NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Does the genetic code assign similar codons to similar amino acids because of chemical interactions between them? Unlike adaptive explanations, which can only explain the relative positions of amino acids in the code, stereochemical explanations could tie codon assignments to absolute, verifiable rules. However, modern translation encodes amino acid sequences without direct codon/amino acid interaction. If there is a relationship between RNA sequences with intrinsic affinity for amino acids and the modern genetic code, we must therefore explain a historical transition in which direct interactions were abandoned. We review the literature and find no evidence that interactions between short sequences (mono-, di- or trinucleotides) and amino acids are strong or specific enough to originate genetic coding. Instead, interactions between amino acids and longer nucleic acid sequences appear to recapture some assignments of the modern code. For example, real codons are concentrated in newly selected amino acid binding sites to a greater extent than codons from similar, but randomized, codes. This implies that some initial coding assignments were made by interaction with macromolecular RNA-like molecules, and have survived. Thus, subsequent selection, such as selection to minimize coding errors, has not erased all primordial chemical relationships. Retention of initial stereochemical codon assignments for three of six amino acids (arginine, isoleucine, and tyrosine, but not glutamine, leucine or phenylalanine) is strongly supported.
Combining data for the six amino acids, significant stereochemical relationships are of more than one type—codons and anticodons are each concentrated in some binding sites. Further work will be required to catalog the relationships between amino acids and binding site sequences, especially if, as now appears, more than one type of interaction has been transmitted to the modern code.
The Codon-Correspondence Hypothesis
The codon-correspondence hypothesis, tested in any stereochemical theory of the origin of the genetic code, may be stated:
For each amino acid, there is a coding sequence for which it has the greatest association. The association between these sequences and amino acids influenced the form and content of the genetic code.
The codon-correspondence hypothesis is compatible with establishment of the genetic code either before or during the RNA world. A direct association between mono-, di- or trinucleotides and their cognate amino acids would suggest that the code arose before complex RNA catalysts, since trinucleotides would likely occur before the reproducible synthesis of longer oligonucleotides. Alternatively, an association between trinucleotides and their cognate amino acids that requires RNA tertiary structure would suggest that the genetic code arose in the RNA world (the earliest evolutionary time at which long RNA-like molecules were available). Larger RNAs loosen the constraint on the role of the coding sequences, which could then support the amino acid binding site but need not comprise it entirely. Amino acid/RNA complexes might have functioned in translation from the beginning, but alternatives abound. Their original functions may have been varied: as coenzyme sites for ribozymes,1 to stabilize RNA double helices,2 or to label tRNA-like genomic tags.3,4
Chemical Associations: A Historical Perspective
The idea that the genetic code might be stereochemically determined predates the elucidation of the code. Gamow's ‘diamond code’, in which amino acids would fit specific pockets bounded by four DNA bases, relied on direct interaction between amino acids and nucleic acids.5 More abstruse possibilities exist: mathematical (and even numerological) schemes for solving the coding problem abounded before the actual codon assignments were fully uncovered (reviewed in refs.6,7).
The structure of the code showed clear patterns. Chemical explanations for such order were sought by two routes. Physicochemical theorists8,9 hoped to measure interaction between bases and amino acids. This might have resulted in chromatographic copartitioning on the early earth, which would be reproducible today by chemical techniques. In contrast, stereochemical theorists10,11 assumed that molecular modeling could reveal molecular complementarities between amino acids and coding triplets.
Stereochemistry/Molecular Models: The first chemical investigations of codon assignments were via molecular modeling. Molecular models have been said to prove that the genetic code was established in quite varied ways. For example, amino acids might pair with codons12 or anticodons10,13 in the tRNA. Codonic mononucleotides and ahelical homopolymeric amino acids may bind each other specifically (this model “correctly predicts the glycine codon GGG”, although it unfortunately fails to predict any other).14 Free glycine and free nucleotides15 may have affinity, or free amino acids may intercalate into adjacent bases in the anticodon doublet through H-bonding between methylene groups and the π-electrons of the bases.16 Specific 2' aminoacylation of the second position anticodon base may have been mediated by the first position anticodon base.17 Amino acids may be able to intercalate between first and second position bases in double-stranded RNA molecules.18 Cavities caused by removal of the second-position codon bases in B-DNA may accept amino acids.19 Perhaps amino acids nestle into a pentanucleotide cup with the anticodon in the center.20 Pairing between amino acid sidechains and cavities in a complex of four nucleotides (C4N) on the acceptor stem of tRNA might occur. Or perhaps amino acids can bind their codons transposed 3'>5'.22,23 A double-stranded complex of the codon and anticodon has also been suggested.18,24
The modeling approach was tarnished early on when a claimed association between codons and amino acids12 relied on models that had been built backwards, 3' to 5'.25 Nevertheless, even the idea that there is a relationship between reversed codons and amino acids has been defended.22,23
Clearly, modeling methods used thus far are not sufficiently constrained. As a result, they allow too many solutions. Additionally, these approaches tend to assume that the entire code was uniquely determined by stereochemical fit (and even that modern variant codes reflect fits induced by different environments).26 If amino acids were added to the code over time and for different reasons, as seems probable,27,28 such explanations are overstatements that may prevent confirmation even if the basic hypothesis is true.
