NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
All nuclear-encoded eukaryotic messenger RNAs possess a 5' cap structure (m7GpppN) and, with a few exceptions, alsPreisso a 3' poly(A) tail. These modifications are added as part of the mRNA processing pathway during or immediately after transcription in the nucleus. Subsequently, they both influence different aspects of mRNA metabolism including splicing, transport, stability and translation. The cap structure has an important role during the initiation phase of translation as it recruits ribosomes and associated factors to the mRNA. The poly(A) tail can also stimulate translation and cooperates with the cap structure in a synergistic fashion. The eukaryotic initiation factor eIF4G plays a central part as a multifunctional adapter, which brings together various components of the translation apparatus. Through simultaneous interactions with the cap-binding protein eIF4E and the poly(A)-binding protein (PABP), eIF4G is able to bridge the two ends of the mRNA. The resulting pseudo-circular structure of the mRNA is thought to have important functional consequences for the translation process. The importance of the poly(A) tail is further underscored by the fact that the regulated variation of its length on maternal mRNAs is an integral part of gene regulation during oocyte maturation and in early embryonic development. Finally, the majority of cellular mRNAs are degraded by processes that are interconnected with translation and are initiated by poly(A) tail shortening.
Introduction
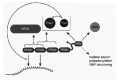
A common view holds that most control mechanisms to regulate eukaryotic gene expression target the primary step, namely transcription in the nucleus. In contrast to this, it is becoming increasingly apparent that controls acting on the level of translation and mRNA stability are also of critical importance. The translation process is usually divided into three phases: (i) initiation,1 (ii) elongation,2 and (iii) termination.3 The initiation phase represents all processes required for the assembly of a complete (80S) ribosome, consisting of a small (40S) and a large (60S) subunit, at the start codon of the mRNA. During the elongation phase, the actual polypeptide synthesis takes place. When the ribosome reaches the stop codon, this signals termination, including the dissociation of the completed polypeptide and the ribosome from the mRNA. Initiation usually is the rate-limiting step of translation and thus the preferred target of regulatory intervention.4
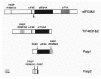
The salient model of cap-dependent translation was originally developed in the mid seventies5 and has since been continuously refined (for a review, see refs. 1, 6, 7). Initially, the 40S ribosomal subunit is recruited near the 5' end of the mRNA. Next, a lateral movement termed “scanning” along the 5' untranslated region (UTR) leads to the recognition of the usually first AUG triplet as the initiator codon. Then follows the formation of an 80S ribosome and the elongation phase begins (Fig. 1). A large number of eukaryotic initiation factors (eIF) contribute to these processes (for an up-to-date review, see ref. 7 and chapter 18 by F. Poulin and N. Sonenberg). The hetero-trimeric factor eIF4F binds in the cytoplasm to the cap structure at the 5' end of the mRNA. eIF4F consists of the cap-binding protein eIF4E,8 and the interacting proteins eIF4G and eIF4A (for a review, see ref. 9). eIF4A is an ATP-dependent RNA helicase and is able, upon stimulation by eIF4B, to unwind secondary structure in the cap-proximal region of the mRNA. In addition, eIF4G binds to the hetero-oligomeric factor eIF3, which associates with the 40S ribosomal subunit.10 The 40S subunit binds, furthermore, the ternary complex comprising initiator-methionyl-tRNA (tRNAMet), eIF2 and GTP, which has an important role in start codon recognition (for a review, see ref. 11). Once the 40S subunit and associated factors have arrived at the AUG then 60S subunit joining occurs, aided by eIF5 and eIF5B. eIF5 stimulates GTP hydrolysis of eIF2 at the start codon prior to ejection of all eIFs and joining of the 60S subunit. eIF5B is itself a ribosome-dependent GTPase.12
The Mechanistic Role of the Poly(A) Tail During Initiation of Translation
As we now know, the cap structure and the poly(A) tail are of similar importance for the initiation of translation on a typical mRNA (for a review, see refs. 13, 14). Despite the fact that both end modifications had been described at about the same time, it took much longer to develop good models for the function of the poly(A) tail.14 By 1990, it was known that adding a poly(A) tail to a test mRNA resulted in a modest 2 to 3-fold stimulatory effect on its translation in mammalian cell extracts (for a review, see ref. 15). In the commonly used rabbit reticulocyte lysate system, this was shown to be due to an enhanced 60S joining.16 Furthermore, a number of studies had documented a tight correlation between cytoplasmic polyadenylation of maternal mRNAs and their