NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The ribosome is a dynamic particle that undergoes an iterative series of conformational changes during translation. Individual structural changes in the ribosome in response to tRNA or mRNA binding, initiation or elongation factor binding, buffer conditions, and antibiotic effects have been observed using a variety of techniques over the years. With new high resolution cryoelectron microscopy and x-ray crystallography structure models available, we are approaching an understanding of how these conformational changes mesh together in the context of the whole ribosome. This review emphasizes conformational changes in the rRNA and their likely roles in various steps of translation.
The notion that the ribosome is dynamic seems intuitive to ribosomologists, yet detailed structural information associated with conformational changes of known function has been rather elusive. To demonstrate a switch, one must show experimentally that a structural change occurs (e.g., an alteration in a base pairing scheme within the rRNA, or a specific change in protein-protein or protein rRNA contacts) then show that the structural change has a tangible effect on some ribosomal function, otherwise the change may simply indicate that the observed region of the ribosome is not constrained. In this review, evidence for several conformational changes within the ribosome, with emphasis on those changes that occur in the rRNA during translation, will be discussed from structural and functional viewpoints. Indeed, the ribosome literature is replete with references to presumed or confirmed conformational changes and a comprehensive review is not possible here. I apologize in advance for omissions. Several excellent reviews on various aspects of conformational dynamics of the ribosome have been published; the reader is encouraged to explore these rich resources and references therein.110
The “Active-Inactive” Interconversion of 30S and 50S Subunits
Perhaps the most extensively studied conformational change in the ribosome occurs during the so-called active-inactive interconversion of 30S subunits.11 This conformational change is concomitant with a change in the tRNA binding properties of 30S subunits. In the inactive form, 30S subunits are incapable of binding aminoacyl tRNA (aa-tRNA) that is not complexed with either initiation factor IF-2 or elongation factor EF-Tu. In the active form, 30S subunits bind near-stoichiometric amounts of tRNA. The interconversion is brought about by manipulation of mono- and divalent cations and temperature during incubation in vitro. The inactive form is achieved by incubation of subunits in conditions of low magnesium (ca. 0.5 mM) and/or low monovalent cation (potassium or ammonium). Inactive 30S subunits can be transformed into the active (tRNA binding-competent) form by replenishing the cation concentration and heating to 37–40°C for several minutes.
One might argue that depleting the system of magnesium or potassium nonspecifically denatures the ribosome and hence inhibits tRNA binding. However, there is substantial evidence that this transition is not an artifact of the experimental conditions, but actually represents a discrete, physiologically relevant conformational change. For example, inactive 30S subunits become active when presented with aa-tRNA in complex with EF-Tu, suggesting that, in vitro, adjusting temperature and salt concentration mediates a conformational change normally carried out by an elongation or initiation factor.11 Furthermore, no gross perturbations of the ribosome structure or loss of ribosomal proteins could be detected during the conversion from active to inactive that might be expected if the ribosome were simply denaturing in these conditions.1113
The structural aspects of this interconversion have been monitored by several methods. Oligodeoxyribonucleotide probes complementary to regions of 16S rRNA have been used to assess the availability of the rRNA for hybridization in active and inactive subunits.14,15 Differences in probe binding were found in the 3' minor domain of 16S rRNA, a region known to interact with tRNA and mRNA called the “decoding domain” (reviewed in ref. 16). Oligonucleotides were targeted to this part of the 16S rRNA because it was known to be important in binding mRNA and tRNA, and it turned out that this region was also affected by the active-inactive transition. In most cases, the availability for probe hybridization was decreased in the inactive subunits, arguing against a general unfolding of the 16S rRNA that would likely increase probe binding. In several cases, oligonucleotide binding was increased in the inactive vs. active subunits, suggesting that a discrete, reciprocating conformational change occurs during the interconversion.15
Oligonucleotides tethered to photolabile crosslinking agents or to chemical nucleases have also substantiated conformational changes at the decoding site during the active-inactive interconversion. Cooperman's group noted major changes in the profile of ribosomal proteins crosslinked by an oligonucleotide bound to the decoding region before and after activation.17 Using the chemical nuclease copper:phenanthroline tethered to an oligonucleotide complementary to the decoding region, Hill's laboratory demonstrated a conformational change in the rRNA environment.18 Whereas nucleotides 923–929 and 1391–1396, which are located in the secondary structure, and nucleotides 1190–1192 which are located far away in helix 34, were cleaved in inactive subunits, these cleavages disappeared upon activation of the subunits, leaving only cleavages in the region of oligonucleotide binding. This difference in cleavage patterns appears to be a result of conformational changes.
Noller and colleagues used chemical probing of the rRNA to monitor this structural conversion. 19 First, this study clearly indicated that there were not gross changes in the secondary structure of the rRNA after cation depletion, arguing against an overall denaturation during inactivation. Second, the nucleotides whose reactivity toward chemical probes were altered were conspicuously clustered in the functionally important decoding region, not randomly distributed throughout the 16S rRNA. Third, they observed that the transition increased the reactivity of some nucleotides and decreased that of others, indicating a concerted change in structure rather than a random loosening or tightening of the subunit. Finally, since these changes in chemical reactivity occurred in regions already known to be instrumental in binding tRNA and mRNA, this study correlated a function of the ribosome (i.e., substrate or message binding) with a specific conformational change. However, although a partial model of the transition was offered, the exact nature of the conformational change was not known except that the reactivity of several nucleotides in the decoding domain were altered during the transition (see Table 1).
Table 1
Summary of chemical probing results for the active-inactive interconversion of the 305 subunit.
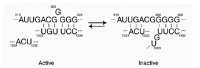
More recently, Wollenzien's group used a combination of site-directed mutagenesis and chemical probing to study this interconversion.20 Mutations in the G926, C1533, and A1394 regions were introduced into synthetic ribosomes based on the results of previous structure probing19 and psoralen crosslinking21 studies and were shown to affect the inactive-active transition as measured by chemical probing and tRNA binding. A model for a structural transition within this region was proposed, and included alternative base pairing schemes in the active and inactive forms. Here again, strong evidence for a conformational switch was presented, this time with a detailed description of base pairing rearrangements (see Fig. 1).
The active-inactive interconversion is not limited to the 30S subunit. The 50S subunit, when depleted of monovalent cations, becomes incapable of binding the peptidyltransferase-inhibiting antibiotic chloramphenicol and renders the subunit incompetent for peptide bond formation.2224 The structural basis for this effect was recently shown to be associated with a conformational change at the peptidyltransferase center in the vicinity of nucleotide A2451.25 In particular, this study showed that the reactivity of several nucleotides toward dimethyl sulfate (DMS) changes upon the active/inactive interconversion. Nucleotide A2451 was shielded in the active and susceptible to attack in the inactive conformation of E. coli ribosomes. The data suggest that the increased reactivity of A2451 with DMS in the inactive conformation is a consequence of the loss of a specifically bound potassium ion. This is especially interesting in light of the recent high resolution crystal structures of the large subunit that demonstrate that nucleotide 2451 is very close to the catalytic active site, and may be a direct participant in peptidyltransfer,26,27 but the mechanism of peptidyltransfer is still a matter of active debate.28,29 Furthermore, this study suggests that the previously reported pH-dependence of the A2451 reactivity with DMS was due to a local conformational change rather than to modulation of the pKa of this nucleotide mediated by interactions with nearby nucleotides.30
Structural Changes Associated with mRNA Binding and Initiation Events
Among the ribosome's first tasks in translation is the binding of an appropriate mRNA. This interaction is promoted in prokaryotes by the mRNA-rRNA interaction known as the Shine-Dalgarno (SD) interaction.31 Highly expressed prokaryotic mRNAs usually have a purine-rich sequence upstream of the AUG initiation codon with complementarity to a conserved pyrimidine-rich region near the 3' end of the rRNA (in the vicinity of nucleotides 1435–1440 of E. coli 16S rRNA) called the anti-Shine-Dalgarno (ASD) region. This interaction is a powerful determinant of the efficiency of translation of a particular message; mRNAs with a weak SD sequence (less than perfect complementarity with the rRNA sequence) are translated with low efficiency, whereas highly translated messages almost invariably have strong SD sequences (reviewed in ref. 32).