Physicochemical Effects/Chromatography: A second line of evidence comes from chromatography. Because chromatographic properties of amino acids show regular variation in the genetic code, any mechanism for the code's origin must account for this organization. Various studies have shown that the code conserves certain properties, such as polarity. The polar requirement of amino acids (the ratio of the log relative mobility to the log mole fraction water in a water-pyridine mixture) orders coding assignments impressively. Amino acids with U in the second position of their codon are hydrophobic while those with A are hydrophilic; those with C are intermediate, and those with G are mixed. Furthermore, codons that share a doublet have almost identical polar requirements even if not otherwise related (e.g., His and Gln; possibly Cys and Trp). Thus the code is ordered with respect to amino acid properties, but such evidence cannot tell us whether the code was optimized to minimize errors due to mutation or established by direct chemical interactions.28
Nor does such chemical order suggest a mechanism for actual codon assignments. Partitioning of amino acids and nucleotides between aqueous and organic phases, as in a primordial oil slick, might have associated AAA codons with Lys and UUU codons with Phe.30 However, none of these molecules are produced in prebiotic syntheses31 and a further hypothesis is required to bring chromatographic partitioning to bear on codon assignment. Analysis with two further chromatographic systems, water/micellar sodium dodecanoate and hexane/dodecylammonium propionate-trapped water, confirmed the previous hydrophobicity scales in a context closer to prebiotic conditions.32 The relative hydrophobicity of the homocodonic amino acids (Phe UUU, Pro CCC, Lys AAA, Gly GGG) and the four nucleotides in an ammonium acetate/ammonium sulfate system showed an anticodonic association, and for dinucleoside monophosphates the association was also with the anticodon, rather than the codon, doublets.33 Multivariate analysis of the properties of dinucleoside monophosphates and amino acids, focusing on hydrophobicity, revealed many strong (p < 0.001) correlations between anticodons and amino acids, but not between codons and amino acids.34
Thus, chromatographic data suggest anticodonic, rather than codonic interactions (note the underlying assumption that molecules with similar properties interact). However, although chemical partitioning on the early earth could conceivably have led to specific cofractionation between particular nucleotides (or oligonucleotides) and prebiotic amino acids, there do not seem to be consistent correlations. Chromatographic separation on various plausibly prebiotic surfaces (silicates, clays, hydroxyapatite, calcium carbonate, etc.) showed that, on a silica surface under an aqueous solution of MgCl2 and (NH4)H2PO4, Ala comigrates with CMP and Gly comigrates with GMP.35 Ala is assigned the GCN codon class, while Gly has the GGN codon class. However, there was no strong separation between GMP and UMP or between AMP and CMP even on silica, and many prebiotic amino acids (Pro, Ile, Leu, Val) fell well outside the range of the nucleotides. The situation was even worse on other surfaces, which did not provide any amino acid-nucleotide concordances. Thus, the data do not support the conclusion that copartitioning of nucleotides and amino acids led to the genetic code,35 especially in the absence of a plausible mechanism for transforming a copartition into modern codon assignments.
Physicochemical Effects/Direct Interactions: The third type of evidence comes from tests for direct interaction between nucleotides and amino acids. Mononucleotides show nonspecific but charge-dependent interactions with polyamino acid chains, as measured by the change in turbidity of the cosolution.14 Affinity chromatography, which tested retardation of the four nucleotide monophosphates by each of nine amino acids (Gly, Lys, Pro, Met, Arg, His, Phe, Trp, Tyr) immobilized by their carboxyl groups, showed no association between binding strength and codon or anticodon assignments.36 Interactions between free amino acids and poly(A), as measured by the chemical shift of the C2 and C8 protons of A, are also “not easily reconcilable with the genetic code”.37 Further affinity chromatography and NMR experiments on the interaction between amino acids and mono-, di- and trinucleotides showed that amino acids did selectively interact with specific bases,38 although the interactions did not parallel the genetic code. Imidazole-activated amino acids esterify the 2'-OH groups of RNA homopolymers with some specificity.39 However, since the two amino acids tested, phenylalanine and glycine, much preferred poly(U) over any other polynucleotide, the results do not support the authors' contention that this mechanism led to the present codon assignments.
The dissociation constants of AMP complexes with the methyl esters of amino acids also show selectivity, ranging about sevenfold from Trp (120 mM) to Ser (850 mM).40 However, neither Trp (UGG) nor Ser (CUN, AGY) have particularly many or few A residues in their codons or anticodons, while the amino acids that do (Lys AAR, Phe UUY) have intermediate dissociation constants (320 and 196 mM, respectively). These data did show a strong negative correlation between the association constant (1/KD) and amino acid hydrophobicity. There are positive correlations between the dissociation constant and the number of codons assigned to the amino acid, and to frequency of the amino acid in proteins.40 Condensation of dipeptides of the form Gly-X in the presence of AMP, CMP, poly(A) and poly(U) was mainly enhanced by the anticodonic nucleotides, where a pattern was apparent.41 Different amino acids differ in their ability to stabilize poly(A)-poly(U) and poly(I)-poly(C) double helices, although the order is similar in each case and so cannot have contributed to the establishment of the genetic code. Finally, D-ribose adenosine biases esters with L-Phe but not D-Phe towards the 3'-OH (the pattern is reversed with L-ribose adenosine). Thus, single nucleotides moderately regio- and stereo-selectively aminoacylate themselves.42
Recent evidence also suggests that self-assembly of purine monolayers differentially affects adsorption of amino acids. The spacing between residues is consistent with peptide bond distances: such self-assembly might have formed a primordial code, although apparently one very different from the modern genetic code.43–45
Summary: Two comprehensive reviews of these and other data46,47 suggested that if the genetic code were established by interactions between simple molecules (not more complicated than dipeptides or trinucleotides) and amino acids, then the greatest specific interaction was between amino acids and their anticodon nucleotides. However, individual experiments were equivocal or correlated with both anticodons and occasionally codons, so no strong direction is evident in the data.
The absence of obvious, strong or reproducible correlations from these highly varied approaches, considered alone or especially in sum, weakens the hypothesis that the code rests on the chemistry of trinucleotide-amino acid interactions. We suggest instead a later origin for the code, involving larger RNAs.