translational activation in vertebrate oocytes and developing embryos (for a review, see ref. 17). Collectively, these observations indicated that the poly(A) tail was somehow involved in the translation process. A groundbreaking study then showed in 1991 that exogenous mRNA can be introduced in eukaryotic cells by electroporation and its translation can be monitored in vivo.18 When the mRNA was equipped with a cap structure or a poly(A) tail, this led to a stimulation of translation. Interestingly, the addition of both end modifications led to a much more than additive effect in cells of mammalian, plant and yeast origin. This translational synergy between cap structure and poly(A) tail became one of the focal points of translation research in the following years (for a review, see refs. 9, 13–15). The study of the underlying molecular mechanisms was initially hindered by the fact that all previously known in vitro systems were unable to reproduce this phenomenon. This changed with the report of a “synergy-competent” cell-free translation system based on extracts from Saccharomyces cerevisae.19
Using this system, it was demonstrated that the poly(A) tail, like the cap structure, was able to support the recruitment of the 40S ribosomal subunit (for a review, see ref. 14). This function of the poly(A) tail as well as the functional synergy with the cap structure depends on the poly(A)-binding protein PABP (in yeast: Pab1p).20 PABP can bind to poly(A) tails with a periodicity of about 25 adenosine residues, although 12 adenosines are sufficient for binding.2123 The discovery of an interaction between Pab1p and the N-terminal part of eIF4G in yeast (Fig. 2) then presented a suitable molecular interaction between the 3' end of the mRNA and the process of translation initiation at the 5' end.24,25 These observations led to the hypothesis of a pseudo-circular mRNA conformation through simultaneous binding of eIF4E and Pab1p to eIF4G (Fig. 1). This was consistent with earlier observations of circular polyribosomes26 and was further substantiated through functional data and direct visualization. A set of experiments using the yeast extract system showed that poly(A)-mediated translation by itself displays no preference for the 5' end of the mRNA. Polypeptide synthesis in the isolated poly(A)-dependent mode frequently starts at internal sites of the mRNA. The cap structure exhibits the additional function to tether the ribosome recruitment potential of the poly(A) tail to the 5' end.27,28 Adding the minimal components eIF4E, eIF4G, PABP, and a capped and polyadenylated mRNA together in vitro, resulted in the formation of pseudo-circular complexes which could be visualized by atomic force microscopy.29
After its discovery in yeast, it did not take long for evidence for a direct interaction between PABP and eIF4G to appear also in plant30 and mammalian cells. Binding of mammalian PABP to eIF4G was demonstrated in two independent studies.31,32 Using the two-hybrid system, it was shown that eIF4G binds to the non-structural rotavirus protein NSP3.31 NSP3 and PABP compete for the same binding site in a previously overlooked N-terminal region of human eIF4G (Fig. 2). Since NSP3 also interacts at the same time with the (non-adenylated) 3' end of rotavirus mRNA it can provide the virus with a translational advantage in two ways. On the one hand, it selectively blocks PABP-dependent cellular translation and on the other hand, it bridges the two ends of rotavirus mRNAs.33 In parallel, analyses of cDNAs for both mammalian isoforms of eIF4G also revealed this new N-terminal region and facilitated a direct demonstration of PABP-binding to this region by co-immunoprecipitation.32 Interestingly, despite the evolutionary conservation of the eIF4G-PABP interaction, there is no apparent sequence homology between the PABP-binding regions of yeast and mammalian eIF4G. Indeed, human PABP cannot bind to yeast eIF4G.34 Additional evidence for such evolutionary divergence is the existence of two human PABP-interacting proteins, Paip1 and Paip2, with documented roles in translation (Figs. 2 and 3) whereas the yeast genome does not contain any homologous genes. Paip1 was discovered in a screen for PABP-binding proteins and is a protein with similarity to the central third of eIF4G. Paip1 interacts with eIF4A and can co-activate cap-dependent translation- despite having no eIF4E binding motif.35 Paip2 is a small acidic protein that acts as a translational repressor, with a preferential effect on translation of polyadenylated mRNAs. It reduces the affinity of PABP for oligo(A) and disrupts the periodicity with which multiple PABP molecules bind to poly(A). Furthermore, Paip1 and Paip2 interact with the same regions on PABP and compete with each other for binding (Fig. 3).36 Recent years have seen the development of “synergy-competent” cell-free translation systems from higher eukaryotes.3739 With these systems in hand, we now have the potential for a biochemical characterization of the translational role of the poly(A) tail in more complex organisms.