After the SD-ASD interaction aids in the recruitment of an mRNA for the ribosome, it may be advantageous to sequester the ASD sequence for two reasons. First, the ribosome must be able to release the 5' end of the message so that it can move toward the 3' end of the mRNA without dragging along the 5' end. Second, when a ribosome is actively engaged in translation of a given mRNA, additional Shine-Dalgarno mediated initiation events with a second mRNA should be prevented. Kössel and colleagues proposed that the ASD sequence might hybridize to the 5' region of the 16S rRNA when the ribosome is actively translating.33 The authors presented sequence comparisons of the 5' and 3' ends of 16S rRNAs from several organisms which were consistent with the idea that these regions could form base pairs, although direct experimental evidence was not presented. More recent compilations of rRNA sequences have not fully supported this interaction inasmuch as canonical base pairing has not been conserved in all species.34 Another helical switch model for the 5' end region was proposed by the Brakier-Gingras group,35 based on a combination of phylogenetic and biochemical data including translational fidelity and streptomycin binding with wild-type and mutant ribosomes.36 In this model, the central pseudoknot, comprised of nucleotides 17–19:918–916, transiently opens and alternate base pairs are formed (nucleotides 12–16 base-pair with 911–915). However, subsequent studies using site-directed mutagenesis of nucleotides implicated in the proposed switch, while not ruling out a conformational change in the region, did not directly support it.37
Another study on the central pseudoknot was performed using the specialized ribosome system. 38 Here it was shown that, at least among ribosomes containing an artificial anti-Shine-Dalgarno sequence, any complementarity between nucleotides 17–19 and 918–916 was necessary and sufficient for normal initiation on the ribosome. If these sequences were engaged in transient alternate base pairing schemes (i.e., a switch), one might expect to find some additional defects that arise in these mutants due to lost complementarity in the other conformation of the switch. However, because the specialized ribosome system utilizes ribosomes containing mutated anti-Shine-Dalgarno sequences, there is potential for interference with the analysis of helical switching involving sequences also in this region. The central pseudoknot is clearly visible in recent crystal structures,39,40 but alternate conformations may be forthcoming in future crystal structures.
Results from the Gualerzi and Brimacombe laboratories suggest that the structure of the ASD region as well as the path of the mRNA through the ribosome changes upon binding of initiation factors (IFs).4143 First it was shown that an oligoribonucleotide mimicking the SD region of a mRNA could effectively compete with a full length mRNA for 30S subunit binding until initiation factors were added. At that point, the SD oligo could no longer compete for binding, suggesting that the SD region had been altered. In the second study, synthetic mRNAs were transcribed using the photolabile nucleotide analog 4-thiouridine. The position of the mRNA on the ribosome could then be monitored by binding the mRNA to the ribosome in the presence or absence of IFs, then irradiating the complexes with UV light and examining the ribosomal components for sites of crosslinking with the mRNA analog. Indeed, two distinct sets of nucleotides in the 16S were crosslinked, depending upon whether or not IFs were present, providing compelling evidence for a positional change of the mRNA relative to the 30S subunit under the influence of initiation factors. The nature of the conformational change was schematized with respect to the proximity of the crosslinked nucleotides and ribosomal proteins.
Another model for a helical switch has been proposed by the Ehresmann group for the ASD region of 16S rRNA, influenced by IF-3.44 According to this model, the 3' end of 16S rRNA pairs with the 830 region of 16S rRNA unless IF-3 is present. IF-3 competes with the ASD for binding near position 830, allowing the ASD to hybridize with mRNA. This model was proposed based on crosslinking data of the IF-3 to the rRNA and on the apparent complementarity of the ASD and the 830 region. In fact, recent cryoelectron microscopy, crystallographic, and crosslinking data confirmed the ASD-830 region proximity and demonstrated a conformational change of the 30S subunit upon IF-3 binding.45,46,46a The cryo-EM study demonstrates a binding site at the subunit interface, with conformational changes seen in the head and platform, while in the crystal structure, the C-terminal domain of IF-3 was localized to the solvent side of the subunit, not at the subunit interface.46 It is obvious that these two localizations of IF3 do not represent the same physiological state of the ribosome, even if both represent physiologically relevant IF3 binding states. However, in both cases IF3 binding to the 30S subunit causes conformational changes that can at least partially explain IF3's functions in translation.
A recent crystal structure of the 30S subunit with IF-1 from Ramakrishnan's group has helped to shed some light on the role of this initiation factor whose supporting role in initiation is known to be essential, but whose mechanism of function has been elusive.47,48 Compared to its structure in an empty 30S subunit, helix 44 of the IF-1 bound subunit adopts a markedly different conformation. IF-1 and protein S12 form separate pockets for nucleotides A1492 and A1493, locking them into a splayed-out conformation, in contrast to their stacked, flipped out conformation that is the hallmark of the paromomycin-bound (or cognate codon-anticodon recognition) state.49,50 The presence of IF-1 at the top of helix 44 not only occludes the A site from tRNA binding, but its effects are propagated down the helix, interfering with normal helical base pairing over a large distance. Helix 44 is observed in the crystal structure to undergo a lateral shift of several angstroms, a phenomenon that was observed previously under various experimental conditions using cryoelectron microscopy (see below).51,52 IF-1 binding also causes shifts in relative subunit domain positioning. The platform and shoulder rotate toward the A site upon factor binding. Intriguingly, binding of several antibiotics also cause a similar rotation of the shoulder and platform, with the additional effect of the head tilting back and away.49 These observations lend credence to the idea that some antibiotics target and block normal concerted movements in the ribosome.
Initiation factor 2 (IF-2) is responsible for installing the initiator tRNAfMet into the P site of the initiating 30S subunit. Although a high resolution crystal structure of this interaction has not yet been published, biochemical data indicate a conformational change in the 30S subunit upon IF-2 binding.53,54 Some of the observed changes in reactivity to chemical probes and crosslinking reagents occurred in the classical decoding and P site domains (notably nucleotides A1493 and G1401), while others were observed in more distal regions of the 16S rRNA, suggesting an alteration of the overall topology.
A different helical switch involving mRNA-rRNA and rRNA-rRNA base pairing, called the “downstream box” interaction, has been debated in the literature5559 and is described in detail in Chapter 16 by Brakier-Gingras et al. Although mutations in this region of rRNA did have effects on cell growth, the ensemble of experiments with mutations in rRNA and mRNA have not supported the idea of a simple helical switch involving rRNA-mRNA base pairing interactions.
Conformational Changes During Ribosome-tRNA Interactions
The possibilities for conformational changes are numerous during the tRNA's transit through the ribosome: initial recognition of ternary complex, selection at the A site, peptide transfer from one tRNA to the next, translocation, and ejection of the spent deacylated tRNA from the E site. Conformational changes within the ribosome have been proposed for most of these steps, supported with various types of experimental evidence.
Extensive analysis of tRNA-rRNA interactions from Noller and colleagues using chemical probing of rRNA structure in the presence and absence of ribosome binding has yielded an extensive list of potential contact points (reviewed in ref. 60). They identified a set of highly conserved nucleotides in 16S and 23S rRNA whose reactivities toward chemical probes changed when the ribosome was bound by tRNAs and antibiotics known to inhibit various steps in translation. By repeating this approach with different types of tRNA under different experimental conditions, a path of changes in nucleotide reactivities caused by tRNA binding could be followed through the ribosome.61 Because some of the changes in chemical reactivity were difficult to explain as a result of direct shielding of nucleotides by tRNA, conformational changes were strongly implicated. For example, it was determined that small acylated oligoribonucleotide tRNA analogs caused almost the same footprints on the rRNA as the intact tRNA.62 Because it is unlikely that the small tRNA analog physically contacted all of the same sites that an intact tRNA would have, it appeared that the changes in reactivity were part of a coordinated conformational change in response to the binding of tRNA or of a tRNA analogue.
Ribosome : aa-tRNA Interactions and Translational Fidelity
Aminoacyl-tRNA selection probably involves at least two major steps, initial selection and proofreading, and each step has potential for conformational alterations. First, as the ternary complex binds the ribosome, a structural change may be necessary to accommodate it. Then, the ribosome-dependent GTP hydrolysis on the EF-Tu may be preceded by, accompanied by, or followed by structural changes in the ribosome, the EF-Tu, the tRNA, or a combination thereof. The EF-Tu-GDP complex then dissociates from the ribosome, leaving the aminoacyl-tRNA at the A site (or at a proofreading site) where it will either be used in the peptidyl transfer reaction or will be rejected prior to the peptidyl transfer reaction; each of these increments could involve conformational changes.