Adaptors and Adaptation
Perhaps the simplest explanation for the observed order in the genetic code11,48–50 is that codon assignments were determined by stereochemical association between oligonucleotides and amino acids.8–10,12 This mechanism would assign similar amino acids to similar codons because of intrinsic affinity, rather than as a result of natural selection among alternative codes. Although the resulting codon assignments might appear adaptive, in that they reduce various errors relative to other possible codes, they would not be an adaptation.
Stereochemical pairing: Several such stereochemical schemes are conceivable. Thus, the primordial sequences with which pairing occurred can either be the actual codons, or some simple transform thereof.9 As detailed in Section 2, interactions have been proposed between amino acids and codons,12 anticodons,10,13 codons read 3'< 5' instead of 5'<3',22,23 a complex of four nucleotides (C4N) formed by the three 5' nucleotides of tRNA with the fourth nucleotide from the 3' end, 21 and a double-stranded complex of the codon and anticodon.18,24
A fundamental problem that all stereochemical models share is that codons and amino acids are never stereochemically linked in modern translation. Thus an implied evolutionary shift has occurred in which direct associations were lost, but their logic was nevertheless transmitted to the present. Such a conservative transition, required to make a stereochemical origin observable, is supported by a strong argument from continuity. The shift to indirect associations must occur in a translation apparatus that is making useful peptides (otherwise the translation apparatus itself could not have been selected). Thus the logic of the older direct interactions must be preserved or the altered translation apparatus will be of no use. After consideration of the evidence, we discuss this transition to indirect coding again.
The existence of adaptors, tRNAs and aminoacyl-tRNA synthetases, in the modern system allows codon assignments to be readily shuffled among amino acids.51 Accordingly, adaptive evolution can erase primordial codon assignments. Thus we would only expect some amino acids to show codon/site associations, especially if others were added to the code later. Consequently, it is remarkable that any associations persist to the present.52
Amino Acid-Binding RNA: Most attention to sequence/binding site associations initially focused on arginine, since arginine binds specifically to two completely distinct classes of natural RNA molecules. The first class is the guanosine-binding site of self-splicing group I introns, which binds arginine as a competitive inhibitor. The guanidinium side-chain of arginine is similar in structure to the Watson-Crick face of G.53 A conserved Arg codon confers this activity, and the binding site is almost invariably composed of several Arg codons in close juxtaposition.54,55 The second class has been extensively studied because of potential medical importance: free arginine can mimic the natural interaction of HIV Tat peptides with TAR RNA.56 In this case, however, no Arg codons are conserved at the binding site.57
Natural amino acid-binding RNAs are few; more significantly, they can provide only anecdotal evidence for codon/binding site interactions because they are almost certainly under strong selection for properties other than binding to the free amino acids. However, SELEX or selection-amplification, a technique for directed molecular evolution,58–60 makes it possible to select those RNA molecules that perform a desired catalytic or binding function from large random pools (see ref. 61 for review). This technological advance makes it possible to find out whether RNA molecules that bind to particular amino acids share any characteristic motifs at their binding sites.
Aptamers have now been isolated from a variety of amino acids (Table 1), including hydrophobic amino acids such as valine,62 phenylalanine/tyrosine,63 isoleucine,64 tyrosine,65 leucine (I. Majerfeld and M. Yarus, unpublished data), and phenylalanine,65a and hydrophilic amino acids such as glutamine (G. Tocchini-Valentini, unpublished data) and citrulline, which is not normally found in proteins.66 However, RNA aptamers for arginine are most abundant in the literature, and have been independently isolated in several different experiments.66–73 Since structural information is available for many of these sequences, it becomes possible to ask whether particular sequences are overrepresented at recently selected binding sites, and, if so, whether these sequences have any relationship to the modern genetic code.
Table 1
Natural and artificial amino acid-binding RNA.
Statistical Evidence for Triplet/Binding Site Associations
The theory that the code arose by stereochemical means is both specific and unique; its predictions are explicit and different from other prevalent theories. Co-evolution theories (that coding was extended along biosynthetic pathways74) are typically agnostic about which trinucleotide-amino acid pairing established the initial codon assignments, but predict that such pairings, if they exist at all, can account for only a small part of the codon catalog. Optimization theories (that coding minimizes errors in expression75) predict no correspondence at all between trinucleotides and amino acid binding sites.
Evolution of Binding Triplets: Assuming that original amino acid binding sites were RNA-like, they could have evolved into any of the components of modern translation: tRNA, rRNA, mRNA, or primitive aminoacyl-tRNA synthetases (subsequently replaced by protein enzymes). Depending on which modern translation component descended from ancient amino acid interactions, we predict different associations between coding nucleotides and amino acids. If binding sites evolved into tRNAs, for instance, the anticodons should be overrepresented in amino acid binding sites, whereas if they evolved into mRNA the codons should be overrepresented.76
The selection of RNA molecules (aptamers) that bind amino acid ligands has made such conjectures testable (Table 1). Because in vitro selection searches a large space of possible sequences for optimal or near-optimal “solutions” to particular binding problems, such directed evolution might be able to recapitulate primordial interactions between amino acids and short RNA sequences. If amino acids interact favorably with coding RNA sequences, this relation might be observed, or even proven. Since aptamers can be selected for each amino acid, and since the specific nucleotides important to binding can be determined, standard statistical tests for association (such as the ϰ2 or G tests) will reveal any consistent relation between binding-site nucleotides and nucleotides in coding sequences.77
Such a search for motifs faces predictable difficulties. RNA is more versatile than might have once been thought, and many oligomers often bind an amino acid. The diversity of RNAs that bind arginine, for example, shows that efforts to emulate a unique primordial RNA for each amino acid would be futile.57 Recurrence of specific sequence motifs in amino acid aptamers, such as codons or anticodons, cannot prove that similar interactions led to the establishment of present codon assignments. However, suppose that coding sequences embody such general interactions that they will still be detectable in the most probable modern binding sites. Proof of any specific pairings at all would show that the specificity existed to originate a genetic code. If specific pairings detected with in vitro selection actually match present codon assignments, then similar processes in ancient translation are supported. If there are frequent, strong associations between present codons or anticodons and amino acids, their involvement in the origin of the code is the only plausible explanation.