The wealth of data on the importance of the eIF4G-PAPB interaction during translation effectively focused much attention on an involvement of the poly(A) tail in 40S subunit recruitment (Fig. 1). There are, however, several indications from experiments in yeast, which suggest that this may not be the whole story. First, early genetic studies had demonstrated that suppressor mutations of a deletion of the essential PAB1 gene in yeast also altered the level of 60S ribosomal subunits,40,41 consistent with an apparent involvement of the poly(A) tail in the 60S subunit joining step (see above).16 Second, there are indications for a certain redundancy of the Pab1p-eIF4G interaction. Several mutations in eIF4G or Pab1p that decrease or abolish poly(A)-dependent translation do not have the same deleterious effect on translational synergy between cap structure and poly(A) tail in vitro and do not result in cell inviability.25 A mutation in Pab1p was isolated that inhibits poly(A)-dependent translation but does not abolish eIF4G-binding.42 Thus, it is quite possible that Pab1p makes additional contacts to the yeast translation machinery. Third, a number of reports suggest that the poly(A)/Pab1p complex stimulates translation by counteracting an inhibitory complex comprising several yeast super killer (Ski) proteins.4347 The main evidence for this is that electroporated, capped, but not adenylated, reporter mRNAs are highly expressed in various ski mutant cells. The SKI genes were identified from mutations allowing the increased production of killer toxin encoded by the M virus, a satellite of the double-stranded RNA L-A virus (for a review, see ref. 48). The L-A virus lacks both 5' cap structure and 3' poly(A) and thus it is not surprising that several SKI genes have roles in mRNA degradation pathways (see below).49,50 Nevertheless, analysis of mRNA stability substantiated the claim that the Ski proteins modulate the poly(A) tail's effect on translation. Finally, a recent study may link the effects of the SKI genes with the 60S subunit joining step of translation initiation.51 Using the same mRNA electroporation assay, it was shown that either a deletion of the FUN12 gene or a defect in TIF5 (encoding the yeast homologs of eIF5B and eIF5, respectively) specifically reduced translation of polyadenylated mRNA. Furthermore, deletion of FUN12 in the context of a deletion of SKI2 and the related gene SLH1 reinstated the repression of non-adenylated mRNA. These results suggest a model, which posits that the poly(A)/Pab1p complex inhibits the function of Ski2p and Slh1p, which in turn inhibit Fun12p and Tif5p, which are required for 60S subunit joining. Another recent study, however, describes an additional function of yeast eIF5 in an earlier step of the initiation pathway, namely in binding of mRNA to 40S subunits through a bridging interaction between eIF3 and the C-terminal part of eIF4G.52 The mechanistic involvement of eIF5 and eIF5B in poly(A)-tail-mediated translation can now be tested in cell-free translation experiments and a future challenge will be to make a more complete model of the role of the poly(A) tail in translation from the above hypotheses.
Structural Information on the Building Blocks of the Bridge between Cap Structure and Poly(A) Tail
The multivalent adapter molecule eIF4G is the centerpiece of a bridge that forms between the ends of the mRNA during translation (for a review, see ref. 53). The binding regions for several interaction partners of eIF4G have been mapped by deletion and mutation analysis (Fig. 2). Apart from the already mentioned factors with a direct role in the translation initiation process, these also include a number of proteins with a modulating influence on translation (i.e. the eIF4E-kinase Mnk-154,55), or the potential to link it to other aspects of mRNA metabolism (i.e. the nuclear cap-binding complex CBC;56,57 or the decapping enzyme Dcp1p58). Collectively, this forms the picture of a modular structure of eIF4G (Fig. 2). The N-terminal third with binding sites for PABP and eIF4E serves to latch on to the mRNA. The central third with binding sites for eIF4A, eIF3, and RNA primarily functions in ribosome recruitment while the C-terminal third with binding sites for eIF4A and Mnk-1 has a regulatory role.59,60 The structure of the phylogenetically conserved middle third of eIF4G61,62 was recently solved. A region of 259 amino acids folds into 10 α-helices, which are arranged into 5 HEAT motifs.63 Proteins with HEAT repeats are commonly involved in the formation of multi-protein complexes.64
The amino acid sequences of all known PABPs exhibit the same domain structure.65 The N-terminal part of the protein consists of 4 RNA-Recognition-Motifs (RRM), joined together by highly conserved linker sequences. The C-terminus forms a further conserved domain for protein-protein contacts (Fig. 3). Systematic biochemical and genetic studies have shown that a fragment consisting of RRM-1 and -2 is responsible for binding to poly(A)66,67 and also interacts with eIF4G.42,65,68 X-ray crystallography of a complex between the PABP-RRM-1 and -2 fragment and poly(A) revealed the molecular details of the interaction between single-stranded poly(A) and the RNA-binding motifs of both RRMs. The surface of the PABP-fragment facing away from the RNA forms a phylogenetically conserved region, which was postulated to contact eIF4G.65 This hypothesis is supported by the results of targeted mutational studies of conserved amino acids in this region.34 Structural analyses of the eIF4E-complex with m7GDP and a short eIF4G-fragment display a certain analogy. eIF4E has a concave side with a small hydrophobic slot for binding the cap structure and a contiguous region for mRNA binding. On the opposite, the convex face of the protein is the contact region with eIF4G.69,70