During tRNA selection, the ribosome must bind incoming tRNAs with minimal nonspecific interactions in order to emphasize the codon-anticodon interaction relative to the overall binding energy. A likely mechanism for this discrimination relies upon differences in binding geometry of the fully complementary versus the noncognate tRNAs. One can envision that a fully complementary codon-anticodon interaction would allow the tRNA to settle completely into a binding pocket and make contacts with the mRNA, rRNA and/or ribosomal proteins not available to noncognate tRNAs. These additional contacts could enhance stability of the tRNA-ribosome complex or may trigger the ribosome to proceed to the next step of translation.
Several models of aminoacyl-tRNA selection invoke two or more functional states of the A site and several lines of evidence suggest that structural changes occur when the tRNA-GTP-EF-Tu complex encounters the ribosome. Recent cryoelectron microscopy data have confirmed our suspicions that there are conformational adjustments on the ribosome upon EF-Tu binding. In particular, significant rearrangements in the 50S subunit were detected in the vicinity of proteins L7/L12 and the A site tRNA binding region upon binding of a kirromycin-stalled ternary complex.63 At the biochemical level, Moazed and Noller demonstrated that the ternary complex interacts with a different set of nucleotides in the rRNA compared to after GTP hydrolysis and factor release.61 The footprint for A site aa-tRNA binding did not appear on the 23S rRNA until after hydrolysis of GTP and release of EF-Tu, suggesting that binding of charged tRNA to the A site is at least a two-step process with an associated conformational change. A similar two-step scenario with intermediate hybrid states of the ribosome was proposed for the translocation event, described below.
The 530 loop's involvement in translational fidelity has been extensively studied, and several lines of evidence suggest that an “open/closed” structural change takes place here during decoding. Neomycin binding causes increased reactivity at C525,64 while streptomycin aids tRNA protection of G530. Based upon chemical probing of ribosome-tRNA complexes in the presence and absence of antibiotics and hyperaccurate S12 alleles, it was proposed that the loop adopts an open conformation during proofreading that correlated with a diminished off rate for EF-Tu GDP65,66 In this state, the tRNA is able to diffuse from the ribosome prior to peptidyl transferase. However, in contrast to this model, one study with chemical and enzymatic probing suggested that an open form of the 530 loop promotes translational errors.67 Conformational adjustment of this loop, especially a syn to anti rearrangement of G530 upon codon recognition seen in recent crystal structures, attests its importance in translational fidelity.50
Kinetic analysis of tRNA and ternary complex interactions with the ribosome by the group of Rodnina and Wintermeyer has made possible the dissection of aa-tRNA selection into several discrete steps with intriguing implications for an induced fit mechanism for tRNA selection (Fig. 2). First, complete kinetic descriptions of aminoacyl-tRNA binding, accommodation, and peptidyl transfer were established using stopped-flow fluorometric techniques to examine tRNA interactions with EF-Tu and with the ribosome, and quench-flow measurements using radioactive detection to examine rates of GTP hydrolysis and peptide bond formation (see ref. 68 and references therein). This analysis of each kinetically identifiable step in tRNA selection formed the foundation for a new model that has clarified our view of how a two-step selection mechanism might function to optimize tRNA selection with respect to speed and accuracy.
Particularly interesting in the kinetic data is the observation that the rates of GTPase activation prior to EF-Tu departure, and accommodation of the aa-tRNA into the peptidyltransferase-active A site when it has been released by EF-Tu, are vastly different between cognate and near-cognate tRNAs.68,69 This demonstrates that not only is a correct codon/anticodon fit necessary for stable initial binding of the ternary complex to the ribosome, but that correct geometry of the fit actually accelerates the next step in the sequence. Such an acceleration of catalytic activity most likely results from a change in the conformation of the ribosome, tRNA, or EF-Tu, or a combination thereof in response to a certain favorable geometry of tRNA-mRNA binding. These data are best explained by an induced fit mechanism that was proposed by Pape et al.69 According to this model, a correct codon-anticodon interaction promotes 16S rRNA to adopt a conformation that then accelerates GTPase activation and accommodation of the aminoacyl-tRNA —CCA end into the peptidyl transferase active site. In fact, this activated 16S conformation was also shown by kinetic methods to be induced by the translational error-inducing antibiotic paromomycin,70 with clear mechanistic implications for this class of antibiotics. These authors suggested that the structural changes are likely to be related to conformational changes observed at nucleotides 1492, 1493 and 530 (E. coli numbering) of the decoding region.9,49,71,72 This argument has been strongly supported by recent crystallographic evidence of an induced fit by cognate tRNA and potentiated by paromomycin.50
The Nierhaus group's allosteric three site model has been useful in helping to rationalize results of diverse tRNA binding experiments and has helped to emphasize the importance of the E site in translation.73 According to this model, the presence of an aminoacyl-tRNA in the A site causes a decrease in the affinity for deacylated tRNA in the E site and vice versa. Such an interconversion of affinities would suggest a simultaneous structural change in each of these sites. Modulation of the A site affinity was proposed to aid in correct aa-tRNA selection. In fact, the allosteric three site model has undergone some revision in recent years. However, a new model, called the α/ϵ model, that is largely compatible with the older allosteric model, also predicts conformational changes resulting in high and low affinity tRNA binding to the A and E sites, with obvious implications for aa-tRNA selection.4,74,75
Evidence for the α/ϵ conformational change was derived from several types of experiments. Originally, different species of radiolabelled tRNAs were used in binding experiments to monitor the occupancy of ribosomes under different conditions. Then biophysical and biochemical structural studies, including iodine cleavage of phosphorothioate containing tRNAs, neutron scattering, and cryoelectron microscopy, were applied to monitor the local environment of the tRNAs as they passed through the ribosome.7577 These studies showed an apparent lack of change of the local environment surrounding the tRNAs in steps prior to or after translocation. Because the structure of the tRNAs themselves do not change during translocation, the authors propose a movable domain wherein the A and P site bound tRNAs are transported together by a conveyor within the ribosome to the P and E sites. In this model, the A site and E site alternate between high affinity and low affinity states in a reciprocal fashion, although the interaction is not allosteric. Rather, the affinity state is determined by the relative positions of the decoding center and the movable domain (see ref. 4 for a detailed description of the α/ϵ model). While the particular nucleotides or proteins responsible for the modulation of A site affinity have not been identified, cryoelectron microscopy has suggested a narrowing of the A site entrance and the appearance of unidentified density that could affect ternary complex binding and aa-tRNA selection.78,79
Evidence for reciprocal, competing conformers related to tRNA selection was demonstrated with chemical probing techniques by Allen and Noller.80 They showed that nucleotides A908 and A909 in helix 27 of 16S rRNA were more reactive to dimethylsulfate in ribosomes isolated from a mutant strains of bacteria that have enhanced rates of translational errors, and were less reactive in strains that have lower levels of translational errors. The mutations responsible for these translational fidelity phenotypes are in ribosomal proteins S4 and S12. Mutations in S12 confer resistance to or dependence upon the translational error inducing drug streptomycin.81 Some of these “restrictive” mutations, so-called because they restrict the suppression of nonsense codons, can also cause ribosomes to translate slowly and with increased translational accuracy.82 Another class of mutations was subsequently discovered that could counteract the hyperaccurate, streptomycin dependent phenotype of the S12 mutations. These second-site mutations were mapped to ribosomal proteins S4 and S5. When segregated from the S12 restrictive mutations, these S4 and S5 mutations were found to increase the level of translational errors and were designated ram mutations, for ribosomal ambiguity.