Binding Site Preferences: That any codon/binding site associations could survive to the present has been questioned.78 However, the association between arginine and its binding sites is exceptionally strong, and has proven remarkably robust to statistical methodology, choice of binding sites, and choice of sequences from selected pools52,76–78 In particular, arginine binding sites show strong associations with arginine codons (Table 2), but not anticodons (Table 3), codon or anticodon sets for other amino acids, other groups of 4+2 codons incorporating a family box plus a doublet, or other short motifs. The relationship remains highly significant even with many plausible modifications. Sequences where the selected binding site overlaps the constant regions are excluded, the data can be corrected for nucleotide bias at binding sites and alternative sequences can be chosen from reported pools without altering the conclusion.
Table 2
Tests for association between amino acid binding sites and their cognate codons.
Table 3
Test for association between binding sites and the cognate codons, anticodons, and codons reversed 3' to 5'.
Arginine may be unique: it acts as a nucleotide mimic,53 perhaps more so than other amino acids. However, significant associations between Tyr aptamer binding sites and codons have been reported,52 and Ile aptamers contain conserved Ile codons at their active sites.64 Data from several other amino acids have become available, allowing a more general test of generality for the association between binding sites and codons. We now extend the analysis to all available amino acids (Table 1) and reassess hypotheses about specific associations.
Testing Triplet/Site Associations: Codons occur more often in binding sites than expected for each of the six amino acids for which data are available, an improbable outcome itself (P = (0.5)6 = 0.016). Individually, the arginine aptamers showed a significant codon/site association only. Tyrosine and isoleucine aptamers showed significant associations between both codons and anticodons: except for the association between tyrosine and its codons, these relationships persist even when corrected for six multiple comparisons (P < 0.01). Glutamine, leucine and phenylalanine have no significant tendency to locate codons or anticodons in their binding sites (when corrected for multiple comparisons). The most sensitive tests combine all data; then we observe highly significant associations overall with both codons and anticodons, even when the single most influential amino acid is excluded from the analysis (P < 10−6 in all cases). Thus there is reason to believe that codons and anticodons are associated with binding sites, and this conclusion does not depend on any one selection or set of binding sites.
On the other hand, controls show that this method can rule out certain possibilities. There was no significant association for any amino acid, or for the set as a whole, with the codons reversed 3' to 5', indicating that this hypothesis can be clearly rejected.
It is possible that the 21 codon (or anticodon) sets are an unfair comparison class, since they range in size from 1 to 6 codons. A less precise, but perhaps more robust, test is to see whether there is a significant association between the amino acid binding sites and the codon (or anticodon) that contains the cognate doublet: this reflects the intuitively plausible idea that the primitive code may have assigned amino acids only to family boxes. However, doublet analysis (Table 4) does not greatly change the outcome. Significant associations are observed for both doublets and codons/anticodons. Thus, again, the results to date suggest both associations between codons and anticodons.
Table 4
Test for association between binding sites and codon doublets (XYN) or anticodon doublets (NY'X'), where X and Y are specified and N is any base.
We can carry these conclusions a step further by freeing them of the assumptions required even for standard statistical tests. If there is an association between the triplets found at amino acid binding sites and the modern genetic code, it should be found only with the actual genetic code and not with randomized versions of it. Accordingly, we generated many alternative codes, and tested for codon/binding site associations. This preserves important aspects of the experimental results, such as the spatial correlations within binding sites (they occur in specific sections of the molecule), and the influence of the occurrence of each triplet on the probability of finding others. In order to eliminate dependence on any particular method for generating variant codes, we used several quite different permutation methods.
An ISO C program randomized the code according to the following schemes:
1. Codon permutation: a codon can randomly and independently take on any identity (including its real one). This keeps the number of codons per amino acid constant, but usually completely disrupts the fine structure of the code (such as wobble relations). This potentially generates 64! = 1.2 × 1089 possible codes.
2. Amino acid permutation: any amino acid can randomly and independently take any existing coding block(s), including those of stop codons. This preserves the structure of the code entirely (the number and size of blocks for codons are preserved, and their relative positions are preserved within the coding table), but amino acids can be given different numbers of codons. At one extreme, Arg, which normally has 6 codons split into a 4-block and a 2-block, might end up with Trp's single codon. This potentially generates 21! = 5.1 × 1019 possible codes.
3. Codon block permutation. Keeping the structure of the code constant, we randomly assorted amino acid identities among groups of codons of the same size. For example, the CGN block assigned to Arg might be swapped with the CCN block normally assigned to Pro, but could not swap with the single UGA codon assigned to Trp. Treating the three Ile codons as a 2-block and a 1-block, this leads to 8!×14!×4! = 8.4 × 1016 codes with 8 4-blocks, 14 2-blocks, and 4 1-blocks. This “n-block” scheme completely preserves the degeneracy of the code, and also conserves the number of codons assigned to each amino acid. Compared to the other randomization schemes, amino acids are far more likely to retain some of their actual codons.
5. Base identity permutation: in addition to the block permutation of method 3, this method randomizes the meaning of the first and second position base . This partially disrupts the code's structure (so that, for example, the UGN codon block need not be split into blocks of 2, 1, and 1), but preserves the degeneracy across a row and down a column. This multiplies the number of codes from method 3 by a factor of (4!×4!)/2 for a total of 2.4 × 1019 codes, and dramatically reduces retention of fragments of the present code.