Very recently, structural data was obtained also for the C-terminal part of PABP.71 This region of the protein contains a conserved sequence of about 60–70 amino acids in length (PABC), the structure of which has been solved by NMR spectroscopy and X-ray crystallography. The NMR studies showed that a region of 74 amino acids folds into a globular domain.72 The X-ray structural analysis was carried out on an ortholog of this domain from the human hyperplastic disc protein and shows a very similar structure.73 The PABC domain consists of 5 α-helices that are arranged in relation to each other in the shape of an arrow. PABC binds specifically through a hydrophobic region to an approximately 12 amino acid peptide motif. This motif is found in a number of proteins that interact with the C-terminal part of PABP, including the aforementioned Paip1 and Paip2, and the eukaryotic release factor eRF3 (Fig. 3).71 In the case of Paip2, the PABC-interacting motif resides near the C-terminus (Fig. 2). In addition, Paip2 exhibits another PABP-binding site in its central region, which interacts specifically with a region encompassing RRM-2 and 3 of PABP (Fig. 3).74 The latter interaction displays the higher affinity, is sufficient to promote the characteristic disruption of the PABP-poly(A) complex, and to repress translation in a poly(A)-responsive in vitro system. As binding between the Paip2 C-terminus and PABC fails to show these effects, it is at present unclear what the physiological function of this interaction might be. It could aid in antagonizing Paip1-binding to PABP and/or regulate other processes than translation initiation. Once again, a parallel can be drawn to the case of the cap-binding protein eIF4E. Binding of translational activator proteins (eIF4G to eIF4E, Paip1 to PABP) is antagonized by translational repressors (the eIF4E-binding proteins, 4E-BPs,9 and Paip2, respectively). The 4E-BPs75 and the eIF4E-binding region of eIF4G76 exist just like the PABC-binding peptide72 in an unfolded state which folds into an ordered structure upon binding to the target protein.
Collectively, this leads to a working model for the function of the PABP/poly(A) complex as follows (Fig. 3):72 the two N-terminal RRMs of PABP are responsible for binding to poly(A) and for contacting the 5' end of the mRNA through binding of eIF4G. This leaves the PABC region free for further protein-protein contacts, which may also influence translation or other aspects of mRNA metabolism. Paip1 represents a further link to translation initiation while the interaction with eRF3 has the potential to bridge to the termination of translation. Additional proteins with regions of homology to PABC or the PABC-interacting peptide have already been identified and could hint at functional links to other cellular processes.72,73
Molecular Concepts to Explain Translational Synergy
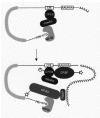
The available data displays a clear connection between the functional cooperativity of the cap structure and the poly(A) tail during translation and a pseudo-circular structure of the mRNA. Potential advantages of this circular structure are easily recognized. The error rate of translation could be reduced since only intact mRNAs act as efficient templates. A well-characterized mRNA decay pathway starts with a deadenylation step, followed by decapping and exonucleolytic degradation of the mRNA body (Fig. 6). The association of both mRNA ends with the translation machinery may therefore stabilize the mRNA (see below).77 In addition to this protective effect, the spatial proximity could also have a direct positive effect on translation. Ribosomes may not dissociate away from the mRNA after termination but instead initiate a new round of translation at the 5' end of the same mRNA molecule. So far, however, there is no direct evidence for this attractive concept of ribosome recycling. This model further suggests a mechanistic distinction between the first and subsequent rounds of translation (Fig. 4). The idea of a specialized first round of translation is also appealing as it would provide opportunities for an mRNP remodeling process on newly exported mRNA, from a nuclear to a cytoplasmic form, and for monitoring by mRNA surveillance systems (see chapter 13 by L Maquat).56,78
Experiments in the yeast system have shown that translational synergy at least partly originates from a competition for limiting components of the translation machinery.28,79 Translation reactions using nuclease-pretreated yeast extracts display robust translation rates when only one of the two end modifications are appended to the test mRNA, but little or no synergy. In untreated extracts, the endogenous cellular mRNAs are still preserved: the competitive conditions here massively favor test mRNAs that exhibit both end modifications.27 These competition effects could arise at the level of the first or the subsequent rounds of translation. Various demonstrations of cooperative binding along the sequence cap-eIF4E-eIF4G-PABP-poly(A)30,8083 suggest that the observed synergy effects occur at least in part on the level of initial recruitment of the mRNA. Regarding the molecular causes for translational synergy, there are still many unanswered questions. The determination of the structure of a complex comprising eIF4E, eIF4G and PABP could make a significant contribution to answer these questions. Another goal is the development of strategies in cell-free systems to distinguish experimentally between ribosome recruitment and recycling.