Allen and Noller proposed that the 908–909 region was involved in an equilibrium between two conformational states in 16S rRNA. The balance of the equilibrium was controlled at least in part by ribosomal proteins S4 and S12 (they did not test the influence of S5), hence mutations in either of these proteins could perturb the equilibrium. Interestingly, streptomycin could also influence the equilibrium similarly to the S4 ram mutations. They proposed that the reactivity of A908–A909 was an indicator of the position of the equilibrium, although with the limited amount of structural data available, they did not propose the specific structures involved. Recent high resolution glimpses at the 30S subunit clearly indicate that S4, S5, and S12 are close to the 900 region and close to the decoding region, making the proposition that nucleotides 908–909 have a direct or indirect role in tRNA selection appear very reasonable.39,40,45,49,83
Another conformational change associated with translational fidelity has been proposed for nucleotides 910–912, also in helix 27.84 Through mutagenesis, chemical structure probing, and complementation with different mutant ribosomal protein alleles, it was proposed that nucleotides 910–912 base pair not only with nucleotides 885–887, as is commonly depicted on the secondary structure maps, but also transiently with nucleotides 888–890 (Fig. 3). It is notable that mutations that favored strictly one or the other of the helix 27 conformations were deleterious or lethal, while mutations that permitted both base-pairing arrangements were tolerated. This suggests that both arrangements are utilized during translation. Mutations in the rRNA that favored the 912:885 conformation exhibited an error-prone phenotype. Furthermore, these rRNA mutations were very deleterious when combined with ram S5 alleles, but were completely compatible with restrictive mutations in S12. On the other hand, the growth of cells harboring rRNA mutations that favored the 912–888 conformation were incompatible with restrictive S12 mutations, but were complemented with ram S5 mutations. Since the mutations affected translational fidelity, these mutations presumably affect A-site tRNA binding or proofreading, although a connection of this three nucleotide switch with translocation cannot be ruled out. Recent data from Ganoza's group, based upon chemical reactivity changes in helix 27 and the decoding region upon binding of the ribosomal ATPase RbbA and the translocation-inhibiting and error-inducing antibiotic hygromycin B, raise the possibility that RbbA may have a role in the dynamics of this helix.85,86
Chemical structure probing of ribosomes containing helix 27 mutations showed differences in reactivities in several regions of the ribosome implicated in decoding, including the decoding center, helix 34, and the 530 loop.84 Mutations in the rRNA that favored the 912–885 conformation affected the chemical reactivities oppositely to those that favored the 912–888 conformation. Cryoelectron microscopy studies on ribosomes carrying these two types of mutations in helix 27 also revealed discrete structural changes in several regions of the 30S subunit, and indeed some changes in the 50S subunit of a 70S ribosome.51 This suggests that a change in the state of helix 27 could be sensed at distal regions of the ribosome, and therefore could be part of a signal relay system.
Consistent with its putative role in aa-tRNA selection, crystal structure models of the 30S subunit place helix 27 at the floor of the A-site.39,40,45,49,83 In these high resolution structure models, helix 27 has been observed only in its 912–885 conformation so far. Crystallization of ribosomes in the 912–888 restrictive form would be most interesting with respect to rigorously defining the proposed signal relay system. These efforts are hampered somewhat by the deleterious nature of mutant ribosomes harboring mutations that favor only the 912–888 conformation.
Liebman and coworkers have constructed a series of site-directed and selected helix 27 mutations in yeast.87 While these mutations affected translational fidelity and sensitivity to error-inducing antibiotics, the pattern of phenotypes did not match the pattern observed in E. coli. Therefore, while this study lends further support to the notion that helix 27 is important in the decoding process, it does not support the E. coli-style helix 27 switching mechanism in yeast. Further studies will be required to determine whether the proposed switch is a conserved feature among different species.
Further support for alternating conformations and affinities during tRNA selection comes from Ehrenberg's group. They performed a series of studies of the kinetics and thermodynamics of tRNA binding to wild-type and mutant ribosomes in the presence or absence of error-inducing antibiotics. First, it was demonstrated that error-prone mutant ribosomes harboring mutations in ribosomal proteins S4 or S5, or ribosomes treated with error-inducing antibiotics bound their aa-tRNAs more tightly in the A-site, and the proofreading step was diminished.88 Furthermore, they showed that the same mutations or antibiotics that caused increased binding to the A site decreased the stability of binding to the P site, as measured by the propensity for the peptidyl-tRNA to drop off the ribosome.89 In fact, the reverse was true when mutant ribosomes with mutations in ribosomal protein S12 conferring a hyperaccurate phenotype were tested. The authors suggested that such reciprocity reflects a movement in the 16S that is opposite for the ram and restrictive phenotypes.
Conformational Changes During Translocation
During translocation, the ribosome moves along the mRNA and the tRNAs move from one functional site to another within the ribosome and the implications for conformational change are obvious. Spirin's group demonstrated using small angle neutron scattering that the radius of gyration differed between pre- and post-translocational ribosomes.90 Noller's hybrid-states model provided an alternative to the classic A-site then P-site view of translocation.61 As determined by the footprints of tRNA, translocation occurs at different times in the 30S and 50S subunits. According to this model, translocation occurs spontaneously in the 50S subunit upon peptidyl transfer (i.e., A site tRNA moves to P site and P site tRNA moves to E site). In the 30S subunit, however, translocation does not occur until or during the hydrolysis of GTP upon EF-G, after peptidyl transfer. In the interim between these two translocation events, one tRNA is in the A site of the 30S subunit and the P site of the 50S subunit (the A/P site), while the other tRNA is in the P site of the 30S subunit and the E site of the large subunit (P/E site).
These distinctive changes in the footprints indicated that a structural shift occurs. However, because of the limited resolution of the technique, the exact nature of the conformational change was not obvious. The crystal structures of the 70S ribosome with tRNAs bound have so far demonstrated only classic A/A, P/P, and E site positioning of the tRNAs, 83,91 although this snapshot cannot be considered the only physiological state of the ribosome. In fact, cryoelectron microscopy has shown a P/E hybrid state for deacylated tRNA under some buffer conditions,92 but did not provide clear evidence of hybrid states during the intersubunit ratcheting motion observed in the pre- and post-translocational states.52 Furthermore, recent crosslinking results93 and cryoelectron microscopy studies79 suggest the existence of an additional exit site (called the F or E2 site) that could add some complexity to existing models of tRNA transit through the ribosome.
Wintermeyer's group proposed that binding of EF-G to the ribosome causes a conformational change in the E site that enhances its affinity for deacylated tRNA, providing a thermodynamic “escape route” for P site bound tRNA.94 Furthermore, the binding of the 3' end of deacylated tRNA to 23S rRNA was proposed to stimulate GTP hydrolysis and translocation. The nature of the linkage between the EF-G binding site on the ribosome and the E site was not known, but the distance between the two regions of the ribosome based on models of the 50S subunit suggest that it is a long-range interaction. In a different study, it was shown that the accessibility of the E site for binding of an oligonucleotide increased when deacylated tRNA was bound to the P site relative to vacant or aa-tRNA bound ribosomes.95 These data suggested there was a structural change in the ribosome that made the E site more available for binding of an oligonucleotide, and by extension, a tRNA, but only in response to the presence of a deacylated tRNA in the P site, a situation ripe for translocation. Such a structural change may also be correlated with improving the energetics of translocation.
In addition to features relevant to aminoacyl-tRNA selection, Nierhaus's α/ϵ model proposes a novel way of looking at translocation that contrasts with the hybrid states model. Data supporting this model include phosphorothioate footprinting, neutron scattering and cryoelectron microscopy data, and have been described above in the A site-tRNA interactions section.7577,79 Of particular relevance to translocation are the cryoelectron microscopy data where the A site density corresponding to the 1492–1493 region in helix 44 is seen to move the distance of one codon toward the P site when one compares the images of ribosomes complexed with EF-G plus a nonhydrolyzable GTP analogue and EF-G-GDP frozen with fusidic acid.77 This could be interpreted as movement of the conveyor from the A to P site, although higher resolution structural analysis of various conformational states of ribosome-tRNA complexes will be necessary to fully describe translocation.
Other tRNA-rRNA Interactions
Crosslinking experiments have provided a wealth of information about the positions of tRNAs within the ribosome, and some of the patterns of crosslinks are highly suggestive of conformational changes in the ribosome (for recent reviews, see refs. 93,96–98). For example, the anticodon of tRNAArgI (derivatized at position 32 with a photoaffinity crosslinking moiety) was shown to form crosslinks to position 1378 of 16S rRNA from both the A and E sites, but not from the P site, suggesting that the position of this region of rRNA can be altered rather dramatically under different tRNA binding conditions.99 These data have been supported by other crosslinking experiments that gave compatible contact patterns, arguing against the possibility that the position of the tRNA in the ribosome was not properly assigned (e.g., ref. 100). Likewise, crosslinking to the peptidyltransferase region of 23S rRNA from two widely separated residues of P site-bound tRNA (position 47, near the “elbow” of tRNA, and a derivatized aminoacyl moiety at the -CCA end of tRNA) is suggestive of another rRNA conformational change in another functionally important part of the rRNA,96 although assignment of an exact position for a given tRNA on the ribosome is a delicate task (see ref. 92 for example).