6. Codon doublet permutation: like method 4, except that any codon doublet independently takes on the meaning of any other codon doublet. This leads to 16!/(8!×6!×2!) = 360360 times as many codes as method 3, for a total of 3.0 × 1022 possible codes. Both this and method 4 preserve the number of codons assigned to each amino acid and their block structure (e.g., Arg will always have a 4-block and a 2-block), but this method does not preserve the relation between blocks of particular sizes as does method 4.
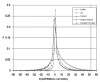
We generated 10 million randomized codes for each of the 5 schemes listed above, and compared codon/site associations in observed amino acid binding sites with those found in the actual code (Fig. 1). The “n-block” model (#3) is uniquely right-skewed, because some of the codons can only swap with a few partners under this model (e.g., there are only 4 blocks containing one codon) so that some of the present structure of the code will often be preserved. Even under this highly constrained model, however, only 0.8% of randomized codes give apparent associations between codons and binding sites better than the actual code. For the other, more completely scrambled models, between 0.11% (method 2) and 0.04% (methods 4 and 5) of all random codes do better than the actual code. Said another way, real codons are more associated with real binding sites than in 99.2 to 99.96% of all randomized codes, even though randomized codes include fragments of the actual code. Using Fisher's method for independent probabilities rather than performing a G test on the summed counts gave similar results (data not shown). Thus, our result is general and not sensitive to choice of alternative codes or sensitive to statistical methodology. It is highly unlikely that we would see as significant an association between codons and binding sites for a genetic code picked at random as that actually seen with the real code. Randomization of anticodon assignments gives similar results, but slightly less significant than for codons. Randomized anticodons are less associated with binding sites than real ones in 99.2 to 99.5% of all codes. This small difference in significance appears also in the statistical tests (Table 3).
These controls argue strongly that the most probable modern RNA-amino acid binding sites capture something of the essential nature of the code. In particular, a stereochemical process involving macromolecular RNA-like binding sites containing codons, and perhaps anticodons, gave rise to the present genetic code. Considering individual amino acids, primordial RNA-like binding sites were probably relevant to the assignment of codons for at least three of six amino acids for which we have data.
Concluding Remarks
We now return to the direct to indirect coding transition implied by every stereochemical model. RNA-amino acid binding sites contain sequences likely to be relevant to the appearance of the code. Thus the logically predicted transition from direct to indirect coding rests first on the ability of coding sequences to serve as structural elements in amino acid binding sites, and then to subsequently serve in normal base pairing. Triplets that became codons might begin as essential elements in binding sites (indirect coding), and later pair with primordial tRNAs (direct coding). Triplets that became anticodons might begin within binding sites (indirect), then employ their more well-known basepairing activity when they begin to act as anticodons (direct coding). The conservative logic of the direct to indirect transition, required by argument from continuity, is implicit as soon as it is known that nucleotide triplets can be essential elements of amino acid binding sites (compare the DRT theory57).
Descendants of the original amino acid-binding sites could play four possible roles: as tRNAs, mRNAs, ribosomes, or aminoacyl-tRNA synthetases. All these activities are known to be possible activities for RNA,79–85 because they exist in modern selected parallels. With present data, it appears that arginine may have been bound in primordial sites containing sequences that became codons in mRNA. We found no strong evidence for association between glutamine, leucine and phenylalanine and their coding sequences. These are negative results based on limited data; however, these codons may have been assigned by other means during later code evolution. Tyrosine and isoleucine present a case we had not anticipated, in which both codons and anticodons are overrepresented (though not because they are paired in the molecules). We cannot confidently specify the descent of the coding sequences for these amino acids. Their binding sequences could have become both tRNA-like and mRNA-like molecules, or these data may be the first indication of the need for a new, more comprehensive theory.
Ideally, with a large sample of independently derived families of aptamers that bind each of the amino acids, it should be possible to test associations between binding sites and individual trinucleotides. If there are, as now appears, to be several classes of amino acids with different relations to coding sequences, such high resolution may be required. It is possible that high-throughput techniques for aptamer isolation will achieve this in the future, but, for the moment, isolating aptamers and determining binding sites is a time-consuming process. Consequently, it may be several years before site/triplet associations are maximally resolved.
However, it is clearly not true that each aptamer binds its target amino acid using only the cognate codons. Amino acid binding sites always require other nucleotides for their construction. Where structures are known, the coding sequences can be in contact with the amino acid or providing less central support for the site—in some cases they are in both places.52 The fact that binding sites with detectable affinities are far more complex than single trinucleotides strongly suggests that the code probably began in an RNA world, after complex RNA molecules were prevalent. Assuming that the RNA world biota were our immediate antecedents, translation was also probably devised in the RNA world.89 An economical interpretation is therefore that coding assignments arose predominantly during initial selection for templated peptide synthesis, rather than via other activities.
These techniques have substantial potential for further analysis. It may be possible to discover why some amino acids have the actual codon assignments they do, and perhaps why some amino acids were incorporated into the code while others, available on the early earth or as metabolic intermediates, were excluded. Furthermore, with complete data in hand it may be possible to define a minimal, stereochemically determined code, and therefore to estimate the relative roles of chemistry and selection in shaping modern codon assignments.
References
- 1.
- Szathmáry E. Coding coenzyme handles: A hypothesis for the origin of the genetic code. Proc Natl Acad Sci USA. 1993;90:9916–9920. [PMC free article: PMC47683] [PubMed: 8234335]
- 2.
- Porschke D. Differential effect of amino acid residues on the stability of double helices formed from polyribonucleotides and its possible relation to the evolution of the genetic code. J Mol Evol. 1985;21:192–198. [PubMed: 6442993]
- 3.