Mechanisms of Translational Control Involving the Poly(A) Tail
Viruses employ a variety of unconventional strategies to usurp the cellular translation machinery (for a review, see ref. 84). eIF4G and its interaction partners are preferred targets for viruses. During infection by certain picorna viruses, the N-terminal third of eIF4G that links the protein to cap structure and poly(A) tail of cellular mRNAs is cleaved from the remainder of the protein by proteolytic attack (Fig. 2).10,85,86 Similarly, the PABC domain is removed from PABP.87,88 These manipulations lead to an inhibition of translation of cellular mRNA in favor of selective translation of the viral mRNA. Another example is the case of the above-mentioned rotaviruses, which employ a translational strategy that directly targets the PABP-eIF4G interaction.31,33
The most recognized area of translational control involving the poly(A) tail, however, is the translational regulation of maternal mRNAs during oocyte maturation and early embryogenesis (for a review, see refs. 13, 89–92). A precise temporal and spatial control of gene expression is particularly important in these early developmental stages. Characteristically, however, there is little or no transcription during this period. Crucial processes during this phase depend therefore on the controlled expression of maternal mRNA molecules, which were stocked in advance in the oocyte cytoplasm. Research into these processes in various genetic model systems of developmental biology is met with an ever-increasing interest. Errors in the post-transcriptional regulation of maternal mRNA expression usually lead to drastic abnormalities in embryonic development.
Typically, these mRNAs are initially stored in the cytoplasm of the developing oocyte in a dormant form with short poly(A) tails (˜20–40 adenosines) until translation needs to be activated. This activation usually correlates with an elongation of their poly(A) tail. Cytoplasmic polyadenylation requires two elements in the 3' UTR of these mRNAs (Fig. 5): the “nuclear” polyadenylation motif AAUAAA and the nearby cytoplasmic polyadenylation element (CPE, consensus sequence: UUUUUAU).93,94 Investigations of these processes during oocyte maturation in the frog Xenopus laevis have yielded interesting insights into the underlying molecular mechanisms. The mos mRNA, for instance, exhibits a CPE and is activated early in the maturation process. A CPE-binding protein (CPEB) is necessary for the concurrent polyadenylation to approximately 150 adenosines. CPEB, a zinc-finger and RRM-containing protein, was first discovered in Xenopus95,96 but is probably present in all metazoans.9799 CPEB is also involved in establishing the preceding inactive state of the mRNAs. This masking activity of CPEB is mediated through the function of a CPEB-interacting protein called maskin.100 Maskin additionally binds to eIF4E in a manner similar to the aforementioned 4E-BPs.9 CPEB and maskin form a stable complex whereas the interaction of maskin with eIF4E is strongly reduced during oocyte maturation. It is plausible that this modulates the formation of functional eIF4F complexes on CPE-containing mRNAs and thus their translation (Fig. 5).90 Furthermore, CPEB is a phosphoprotein and a specific phosphorylation of CPEB at serine 174 by the kinase Eg2 is required for the activation of mos mRNA.101 This phosphorylation stimulates the interaction of CPEB with the AAUAAA-binding protein CPSF (for Cleavage and Polyadenylation Specificity Factor) and in this way can recruit the cytoplasmic polyadenylation machinery to the mRNA.102 How does elongation of the poly(A) tail stimulate translation? One possibility relies on the observation that a cap ribose methylation can occur in response to the ongoing process of poly(A) tail elongation. This modification of the 5' end of the mRNA can enhance translation initiation.103 However, not all CPE-containing mRNAs are modified in this way.104 Another possibility is based on the ability of elongated poly(A) tails to recruit multiple PABP molecules to the mRNA and thus enhance the recruitment of eIF4G/eIF4F to the cap structure, probably at the expense of the maskin-eIF4E interaction (Fig. 5). The common form of PABP is only of low abundance in oocytes105 but its role may be in part carried out by development-specific isoforms.106 Two studies provide evidence that the interaction of some form of PABP with eIF4G is important for translation in oocytes and for the maturation process. PABP was targeted to test mRNAs in Xenopus oocytes using a tethered-function approach and shown to stimulate their translation- apparently through an interaction with eIF4G.107 Expression of a mutant form of eIF4G, defective in PABP-binding, had a dominant-negative effect on translation of polyadenylated mRNA and drastically inhibited oocyte maturation.108
In mammalian oocytes, CPEB seems to serve a similar function as in Xenopus, namely to execute the switch between CPE-mediated mRNA repression to polyadenylation-induced activation during maturation.109112 The generation of CPEB knockout mice has allowed a number of very interesting observations. Both male and female CPEB null mice are viable and show no obvious growth abnormalities but severe fertility defects.113 Detailed analyses revealed that both male and female germ cell differentiation is disrupted at a stage of meiosis prior to meiotic maturation- as early as the pachytene stage of meiotic prophase I. This shows that CPEB function is required earlier in germ cell development than was predicted from the known CPEB target mRNAs. Further, CPEB -/- germ cells harbored fragmented chromatin suggesting a defect in homologous chromosome adhesion or synapsis. Indeed, the mRNAs for two proteins involved in synaptonemal complex formation, SCP1 and SCP3, contain functional CPE-motifs and their translational regulation is disrupted by the CPEB deletion.113 Apart from the reproductive organs, CPEB is also expressed in the hippocampus and is enriched at synapses of neurons in culture.114,115 This offers a very interesting extension of this research area, which has not yet been explored with the CPEB knockout mice. Controlled translation enables particularly fast and large changes in the synthesis of proteins, as they are required for synaptic plasticity. Indeed, it was found that the mRNA for calmodulin-dependent protein kinase II (a-CaMKII), an essential factor for activity-dependent synaptic plasticity, contains CPE-motifs in the 3' UTR. During synaptic stimulation, a-CaMKII mRNA is polyadenylated and translationally activated. It is therefore possible that CPEB-mediated local control of translation is involved in synaptic plasticity and memory.