Conformational Changes Related to Antibiotic Binding
Ribosome-binding antibiotics block specific steps of translation and are therefore useful for both their clinical applications and as probes of ribosomal function. Numerous accounts exist in the literature of a change in conformation of the ribosome upon antibiotic binding. With new high resolution structural information on the ribosome and ribosome-antibiotic complexes, many more conformational changes have been detected. For example, streptomycin was shown in the 1970's to promote the conversion of inactive to active 30S ribosomal subunits,101 and was shown shortly thereafter by biophysical methods to alter the structure of the 30S subunit.102 More recently, Jerinic and Joseph demonstrated that the toeprint of ribosomes on a mRNA was altered when streptomycin or other aminoglycoside antibiotics was added.103 This effect could be modulated by the presence of restrictive or error-prone S12 or S4 alleles. The crystal structure model of the 30S-streptomycin complex did not reveal any major structural changes compared to the vacant subunit, although this does not preclude the existence of other conformers outside these crystal conditions.49 The localization of streptomycin to the helix 27 region in the crystal structure together with the biochemical and genetic data discussed above suggest that streptomycin affects an equilibrium of competing conformers in this region.
Paromomycin, another aminoglycoside antibiotic, has also been shown to alter 30S subunit and rRNA structure. At the fine structure level, Puglisi's group has shown by NMR analysis that paromomycin alters the structure of a model RNA mimicking the decoding region of rRNA in a way reminiscent of a cognate codon recognition event.71,72 Specifically, nucleotides A1492 and A1493 flip out from the helix in response to paromomycin binding. In the 30S crystal structures without or with paromomycin, these nucleotides are disordered or flipped out, respectively.39,49 Again, it is likely that more conformers of ribosomes will be crystallized in the future and further conformational changes will be seen.
Cocrystal structures with the antibiotics tetracycline and spectinomycin strongly suggest that a mechanism of action of these antibiotics is to interfere with normal ribosomal movements.45,49 Spectinomycin is proposed to prevent or alter a normal movement of the 30S head relative to the body, possibly during translocation.49 The spectinomycin binding site in helix 34 is strategically located to prevent such movement. On the other hand, tetracycline's major binding site on the 30S subunit interferes with tRNA entrance to the A-site, while the several alternative sites appear to be able to block other parts of a hypothesized signal relay system.45,104
Pactamycin, a potent antibiotic that exerts its effects on prokaryotes, archae, and eukaryotes, was proposed by Mankin, based upon chemical probing and isolation of pactamycin resistant mutants, to inhibit normal conformational changes in the small subunit that may be associated with initiation of translation.105 Crystallographic data on pactamycin binding did not demonstrate a major conformational change, but indicated that helices 23a and 24a may be frozen together by pactamycin, and that pactamycin may interfere with the Shine-Dalgarno/ anti-Shine-Dalgarno interaction or the normal path of mRNA at the E site.104 A separate crystallographic study by Franceschi, Yonath and coworkers of the edeine-30S complex demonstrated a remarkable agreement on the mechanism of action of these two antibiotics.45 Edeine, a universal antibiotic related to pactamycin, was shown to bind the 30S subunit in the same strategic location as pactamycin, effectively tying together regions involved in mRNA, tRNA, and IF-3 binding (helices 23, 24, 28, 44, 45). In fact, by bringing helices 23 and 24 closer together, edeine promoted the formation of a new base pair. Because cryoelectron microscopy studies have implicated movements of these helices relative to each other during translation,51,77 edeine and pactamycin very likely exert their effects not solely by blocking binding of tRNA, mRNA, or IF-3, but by blocking necessary conformational changes of the ribosome.
Finally, hygromycin B, an aminoglycoside antibiotic that inhibits translocation and causes translational errors, has been localized in co-complexes with 30S subunits near the top of helix 44.104 Hygromycin B did not cause a large conformational change in the 30S subunit, but because helix 44 has been shown in cryoelectron microscopy studies to oscillate,52 and because this region constitutes part of the decoding center, hygromycin B likely acts by inhibiting normal movement of helix 44 during decoding and/or translocation. Whereas the crystal structures of most of these antibiotic/subunit complexes did not reveal large perturbations in the subunit structure, crosslinking and other biochemical experiments suggest that these antibiotics can and do cause conformational changes in ribosomes in solution (see ref. 106 and references therein).
Evidence for Other Conformational Switches
Several switches were proposed in the mid-1980's by examining the double-helical products of partially nuclease-digested ribosomes. This methodology was primarily used to obtain biochemical support for helices proposed in early secondary structure models of the rRNA. Adapted to finding switches, the idea was that if a piece of rRNA normally formed base pairs with two (or more) other sequences, then after digestion, electrophoretic separation, and sequencing of digestion products, it should be possible to identify both helical arrangements. In the digestion products of several studies, some pieces of rRNA were consistently found base paired to two different sequences of rRNA (e.g., see refs. 107–109). Using the rather limited rRNA sequence data available at the time, attempts were made to ensure that only phylogenetically conserved switches were considered. However, these switches have not been tested experimentally.
Several chemical probing studies have been used to examine which nucleotides in rRNA are transiently exposed or protected during translation. Laughrea used in vivo chemical footprinting to examine reactivity of rRNA residues to dimethyl sulfate during translation in actively growing cells.110 These data showed several transient exposures of nucleotides that were abolished when translation was interrupted with chloramphenicol. Specifically, nucleotides in helices 26, 33, and 44 were shown to be transiently exposed during translation. In an in vitro study, Barta and colleagues111 examined susceptibility of rRNA to cleavage by lead(II) ions in pre and post-translocational state. Strikingly, they found that despite all of the evidence for conformational changes in the small subunit, they detected very little change in the cleavage pattern for 16S rRNA before and after translocation. Equally remarkable was the observation that significant conformational changes did occur in 23S rRNA in domains V and VI, near the peptidyltransferase center and the sarcin-ricin factor binding site. This was surprising because, compared to the rather fluid and changeable 30S subunit, there historically has been some temptation to think of the 50S as a relatively static entity.
Another kind of conformational multiplicity has been observed in cryoelectron microscopy images of free 30S subunits compared to those complexed with 50S subunits.112 The major site of structural heterogeneity was at the head/neck region; some of this heterogeneity was lost upon association of the small subunit to the large subunit. It would be interesting in this context to see if antibiotics that affect relative domain positions in the 30S subunit45,49 might limit some of these conformers.
Gating of both the mRNA through the 30S subunit52,112 and of the nascent peptide through the exit tunnel(s) in the 50S subunit113 have been observed by cryoelectron microscopy. In the case of the mRNA channel, a pore is seen to alternately open and close around the mRNA, while for the nascent peptide exit tunnel, the gate may be used to direct a peptide through alternate pathways, or to alternately stop and release the growing peptide chain.
Ribosome Assembly
Whereas the assembly of ribosomes is not per se part of translation, an intriguing helical switching mechanism during 30S subunit assembly has been proposed that merits discussion here.114 A C to U mutation at position 23 (“C23U”) in16S rRNA was shown to confer cold-sensitivity. Growth of this mutant was inhibited below 26°C but not at 37–42°C. When shifted to the nonpermissive temperature, these cells accumulated malformed 30S ribosomal subunits. The mutation was mapped to a helical region in the 5' domain of 16S rRNA, thus it was surprising that it caused cold sensitivity since its effect would apparently be to destabilize a helix that might be further destabilized by heat. Their explanation, supported by genetic and biochemical data, was that nucleotide G11, while depicted as base paired to position C23 in the secondary structure model of 16S rRNA, must also form a base pair with a nucleotide (U-5) in the upstream precursor sequence of immature 16S rRNA during ribosome assembly. Thus, a mutation at position 23 would perturb an equilibrium between two competing helical conformations and interfere with subunit assembly. In addition to this specific helical switching event, a complex and intriguing series of conformational changes in 16S rRNA structure during sequential assembly events has been documented by the Noller group (e.g., see ref. 115). This large body of work is beyond the scope of the present review.
Concluding Remarks
The idea that the ribosome, and in particular, the ribosomal RNA changes conformation during translation has apparently gained acceptance over the last fifteen years. In a 1986 article, Richard Brimacombe wrote: “...many ribosomologists, while tacitly accepting the concept that the secondary structure of mRNA has to be entirely unfolded loop by loop during the translation process, show a surprising resistance to the idea that secondary structural changes can occur in the ribosomal RNA during the same process.”116 Whereas structural evidence for any conformational change in the ribosome may have stirred much excitement in the early 1980's, there is now ample evidence that the ribosome is dynamic. The question of a moveable ribosome has now shifted from ‘whether’ to ‘how, when, and why?’ This field is bristling with excitement as these questions are answered with a balance of experimental techniques. Comparisons of high resolution structure determinations of ribosomes in different states will give us the fine details as no other technique can, while solution biochemical and biophysical approaches combined with genetics, comparative and computational analyses will continue to offer insights not readily accessible in the confines of a crystal lattice.