- Maizels N, Weiner AM. Peptide-specific ribosomes, genomic tags, and the origin of the genetic code. Cold Spring Harb Symp Quant Biol. 1987;52:743–749. [PubMed: 3454285]
- 4.
- Maizels N, Weiner AM. The genomic tag hypothesis: modern viruses as molecular fossils of ancient strategies for genomic replication In: Gesteland RF and Atkins JF, eds. The RNA world New York: Cold Spring Harbor Laboratory Press, 1993577–602.
- 5.
- Gamow G. Possible mathematical relation between deoxyribonucleic acid and protein. Kgl Dansk Videnskab Selskab Biol Medd. 1954;22:1–13.
- 6.
- Woese CR. The genetic code: the molecular basis for genetic expression New York: Harper & Row, 1967. [PMC free article: PMC297137]
- 7.
- Ycas M. The biological code In: Neuberger A, Tatum EL, eds. NorthHolland Research Monographs: Frontiers of Biology Vol. 12 Amsterdam: North-Holland Publishing Company, 1969.
- 8.
- Woese CR, Dugre DH, Dugre SA. et al. On the fundamental nature and evolution of the genetic code. Cold Spring Harb Symp Quant Biol. 1966;31:723–736. [PubMed: 5237212]
- 9.
- Woese CR, Dugre DH, Saxinger WC. et al. The molecular basis for the genetic code. Proc Natl Acad Sci U S A. 1966;55:966–974. [PMC free article: PMC224258] [PubMed: 5219702]
- 10.
- Dunnill P. Triplet nucleotide-amino-acid-pairing: a stereochemical basis for the division between protein and non-protein amino-acids. Nature. 1966;210:1265–1267. [PubMed: 5967806]
- 11.
- Pelc SR. Correlation between coding triplets and amino acids. Nature. 1965;207:597–599. [PubMed: 5883631]
- 12.
- Pelc SR, Welton M G E. Stereochemical relationship between coding triplets and aminoacids. Nature. 1966;209:868–872. [PubMed: 5922773]
- 13.
- Ralph RK. A suggestion on the origin of the genetic code. Biochem Biophys Res Comm. 1968;33:213–218. [PubMed: 5722216]
- 14.
- Lacey J C Jr, Pruitt KM. Origin of the genetic code. Nature. 1969;223:799–804. [PubMed: 5799020]
- 15.
- Rendell MS, Harlos JP, Rein R. Specificity in the genetic code: the role of nucleotide baseamino acid interaction. Biopolymers. 1971;10:2083–2094. [PubMed: 5118645]
- 16.
- Melcher G. Stereospecificity of the genetic code. J Mol Evol. 1974;3:121–140. [PubMed: 4407468]
- 17.
- Nelsesteuen GL. Amino acid-directed nucleic acid synthesis. J Mol Evol. 1978;11:109–120. [PubMed: 671559]
- 18.
- Hendry LB, Whitham FH. Stereochemical recognition in nucleic acidamino acid interactions and its implications in biological coding: a model approach. Perspect Biol Med. 1979;22:333–345. [PubMed: 471692]
- 19.
- Hendry LB, Bransome E D Jr, Hutson MS. et al. First approximation of a stereochemical rationale for the genetic code based on the topography and physichemical properties of “cavities” constructed from models of DNA. Proc Natl Acad Sci USA. 1981;78:7440–7444. [PMC free article: PMC349283] [PubMed: 6950386]
- 20.
- Balasubramanian R. Origin of life: A hypothesis for the origin of adaptor-mediated ordered synthesis of proteins and an explanation for the choice of terminating codons in the genetic code. Bio Systems. 1982;15:99–104. [PubMed: 7104476]
- 21.
- Shimizu M. Molecular basis for the genetic code. J Mol Evol. 1982;18:297–303. [PubMed: 7120424]
- 22.
- RootBernstein RS. Amino acid pairing. J Theor Biol. 1982;94:885–894. [PubMed: 7078229]
- 23.
- RootBernstein RS. On the origin of the genetic code. J Theor Biol. 1982;94:895–904. [PubMed: 7078230]
- 24.
- Alberti S. The origin of the genetic code and protein synthesis. J Mol Evol. 1997;45:352–358. [PubMed: 9321414]
- 25.
- Crick F H C. An error in model building. Nature. 1967;213:798. [PubMed: 6031804]
- 26.
- Mellersh A. A model for the prebiotic synthesis of peptides and the genetic code. Orig Life Evol Biosph. 1993;23:261–274.
- 27.
- Crick F H C. The origin of the genetic code. J Mol Biol. 1968;38:367–379. [PubMed: 4887876]
- 28.
- Knight RD, Freeland SJ, Landweber LF. Selection, history and chemistry: the three faces of the genetic code. Trends Biochem Sci. 1999;24:241–7. [PubMed: 10366854]
- 29.
- Woese CR. Evolution of the genetic code. Naturwissenschaften. 1973;60:447–59. [PubMed: 4588588]
- 30.
- Nagyvary J, Fendler JH. Origin of the genetic code: a physical-chemical model of primitive codon assignments. Orig Life. 1974;5:357–362. [PubMed: 4414951]
- 31.
- Miller SL. Which organic compounds could have occurred on the prebiotic earth? Cold Spring Harb Symp Quant Biol. 1987;52:17–27. [PubMed: 3454260]
- 32.
- Fendler JH, Nome F, Nagyvary J. compartmentalization of amino acids in surfactant aggregates. J Mol Evol. 1975;6:215–232. [PubMed: 1206727]
- 33.
- Weber AL, Lacey J C Jr. Genetic code correlations: amino acids and their anticodon nucleotides. J Mol Evol. 1978;11:199–210. [PubMed: 691071]
- 34.