Translation and mRNA Degradation
The cap structure and the poly(A) tail also play important roles in mRNA degradation. It is thus not surprising that there are numerous indications for an intimate connection between translation and mRNA turnover (for a review, see refs. 78, 116, 117). Four distinct pathways of mRNA decay are known in eukaryotes and have been primarily studied in Saccharomyces cerevisiae (see chapter 14 by P Mitchell). Two pathways start with deadenylation as the initial step and are thought to occur on many if not all mRNA species. In the predominant pathway (Fig. 6), this is followed by a removal of the cap structure, termed “decapping”, and 5' to 3' exonucleolytic degradation of the body of the mRNA. Alternatively, mRNAs can be degraded in 3' to 5' direction following deadenylation. Nonsense-mediated decay (NMD) is a more specific mechanism, which ensures that aberrant mRNA molecules are rapidly decapped and degraded 5' to 3' independently of deadenylation (see chapter 13 by L. Maquat).118122 Finally, some mRNAs are known to contain cleavage sites for specific endonucleases that can trigger their degradation.
There are good indications that the predominant mRNA turnover pathway (Fig. 6) is regulated by the status of the translation initiation machinery on the mRNA. Analyses of yeast strains containing mutations in the translation initiation factors eIF4E, eIF4G, eIF4A or a subunit of eIF3 show increased rates of deadenylation and subsequent decapping of both unstable MFA2 and stable PGK1 mRNAs.77 By contrast, inhibiting the elongation step of translation slows down deadenylation and decapping, perhaps indicating a window of opportunity for mRNA degradation in the translation cycle.117 An attractive model proposes that deadenylation rates reflect the degree of accessibility of the poly(A) tail for the deadenylase. Likewise, decapping rates may reflect a competition between eIF4E and the decapping enzyme Dcp1p for the cap structure. Several observations provide initial molecular explanations for these models. eIF4E has a much lower intrinsic affinity for cap than Dcp1p.123 During active translation initiation, this is probably compensated for by an enhancement of eIF4E-binding to the cap structure, through the interaction with eIF4G.80,124 Dcp1p also binds directly to eIF4G and Pab1p, either as individual proteins or as part of the eIF4F-Pab1p complex.58 Pab1p can modulate the interactions of eIF4G and eIF4E with the mRNA24 while eIF4G can stimulate Dcp1p activity. The latter effect is blocked by eIF4E.58 It has also been shown that Pab1p is required for the poly(A) tail to function as an inhibitor of decapping.125,126 Thus, the emerging picture is one of a dynamic complex that bridges between the mRNA ends and is involved in the switch from translation initiation to mRNA degradation. Loss of Pab1p-binding to the deadenylated 3' end of the mRNA could trigger a rearrangement at the 5' end that allows Dcp1 access to the cap structure.58
Components of the mRNA Degradation Machinery
The most complete set of factors involved in mRNA decay has been identified in yeast and many of them have recognized homologs in other eukaryotes (for a review, see refs. 117, 127). The DCP1 gene encodes the decapping enzyme.128,129 Dcp2p is another factor required for decapping, probably by regulating Dcp1p activity.130 The related proteins Edc1p and Edc2p also stimulate decapping.131 Xrn1p is the exoribonuclease responsible for 5' to 3' degradation of the body of the mRNA.132,133 3' to 5' degradation is carried out by the exosome complex and the accessory proteins Ski2p, 3, 8 (see above and chapter 14 by P. Mitchell).127,134,135 Recently, Ccr4p and Caf1p/Pop2p have been identified as components of the elusive major yeast cytoplasmic deadenylase.136,137 Both proteins have nuclease domains, Ccr4p being a member of a magnesium-dependent endonuclease-related family while Caf1p/Pop2p belongs to the RNase D family of 3' to 5' exonucleases. While Caf1p/Pop2p displays significant similarity to the human deadenylase PARN,138,139 it does not appear to represent a PARN homologue in yeast. Both Ccr4p and Caf1p/Pop2p are highly conserved in eukaryotic cells.