Acknowledgments
We would like to thank Albert Dahlberg for helpful discussions during the preparation of this manuscript and to acknowledge support for research in our laboratory by the NIH (grant number GM35717 to W.E.Hill).
References
- 1.
- Serdyuk IN, Spirin AS. Structural Dynamics of the translating ribosomeIn: Hardesty B, Kramer G, eds.Structure, Function, and Genetics of RibosomesNew York: Springer-Verlag,1986425–437.
- 2.
- Burma DP, Srivastava S, Srivastava AK. et al. Conformational change of 50S ribosomes during protein synthesisIn: Hardesty B, Kramer G, eds.Structure, Function, and Genetics of RibosomesNew York: Springer-Verlag,1986438–453.
- 3.
- Laughrea M. Structural dynamics of translating ribosomes: 16S ribosomal RNA bases that may move twice during translocation. Mol Microbiol. 1994;11:999–1007. [PubMed: 8022290]
- 4.
- Spahn CM, Nierhaus KH. Models of the elongation cycle: an evaluation. Biol Chem. 1998;379:753–72. [PubMed: 9705140]
- 5.
- Wilson KS, Noller HF. Molecular movement inside the translational engine. Cell. 1998;92:337–49. [PubMed: 9476894]
- 6.
- Rodnina MV, Savelsbergh A, Wintermeyer W. Dynamics of translation on the ribosome: molecular mechanics of translocation. FEMS Microbiol Rev. 1999;23:317–33. [PubMed: 10371036]
- 7.
- Agrawal RK, Frank J. Structural studies of the translational apparatus. Curr Opin Struct Biol. 1999;9:215–21. [PubMed: 10322215]
- 8.
- van Heel M. Unveiling ribosomal structures: the final phases. Curr Opin Struct Biol. 2000;10:259–264. [PubMed: 10753818]
- 9.
- Rodnina MV, Wintermeyer W. Ribosome fidelity: tRNA discrimination, proofreading and induced fit. Trends Biochem Sci. 2001;26:124–30. [PubMed: 11166571]
- 10.
- Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: Kinetic and Structural Mechanisms. Annu Rev Biochem. 2001;70:415–435. [PubMed: 11395413]
- 11.
- Zamir A, Miskin R, Elson D. Inactivation and reactivation of ribosomal subunits: amino acyl- transfer RNA binding activity of the 30 s subunit of Escherichia coli. J Mol Biol. 1971;60:347–64. [PubMed: 4938735]
- 12.
- Kearney KR, Moore PB. X-ray solution-scattering studies of active and inactive Escherichia coli ribosomal subunits. J Mol Biol. 1983;170:381–402. [PubMed: 6355486]
- 13.
- Ginzburg I, Miskin R, Zamir A. N-ethyl maleimide as a probe for the study of functional sites and conformations of 30 S ribosomal subunits. J Mol Biol. 1973;79:481–94. [PubMed: 4585975]
- 14.
- Backendorf C, Ravensbergen CJ, Van der Plas J. et al. Basepairing potential of the 3' terminus of 16S RNA: dependence on the functional state of the 30S subunit and the presence of protein S21. Nucleic Acids Res. 1981;9:1425–44. [PMC free article: PMC326767] [PubMed: 7232220]
- 15.
- Weller JW, Hill WE. Probing dynamic changes in rRNA conformation in the 30S subunit of the Escherichia coli ribosome. Biochemistry. 1992;31:2748–57. [PubMed: 1547215]
- 16.
- Zimmermann RA. The decoding domainIn: Zimmermann RA, Dahlberg AE, eds.Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein BiosynthesisBoca Raton: CRC Press,1996 .
- 17.
- Muralikrishna P, Cooperman BS. A photolabile oligodeoxyribonucleotide probe of the decoding site in the small subunit of the Escherichia coli ribosome: identification of neighboring ribosomal components. Biochemistry. 1994;33:1392–8. [PubMed: 8312257]
- 18.
- Muth GW, Hennelly SP, Hill WE. Using a targeted chemical nuclease to elucidate conformational changes in the E. coli 30S ribosomal subunit. Biochemistry. 2000;39:4068–74. [PubMed: 10747796]
- 19.
- Moazed D, Van Stolk BJ, Douthwaite S. et al. Interconversion of active and inactive 30 S ribosomal subunits is accompanied by a conformational change in the decoding region of 16 S rRNA. J Mol Biol. 1986;191:483–93. [PubMed: 2434656]
- 20.
- Ericson G, Minchew P, Wollenzien P. Structural changes in base-paired region 28 in 16 S rRNA close to the decoding region of the 30 S ribosomal subunit are correlated to changes in tRNA binding. J Mol Biol. 1995;250:407–19. [PubMed: 7542348]
- 21.
- Ericson G, Wollenzien P. An RNA secondary structure switch between the inactive and active conformations of the Escherichia coli 30 S ribosomal subunit. J Biol Chem. 1989;264:540–5. [PubMed: 2642480]
- 22.
- Miskin R, Zamir A, Elson D. The inactivation and reactivation of ribosomal-peptidyl transferase of E. coli. Biochem Biophys Res Commun. 1968;33:551–7. [PubMed: 4880391]
- 23.
- Vogel Z, Vogel T, Zamir A. et al. Correlation between the peptidyl transferase activity of the 50 S ribosomal subunit and the ability of the subunit to interact with antibiotics. J Mol Biol. 1971;60:339–46. [PubMed: 4938734]
- 24.
- Zamir A, Miskin R, Vogel Z. et al. The inactivation and reactivation of Escherichia coli ribosomes. Methods Enzymol. 1974;30:406–26. [PubMed: 4605275]
- 25.
- Bayfield MA, Dahlberg AE, Schulmeister U. et al. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc Natl Acad Sci USA. 2001;98:10096–101. [PMC free article: PMC56921] [PubMed: 11517305]
- 26.
- Nissen P, Hansen J, Ban N. et al. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–30. [PubMed: 10937990]
- 27.
- Ban N, Nissen P, Hansen J. et al. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–20. [PubMed: 10937989]
- 28.
- Polacek N, Gaynor M, Yassin A. et al. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature. 2001;411:498–501. [PubMed: 11373685]
- 29.
- Thompson J, Kim DF, O'Connor M. et al. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc Natl Acad Sci USA. 2001;98:9002–7. [PMC free article: PMC55363] [PubMed: 11470897]
- 30.
- Muth GW, Ortoleva-Donnelly L, Strobel SA. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science. 2000;289:947–50. [PubMed: 10937997]
- 31.
- Shine J, Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–6. [PMC free article: PMC388224] [PubMed: 4598299]
- 32.
- Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J Mol Biol. 1988;204:79–94. [PubMed: 2464068]
- 33.
- Kossel H, Hoch B, Zeltz P. Alternative base pairing between 5'- and 3'-terminal sequences of small subunit RNA may provide the basis of a conformational switch of the small ribosomal subunit. Nucleic Acids Res. 1990;18:4083–8. [PMC free article: PMC331163] [PubMed: 2198532]
- 34.
- Maidak BL, Cole JR, Parker CT Jr. et al. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 1999;27:171–3. [PMC free article: PMC148126] [PubMed: 9847171]
- 35.
- Leclerc D, Brakier-Gingras L. A conformational switch involving the 915 region of Escherichia coli 16 S ribosomal RNA. FEBS Lett. 1991;279:171–4. [PubMed: 2001727]
- 36.
- Brakier-Gingras L, Pinard R, Dragon F. Pleiotropic effects of mutations at positions 13 and 914 in Escherichia coli 16S ribosomal RNA. Biochem Cell Biol. 1995;73:907–13. [PubMed: 8722006]
- 37.
- Pinard R, Payant C, Melancon P. et al. The 5' proximal helix of 16S rRNA is involved in the binding of streptomycin to the ribosome. FASEB J. 1993;7:173–6. [PubMed: 7678560]
- 38.
- Brink MF, Verbeet MP, de Boer HA. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993;12:3987–96. [PMC free article: PMC413681] [PubMed: 7691600]
- 39.
- Wimberly BT, Brodersen DE, Clemons WM Jr. et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–39. [PubMed: 11014182]
- 40.