- Jungck JR. The genetic code as a periodic table. J Mol Evol. 1978;11:211–224. [PubMed: 691072]
- 35.
- Lehmann U. Chromatographic separation as selection process for prebiotic evolution and the origin of the genetic code. Bio Systems. 1985;17:193–208. [PubMed: 3995160]
- 36.
- Saxinger C, Ponnamperuma C. Experimental investigation on the origin of the genetic code. J Mol Evol. 1971;1:63–73. [PubMed: 5173657]
- 37.
- Raszka M, Mandel M. Is there a physical chemical basis for the present genetic code? J Mol Evol. 1972;2:38–43. [PubMed: 4668863]
- 38.
- Saxinger C, Ponnamperuma C. Interactions between amino acids and nucleotides in the prebiotic milieu. Orig Life. 1974;5:189–200. [PubMed: 4842070]
- 39.
- Lacey J C Jr, Weber AL, White W E Jr. A model for the coevolution of the genetic code and the process of protein synthesis: review and assessment. Orig Life. 1975;6:273–283. [PubMed: 1153188]
- 40.
- Reuben J, Polk FE. Nucleotide-amino acid interactions and their relation to the genetic code. J Mol Evol. 1980;15:103–112. [PubMed: 7401174]
- 41.
- Podder SK, Basu HS. Specificity of protein-nucleic acid interaction and the biochemcial evolution. Orig Life. 1984;14:477–484. [PubMed: 6462683]
- 42.
- Lacey J C Jr, Wickramasinghe N S M D, Cook GW. et al. Couplings of character and of chirality in the origin of the genetic system. J Mol Evol. 1993;37:233–239. [PubMed: 7693954]
- 43.
- Sowerby SJ, Cohn CA, Heckl WM. et al. Differential adsorption of nucleic acid bases: relevance to the origin of life. Proc Natl Acad Sci USA. 2001;98:820–822. [PMC free article: PMC14666] [PubMed: 11158553]
- 44.
- Sowerby SJ, Heckl WM. The role of self-assembled monolayers of the purine and pyrimidine bases in the emergence of life. Orig Life Evol Biosph. 1998;28:283–310. [PubMed: 9611768]
- 45.
- Sowerby SJ, Stockwell PA, Heckl WM. et al. Self-programmable, self-assembling two-dimensional genetic matter. Orig Life Evol Biosph. 2000;30:81–99. [PubMed: 10836266]
- 46.
- Lacey J C Jr, Mullins D W Jr. Experimental studies related to the origin of the genetic code and the process of protein synthesis—a review. Orig Life. 1983;13:3–42. [PubMed: 6350974]
- 47.
- Lacey J C Jr. Experimental studies on the origin of the genetic code and the process of protein synthesis: a review update. Orig Life Evol Biosph. 1992;22:243–275. [PubMed: 1454353]
- 48.
- Epstein CJ. Role of the aminoacid ‘code’ and of selection for conformation in the evolution of proteins. Nature. 1966;210:25–28. [PubMed: 5956344]
- 49.
- Volkenstein MV. Coding of polar and non-polar amino-acids. Nature. 1965;207:294–295. [PubMed: 5886220]
- 50.
- Woese CR. Order in the genetic code. Proc Natl Acad Sci USA. 1965;54:71–75. [PMC free article: PMC285798] [PubMed: 5216368]
- 51.
- Saks ME, Sampson JR, Abelson J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–1670. [PubMed: 9497276]
- 52.
- Yarus M. RNA-ligand chemistry: a testable source for the genetic code. RNA. 2000;6:475–484. [PMC free article: PMC1369929] [PubMed: 10786839]
- 53.
- Yarus M. A specific amino acid binding site composed of RNA. Science. 1988;240:1751–1758. [PubMed: 3381099]
- 54.
- Yarus M. Specificity of arginine binding by the Tetrahymena intron. Biochemistry. 1989;28:980–988. [PubMed: 2653441]
- 55.
- Yarus M. An RNA-amino acid complex and the origin of the genetic code. New Biologist. 1991;3:183–189. [PubMed: 2065012]
- 56.
- Tao J, Frankel AD. Specific binding of arginine to TAR RNA. Proc Natl Acad Sci USA. 1992;89:2723–2726. [PMC free article: PMC48734] [PubMed: 1557378]
- 57.
- Yarus M. Amino Acids as RNA Ligands: a Direct-RNA-Template Theory for the Code's Origin. J Mol Evol. 1998;47:109–117. [PubMed: 9664701]
- 58.
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. [PubMed: 1697402]
- 59.
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. [PubMed: 2200121]
- 60.
- Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single stranded DNA. Nature. 1990;344:467–468. [PubMed: 1690861]
- 61.
- Ciesiolka J, Illangasekare M, Majerfeld I. et al. Affinity selection-amplification from randomized ribooligonucleotide pools. Meth Enzymol. 1996;267:315–335. [PubMed: 8743325]
- 62.
- Majerfeld I, Yarus M. An RNA pocket for an aliphatic hydrophobe. Nat Struct Biol. 1994;1:287–292. [PubMed: 7545073]
- 63.
- Zinnen S, Yarus M. An RNA pocket for the planar aromatic side chains of phenylalanine and tryptophan. Nucleic Acid Symp Ser. 1995;(33):148–151. [PubMed: 8643354]
- 64.
- Majerfeld I, Yarus M. Isoleucine: RNA sites with essential coding sequences. RNA. 1998;4:471–478. [PMC free article: PMC1369632] [PubMed: 9630252]
- 65.
- Mannironi C, Scerch C, Fruscoloni P. et al. Molecular recognition of amino acids by RNA aptamers: the evolution into an L-tyrosine binder of a dopamine-binding RNA motif. RNA. 2000;6:520–527. [PMC free article: PMC1369933] [PubMed: 10786843]
- 65a.