Additionally, a group of seven Sm-like proteins (Lsm1p-Lsm7p) and the associated factor Pat1p/ Mrt1p activate decapping in the major mRNA degradation pathway.140143 Sm and Sm-like proteins can form heptameric complexes in the shape of a ring.144 Consistent with their role in mRNA degradation, several Lsm proteins (Lsm1,2,3,5,6,7) co-purified biochemically with the Xrn1p exonuclease.142 Lsm1-7 co-immunoprecipitate with Dcp1, Pat1/Mrt1 and mRNA.143 Two-hybrid screens suggest interactions between various Lsm proteins (Lsm1p-Lsm8p), and components of the mRNA degradation pathway: Pat1p/Mrt1p, Dcp1, Dcp2p, and Xrn1p.145 While the above results may not all represent bona fide protein-protein interactions, together they make a strong case for a role of the Lsm1-7 complex in mRNA degradation.
Perspectives
Research into the roles of the poly(A) tail in mRNA metabolism has made great progress in the last ten years. A number of interactions between the poly(A) tail and the mRNA translation machinery have been identified and we have good working models for their function. The presence of further PABP-interacting proteins in higher cells are one indication for a more complex network of contacts with the 5' end than suggested by initial experiments in the yeast model system. The biochemical characterization of these processes in poly(A)-dependent cell-free translation systems from higher eukaryotes can make valuable contributions here. It is possible that this may also yield more substantial insights on other functions of mRNA circularization in mRNA metabolism. Studies on the mechanism of translational activation in early developmental stages also promise to further our understanding of mRNA-specific translation control mediated by elements in the 3' UTR and how they functionally interact with the poly(A) tail. Finally, there is a strong case for multiple interrelations between mRNA translation and degradation mechanisms. Much progress in both areas will depend critically on an appreciation of just how the two processes affect each other.
Acknowledgements
The author wishes to thank M. W. Hentze for the longstanding and productive cooperation, and K. Brennan for suggestions on the manuscript. This work was funded by grants from the Deutsche Forschungsgemeinschaft (PR616/1-1&2).
References
- 1.
- Hershey JWB, Merrick WC. Pathway and Mechanism of Initiation of Protein synthesisIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene Expression Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press,200033–88.
- 2.
- Merrick WC, J N. The protein biosynthesis elongation cycleIn: Sonenberg N, Mathews MB, Hershey JWB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,200089–126.
- 3.
- Welch EM, Wang W, Peltz SW. Translation termination: it's not the end of the storyIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press.,2000467–486.
- 4.
- Mathews MB, Sonenberg N, Hershey JBW. Origins and principles of translational controlIn: Sonenberg N, Hershey JBW, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,20001–31.
- 5.
- Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–53. [PubMed: 1017010]
- 6.
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. [PubMed: 10395892]
- 7.
- Pestova TV, Kolupaeva VG, Lomakin IB. et al. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029–36. [PMC free article: PMC34618] [PubMed: 11416183]
- 8.
- Sonenberg N, Morgan MA, Merrick WC. et al. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5'-terminal cap in mRNA. Proc Natl Acad Sci USA. 1978;75:4843–7. [PMC free article: PMC336217] [PubMed: 217002]
- 9.
- Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. [PubMed: 10872469]
- 10.
- Lamphear BJ, Kirchweger R, Skern T. et al. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. [PubMed: 7665619]
- 11.
- Hinnebusch AG. Mechanism and regulation of Initiator methionyl-tRNA Binding to RibosomesIn: Sonenberg N, Hershey JW, Mathews B, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000185–245.
- 12.
- Pestova TV, Lomakin IB, Lee JH. et al. The ribosomal subunit joining reaction in eukaryotes requires eIF5B. Nature. 2000;403:332–335. [PubMed: 10659855]
- 13.
- Preiss T, Hentze MW. From factors to mechanisms: translation and translational control in eukaryotes. Curr Opin Genet Dev. 1999;9:515–21. [PubMed: 10508691]
- 14.