- Schluenzen F, Tocilj A, Zarivach R. et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–23. [PubMed: 11007480]
- 41.
- Canonaco MA, Gualerzi CO, Pon CL. Alternative occupancy of a dual ribosomal binding site by mRNA affected by translation initiation factors. Eur J Biochem. 1989;182:501–6. [PubMed: 2666129]
- 42.
- La Teana A, Gualerzi CO, Brimacombe R. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA. 1995;1:772–82. [PMC free article: PMC1369318] [PubMed: 7493323]
- 43.
- Petrelli D, LaTeana A, Garofalo C. et al. Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J. 2001;20:4560–9. [PMC free article: PMC125572] [PubMed: 11500382]
- 44.
- Ehresmann C, Moine H, Mougel M. et al. Cross-linking of initiation factor IF3 to Escherichia coli 30S ribosomal subunit by trans-diamminedichloroplatinum(II): characterization of two cross-linking sites in 16S rRNA; a possible way of functioning for IF3. Nucleic Acids Res. 1986;14:4803–21. [PMC free article: PMC311493] [PubMed: 2425339]
- 45.
- McCutcheon JP, Agrawal RK, Philips et al. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc Natl Acad Sci USA. 1999;96:4301–4306. [PMC free article: PMC16327] [PubMed: 10200257]
- 46.
- Pioletti M, Schlunzen F, Harms J. et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–39. [PMC free article: PMC125237] [PubMed: 11296217]
- 46a.
- Shapkina TG, Dolan MA, Babin P. et al. Initiation factor 3-induced structural changes in the 30 S ribosomal subunit and in complexes containing tRNA(f)(Met) and mRNA. J Mol Biol. 2000;299:615–28. [PubMed: 10835272]
- 47.
- Carter AP, Clemons WM Jr, Brodersen DE. et al. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. [PubMed: 11228145]
- 48.
- Gualerzi CO, Brandi L, Caserta E. et al. Translation initiation in bacteriaIn: Garrett RA, Douthwaite S, Liljas A, eds. The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions Washington, DC: ASM Press, 2000477–494.
- 49.
- Carter AP, Clemons WM, Brodersen DE. et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–8. [PubMed: 11014183]
- 50.
- Ogle JM, Brodersen DE, Clemons WM Jr. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. [PubMed: 11340196]
- 51.
- Gabashvili IS, Agrawal RK, Grassucci R. et al. Major rearrangements in the 70S ribosomal 3D structure caused by a conformational switch in 16S ribosomal RNA. EMBO J. 1999;18:6501–7. [PMC free article: PMC1171713] [PubMed: 10562562]
- 52.
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:18–22. [PubMed: 10917535]
- 53.
- Wakao H, Romby P, Ebel JP. et al. Topography of the Escherichia coli ribosomal 30S subunit-initiation factor 2 complex. Biochimie. 1991;73(78):991–1000. [PubMed: 1720674]
- 54.
- Wakao H, Romby P, Laalami S. et al. Binding of initiation factor 2 and initiator tRNA to the Escherichia coli 30S ribosomal subunit induces allosteric transitions in 16S rRNA. Biochemistry. 1990;29:8144–51. [PubMed: 1702020]
- 55.
- Sprengart ML, Fatscher HP, Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16srRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990;18:1719–23. [PMC free article: PMC330588] [PubMed: 2186363]
- 56.
- Sprengart ML, Fuchs E, Porter AG. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996;15:665–74. [PMC free article: PMC449985] [PubMed: 8599950]
- 57.
- O'Connor M, Asai T, Squires CL. et al. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc Natl Acad Sci USA. 1999;96:8973–8. [PMC free article: PMC17717] [PubMed: 10430880]
- 58.
- La Teana A, Brandi A, O'Connor M. et al. Translation during cold adaptation does not involve mRNA-rRNA base pairing through the downstream box. RNA. 2000;6:1393–402. [PMC free article: PMC1370010] [PubMed: 11073215]
- 59.
- Moll I, Huber M, Grill S, Sairafi P. et al. Evidence against an Interaction between the mRNA downstream box and 16S rRNA in translation initiation. J Bacteriol. 2001;183:3499–505. [PMC free article: PMC99648] [PubMed: 11344158]
- 60.
- Noller HF, Moazed D, Stern S. et al. Structure of rRNA and its functional interactions in translationIn: Hill WE, Dahlberg AE, Garrett RA, eds.The Ribosome: Structure, Function, & E`volutionWashington, D.C.: American Society for Microbiology,199073–92.
- 61.
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–8. [PubMed: 2682263]
- 62.
- Moazed D, Noller HF. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc Natl Acad Sci USA. 1991;88:3725–8. [PMC free article: PMC51525] [PubMed: 2023922]
- 63.
- Stark H, Rodnina MV, Rinke-Appel J. et al. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature. 1997;389:403–6. [PubMed: 9311785]
- 64.
- Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–94. [PubMed: 2953976]
- 65.
- Powers T, Noller HF. Evidence for functional interaction between elongation factor Tu and 16S ribosomal RNA. Proc Natl Acad Sci USA. 1993;90:1364–8. [PMC free article: PMC45873] [PubMed: 8433994]
- 66.
- Powers T, Noller HF. Selective perturbation of G530 of 16 S rRNA by translational miscoding agents and a streptomycin-dependence mutation in protein S12. J Mol Biol. 1994;235:156–72. [PubMed: 8289238]
- 67.
- Van Ryk DI, Dahlberg AE. Structural changes in the 530 loop of Escherichia coli 16S rRNA in mutants with impaired translational fidelity. Nucleic Acids Res. 1995;23:3563–70. [PMC free article: PMC307238] [PubMed: 7567470]
- 68.
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7. [PMC free article: PMC1171092] [PubMed: 9857203]
- 69.
- Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–7. [PMC free article: PMC1171457] [PubMed: 10393195]
- 70.
- Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol. 2000;7:104–7. [PubMed: 10655610]
- 71.
- Yoshizawa S, Fourmy D, Puglisi JD. Recognition of the codon-anticodon helix by ribosomal RNA. Science. 1999;285:1722–5. [PubMed: 10481006]
- 72.
- Fourmy D, Yoshizawa S, Puglisi JD. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J Mol Biol. 1998;277:333–45. [PubMed: 9514734]
- 73.
- Rheinberger HJ, Nierhaus KH. Testing an alternative model for the ribosomal peptide elongation cycle. Proc Natl Acad Sci USA. 1983;80:4213–7. [PMC free article: PMC384007] [PubMed: 6348767]
- 74.
- Nierhaus KH, Beyer D, Dabrowski M. et al. The elongating ribosome: structural and functional aspects. Biochem Cell Biol. 1995;73:1011–21. [PubMed: 8722016]
- 75.
- Dabrowski M, Spahn CM, Schafer MA. et al. Protection patterns of tRNAs do not change during ribosomal translocation. J Biol Chem. 1998;273:32793–800. [PubMed: 9830024]
- 76.
- Nierhaus KH, Wadzack J, Burkhardt N. et al. Structure of the elongating ribosome: arrangement of the two tRNAs before and after translocation. Proc Natl Acad Sci USA. 1998;95:945–50. [PMC free article: PMC18634] [PubMed: 9448265]
- 77.
- VanLoock MS, Agrawal RK, Gabashvili IS. et al. Movement of the decoding region of the 16 S ribosomal RNA accompanies tRNA translocation. J Mol Biol. 2000;304:507–15. [PubMed: 11099376]
- 78.
- Agrawal RK, Lata RK, Frank J. Conformational variability in Escherichia coli 70S ribosome as revealed by 3D cryo-electron microscopy. Int J Biochem Cell Biol. 1999;31:243–54. [PubMed: 10216957]
- 79.
- Agrawal RK, Spahn CM, Penczek P. et al. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J Cell Biol. 2000;150:447–60. [PMC free article: PMC2175196] [PubMed: 10931859]
- 80.
- Allen PN, Noller HF. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16 S ribosomal RNA. J Mol Biol. 1989;208:457–68. [PubMed: 2477554]
- 81.
- Gorini L. Streptomycin and misreading of the genetic codeIn: Nomura M, Tissieres A, Lengyel P, eds.RibosomesCold Spring Harbor: Cold Spring Harbor Laboratory Press,1974791–803.
- 82.
- Bohman K, Ruusala T, Jelenc PC. et al. Kinetic impairment of restrictive streptomycin-resistant ribosomes. Mol Gen Genet. 1984;198:90–9. [PubMed: 6394968]
- 83.