- Illangasekare M, Yarus M. Phenylalanine-binding RNAs and genetic code evolution. J Mol Evol. 2002;54:298–311. [PubMed: 11847556]
- 66.
- Famulok M. Molecular recognition of amino acids by RNA-aptamers: an L-citrulline binding RNA motif and its evolution into an L-arginine binder. J Am Chem Soc. 1994;116:1698–1706.
- 67.
- Connell GJ, Illangsekare M, Yarus M. Three small ribo-oligonucleotides with specific arginine sites. Biochemistry. 1993;32:5497–5502. [PubMed: 8504070]
- 68.
- Connell GJ, Yarus M. RNAs with dual specificity and dual RNAs with similar specificity. Science. 1994;264:1137–1141. [PubMed: 7513905]
- 69.
- Yarus M. An RNA-amino acid affinity, in The RNA World Gesteland RF, Atkins JF, eds. New York: Cold Spring Harbor Laboratory Press, 1993205–217. [PMC free article: PMC158964]
- 70.
- Tao J, Frankel AD. Arginine-binding RNAs resembling TAR identified by in vitro selection. Biochemistry. 1996;35:2229–2238. [PubMed: 8652564]
- 71.
- Burgstaller P, Kochoyan M, Famulok M. Structural probing and damage selection of citrulline- and arginine-specific RNA aptamers identify base positions required for binding. Nucleic Acids Res. 1995;23:4769–4776. [PMC free article: PMC307463] [PubMed: 8532517]
- 72.
- Geiger A, Burgstaller P, von der Eltz H. et al. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996;24:1029–1036. [PMC free article: PMC145747] [PubMed: 8604334]
- 73.
- Yang Y, Kochoyan M, Burgstaller P. et al. Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy. Science. 1996;272:1343–1346. [PubMed: 8650546]
- 74.
- Wong J T F. A coevolution theory of the genetic code. Proc Natl Acad Sci USA. 1975;72:1909–1912. [PMC free article: PMC432657] [PubMed: 1057181]
- 75.
- Sonneborn TM. Degeneracy of the genetic code: extent, nature, and genetic implications In: Bryson V and Vogel HJ, eds. Evolving Genes and Proteins New York: Academic Press. 19653772–97. [PubMed: 17799782]
- 76.
- Knight RD, Landweber LF. Guilt by association: the arginine case revisited. RNA. 2000;6:499–510. [PMC free article: PMC1369931] [PubMed: 10786841]
- 77.
- Knight RD, Landweber LF. Rhyme or reason: RNA-arginine interactions and the genetic code. Chem Biol. 1998;5:R215–R220. [PubMed: 9751648]
- 78.
- Ellington AD, Khrapov M, Shaw CA. The scene of a frozen accident. RNA. 2000;6:485–498. [PMC free article: PMC1369930] [PubMed: 10786840]
- 79.
- Illangasekare M, Sanchez G, Nickles T. et al. Aminoacyl-RNA synthesis catalyzed by an RNA. Science. 1995;267:643–647. [PubMed: 7530860]
- 80.
- Illangasekare M, Yarus M. Specific, rapid synthesis of Phe-RNA by RNA. Proc Natl Acad Sci U S A. 1999;96:5470–5475. [PMC free article: PMC21883] [PubMed: 10318907]
- 81.
- Illangasekare M, Yarus M. A tiny RNA that catalyzes both aminoacyl-RNA and peptidyl-RNA synthesis. RNA. 1999;5:1482–1489. [PMC free article: PMC1369869] [PubMed: 10580476]
- 82.
- Welch M, Majerfeld I, Yarus M. 23S rRNA similarity from selection for peptidyl transferase mimicry. Biochemistry. 1997;36:6614–6623. [PubMed: 9184141]
- 83.
- Nissen P, Hansen J, Ban N. et al. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. [PubMed: 10937990]
- 84.
- Yarus M, Welch M. Peptidyl transferase: ancient and exiguous. Chem Biol. 2000;7:R187–R190. [PubMed: 11033085]
- 85.
- Kumar RK, Yarus M. RNA-catalyzed amino acid activation. Biochemistry. 2001;40:6998–7004. [PubMed: 11401543]
- 86.
- Yarus M, Majerfield I. Co-optimization of ribozyme substrate stacking and L-arginine binding. J Mol Biol. 1992;225:945–949. [PubMed: 1613800]
- 87.
- Famulok M, Szostak JW. Stereospecific recognition of tryptophan agarose by in vitro selected RNA. J Am Chem Soc. 1992;114:3990–3991.
- 88.
- Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research 3rd ed. New York: W. H. Freeman and Company, 1995. [PMC free article: PMC173454]
- 89.
- Yarus M. On translation by RNAs alone. Cold Spring Harb Symp Quant Biol. 2001;66:207–215. [PubMed: 12762023]
- Tests of a Stereochemical Genetic Code - Madame Curie Bioscience DatabaseTests of a Stereochemical Genetic Code - Madame Curie Bioscience Database
- Origins and Evolution of Cotranslational Transport to the ER - Madame Curie Bios...Origins and Evolution of Cotranslational Transport to the ER - Madame Curie Bioscience Database
- Thermo-Chemo-Radiotherapy Association: Biological Rationale, Preliminary Observa...Thermo-Chemo-Radiotherapy Association: Biological Rationale, Preliminary Observations on Its Use on Malignant Brain Tumors - Madame Curie Bioscience Database
- IgA and Mucosal Homeostasis - Madame Curie Bioscience DatabaseIgA and Mucosal Homeostasis - Madame Curie Bioscience Database
- Epithelial-Mesenchymal Transformation in the Embryonic Heart - Madame Curie Bios...Epithelial-Mesenchymal Transformation in the Embryonic Heart - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...