and Prevention Systems
- Surveillance Systems
- Reporting and Communication Systems
- Integrated Surveillance, Communication, and Command and Control Systems
- Quality Evaluations
- Technical Information About the IT/DSSs
- Summary: Answers to the Key Questions
- Results - Bioterrorism Preparedness and ResponseResults - Bioterrorism Preparedness and Response
- Preface - Pharmacotherapy for Alcohol DependencePreface - Pharmacotherapy for Alcohol Dependence
Your browsing activity is empty.
Activity recording is turned off.
See more...