- Yusupov MM, Yusupova GZ, Baucom A. et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–96. [PubMed: 11283358]
- 84.
- Lodmell JS, Dahlberg AE. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science. 1997;277:1262–7. [PubMed: 9271564]
- 85.
- Kiel MC, Ganoza MC. Functional interactions of an Escherichia coli ribosomal ATPase. Eur J Biochem. 2001;268:278–86. [PubMed: 11168361]
- 86.
- Ganoza MC, Kiel MC. A Ribosomal ATPase Is a Target for Hygromycin B Inhibition on Escherichia coli Ribosomes. Antimicrob Agents Chemother. 2001;45:2813–9. [PMC free article: PMC90736] [PubMed: 11557474]
- 87.
- Velichutina IV, Dresios J, Hong JY. et al. Mutations in helix 27 of the yeast Saccharomyces cerevisiae 18S rRNA affect the function of the decoding center of the ribosome. RNA. 2000;6:1174–84. [PMC free article: PMC1369991] [PubMed: 10943896]
- 88.
- Karimi R, Ehrenberg M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper- accurate and error-prone ribosomes. Eur J Biochem. 1994;226:355–60. [PubMed: 8001552]
- 89.
- Karimi R, Ehrenberg M. Dissociation rates of peptidyl-tRNA from the P-site of E.coli ribosomes. EMBO J. 1996;15:1149–54. [PMC free article: PMC450013] [PubMed: 8605885]
- 90.
- Serdyuk I, Baranov V, Tsalkova T. et al. Structural dynamics of translating ribosomes. Biochimie. 1992;74:299–306. [PubMed: 1379074]
- 91.
- Cate JH, Yusupov MM, Yusupova GZ. et al. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–104. [PubMed: 10497122]
- 92.
- Agrawal RK, Penczek P, Grassucci RA. et al. Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J Biol Chem. 1999;274:8723–9. [PubMed: 10085112]
- 93.
- Wower J, Kirillov SV, Wower IK. et al. Transit of tRNA through the Escherichia coli ribosome. Cross-linking of the 3' end of tRNA to specific nucleotides of the 23 S ribosomal RNA at the A, P, and E sites. J Biol Chem. 2000;275:37887–94. [PubMed: 10961994]
- 94.
- Lill R, Robertson JM, Wintermeyer W. Binding of the 3' terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–8. [PMC free article: PMC402085] [PubMed: 2583120]
- 95.
- Lodmell JS, Tapprich WE, Hill WE. Evidence for a conformational change in the exit site of the Escherichia coli ribosome upon tRNA binding. Biochemistry. 1993;32:4067–72. [PubMed: 8385994]
- 96.
- Mueller F, Doring T, Erdemir T. et al. Getting closer to an understanding of the three-dimensional structure of ribosomal RNA. Biochem Cell Biol. 1995;73:767–73. [PubMed: 8721993]
- 97.
- Wower J, Wower IK, Kirillov SV. et al. Peptidyl transferase and beyond. Biochem Cell Biol. 1995;73:1041–7. [PubMed: 8722019]
- 98.
- Nagano K, Nagano N. Transfer RNA docking pair model in the ribosomal pre- and post- translocational states. Nucleic Acids Res. 1997;25:1254–64. [PMC free article: PMC146551] [PubMed: 9092637]
- 99.
- Doring T, Mitchell P, Osswald M. et al. The decoding region of 16S RNA; a cross-linking study of the ribosomal A, P and E sites using tRNA derivatized at position 32 in the anticodon loop. EMBO J. 1994;13:2677–85. [PMC free article: PMC395142] [PubMed: 7516877]
- 100.
- Wower J, Scheffer P, Sylvers LA. et al. Topography of the E site on the Escherichia coli ribosome. EMBO J. 1993;12:617–23. [PMC free article: PMC413245] [PubMed: 8440251]
- 101.
- Miskin R, Zamir A. Effect of streptomycin on ribosome interconversion, a possible basis for the action of the antibiotic. Nat New Biol. 1972;238:78–80. [PubMed: 4505414]
- 102.
- Noreau J, Grise-Miron L, Brakier-Gingras L. Comparison of the action of streptomycin and neomycin on the structure of the bacterial ribosome. Biochim Biophys Acta. 1980;608:72–81. [PubMed: 6248117]
- 103.
- Jerinic O, Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J Mol Biol. 2000;304:707–13. [PubMed: 11124020]
- 104.
- Brodersen DE, Clemons WM Jr, Carter AP. et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–54. [PubMed: 11163189]
- 105.
- Mankin AS. Pactamycin resistance mutations in functional sites of 16 S rRNA. J Mol Biol. 1997;274:8–15. [PubMed: 9398510]
- 106.
- Noah JW, Dolan MA, Babin P. et al. Effects of tetracycline and spectinomycin on the tertiary structure of ribosomal RNA in the Escherichia coli 30 S ribosomal subunit. J Biol Chem. 1999;274:16576–81. [PubMed: 10347223]
- 107.
- Atmadja J, Brimacombe R, Maden BE. Xenopus laevis 18S ribosomal RNA: experimental determination of secondary structural elements, and locations of methyl groups in the secondary structure model. Nucleic Acids Res. 1984;12:2649–67. [PMC free article: PMC318697] [PubMed: 6424099]
- 108.
- Glotz C, Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980;8:2377–95. [PMC free article: PMC324088] [PubMed: 6160459]
- 109.
- Spitnik-Elson P, Elson D, Avital S. et al. Long range RNA-RNA interactions in the 30 S ribosomal subunit of E. coli. Nucleic Acids Res. 1985;13:4719–38. [PMC free article: PMC321822] [PubMed: 2410855]
- 110.
- Laughrea M, Tam J. In vivo chemical footprinting of the Escherichia coli ribosome. Biochemistry. 1992;31:12035–41. [PubMed: 1280995]
- 111.
- Polacek N, Patzke S, Nierhaus KH. et al. Periodic conformational changes in rRNA: monitoring the dynamics of translating ribosomes. Mol Cell. 2000;6:159–71. [PubMed: 10949037]
- 112.
- Gabashvili IS, Agrawal RK, Grassucci R. et al. Structure and structural variations of the Escherichia coli 30 S ribosomal subunit as revealed by three-dimensional cryo-electron microscopy. J Mol Biol. 1999;286:1285–91. [PubMed: 10064696]
- 113.
- Gabashvili IS, Gregory ST, Valle M. et al. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol Cell. 2001;8:181–8. [PubMed: 11511371]
- 114.
- Dammel CS, Noller HF. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–70. [PubMed: 7681419]
- 115.
- Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J Mol Biol. 1993;232:362–74. [PubMed: 8345517]
- 116.
- Brimacombe R. The three dimensional organization of Escherichia coli ribosomal RNAIn: Van Knippenberg PH, Hilbers CW, eds.Structure and Dynamics of RNANew York: Plenum Press,1986239–252.
- The “Active-Inactive” Interconversion of 30S and 50S Subunits
- Structural Changes Associated with mRNA Binding and Initiation Events
- Conformational Changes During Ribosome-tRNA Interactions
- Ribosome : aa-tRNA Interactions and Translational Fidelity
- Conformational Changes During Translocation
- Other tRNA-rRNA Interactions
- Conformational Changes Related to Antibiotic Binding
- Evidence for Other Conformational Switches
- Ribosome Assembly
- Concluding Remarks
- Acknowledgments
- References
- Conformational Dynamics within the Ribosome - Madame Curie Bioscience DatabaseConformational Dynamics within the Ribosome - Madame Curie Bioscience Database
- Neuroprotective Strategies in Animal and in Vitro Models of Neuronal Damage: Isc...Neuroprotective Strategies in Animal and in Vitro Models of Neuronal Damage: Ischemia and Stroke - Madame Curie Bioscience Database
- Regulation of Voltage-Sensitive Ca2+ Channels in Bipolar Cells by Divalent Catio...Regulation of Voltage-Sensitive Ca2+ Channels in Bipolar Cells by Divalent Cations and Polyamines - Madame Curie Bioscience Database
- DNA Primer Extension by Telomerase - Madame Curie Bioscience DatabaseDNA Primer Extension by Telomerase - Madame Curie Bioscience Database
- Caspase-Independent Cell Death Mechanisms - Madame Curie Bioscience DatabaseCaspase-Independent Cell Death Mechanisms - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...