- Sachs AB. Physical and functional interactions between the mRNA cap structure and the poly(A) tailIn: Sonenberg N, Hershey JBW, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000447–465.
- 15.
- Jacobson A. Poly(A) metabolism and translation: the closed-loop modelIn: Hershey JWB, Mathews MB, Sonenberg N, eds.Translational controlCold Spring Harbor: Cold Spring Harbor Laboratory Press,1996451–480.
- 16.
- Munroe D, Jacobson A. mRNA poly(A) tail, a 3' enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. [PMC free article: PMC360780] [PubMed: 1972543]
- 17.
- Wickens M. In the beginning is the end: regulation of poly(A) addition and removal during early development. Trends Biochem Sci. 1990;15:320–4. [PubMed: 2204159]
- 18.
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–16. [PubMed: 1682219]
- 19.
- Iizuka N, Najita L, Franzusoff A. et al. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. [PMC free article: PMC359267] [PubMed: 7935446]
- 20.
- Tarun SZ Jr, Sachs AB. A common function for mRNA 5' and 3' ends in translation initiation in yeast. Genes and Dev. 1995;9:2997–3007. [PubMed: 7498795]
- 21.
- Baer BW, Kornberg RD. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci USA. 1980;77:1890–2. [PMC free article: PMC348614] [PubMed: 6929525]
- 22.
- Baer BW, Kornberg RD. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96:717–21. [PMC free article: PMC2112416] [PubMed: 6833379]
- 23.
- Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. [PMC free article: PMC367964] [PubMed: 3313012]
- 24.
- Tarun SZ Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article: PMC452544] [PubMed: 9003792]
- 25.
- Tarun SZ Jr, Wells SE, Deardorff JA. et al. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Nat Acad Sci USA. 1997;94:9046–9051. [PMC free article: PMC23022] [PubMed: 9256432]
- 26.
- Christensen AK, Kahn LE, Bourne CM. Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Am J Anat. 1987;178:1–10. [PubMed: 3825959]
- 27.
- Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. [PubMed: 9548259]
- 28.
- Preiss T, Muckenthaler M, Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. [PMC free article: PMC1369706] [PubMed: 9814754]
- 29.
- Wells SE, Hillner PE, Vale RD. et al. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–40. [PubMed: 9702200]
- 30.
- Le H, Tanguay RL, Balasta ML. et al. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–55. [PubMed: 9195926]
- 31.
- Piron M, Vende P, Cohen J. et al. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–21. [PMC free article: PMC1170909] [PubMed: 9755181]
- 32.
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–9. [PMC free article: PMC1171091] [PubMed: 9857202]
- 33.
- Vende P, Piron M, Castagne N. et al. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3' end. J Virol. 2000;74:7064–71. [PMC free article: PMC112224] [PubMed: 10888646]
- 34.
- Otero LJ, Ashe MP, Sachs AB. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 1999;18:3153–63. [PMC free article: PMC1171396] [PubMed: 10357826]
- 35.
- Craig AW, Haghighat A, Yu AT. et al. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–3. [PubMed: 9548260]
- 36.
- Khaleghpour K, Svitkin YV, Craig AW. et al. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell. 2001;7:205–16. [PubMed: 11172725]
- 37.
- Gebauer F, Corona DF, Preiss T. et al. Translational control of dosage compensation in Drosophila by Sex- lethal: cooperative silencing via the 5' and 3' UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 1999;18:6146–54. [PMC free article: PMC1171678] [PubMed: 10545124]
- 38.
- Michel YM, Poncet D, Piron M. et al. Cap-poly(A) synergy in mammalian cell-free extracts: Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem. 2000;275:32268–32276. [PubMed: 10922367]
- 39.
- Bergamini G, Preiss T, Hentze MW. Picornavirus IRESes and the poly(A) tail jointly promote capindependent translation in a mammalian cell-free system. RNA. 2000;6:1781–90. [PMC free article: PMC1370048] [PubMed: 11142378]
- 40.
- Sachs AB, Davis RW. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–9. [PubMed: 2408148]
- 41.
- Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–67. [PubMed: 2673535]
- 42.
- Kessler SH, Sachs AB. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:51–57. [PMC free article: PMC121449] [PubMed: 9418852]
- 43.
- Widner WR, Wickner RB. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–41. [PMC free article: PMC359991] [PubMed: 8321235]
- 44.
- Masison DC, Blanc A, Ribas JC. et al. Decoying the cap- mRNA degradation system by a double-stranded RNA virus and poly(A)- mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–71. [PMC free article: PMC230507] [PubMed: 7739557]
- 45.
- Benard L, Carroll K, Valle RC. et al. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–96. [PMC free article: PMC110648] [PubMed: 9566888]
- 46.
- Benard L, Carroll K, Valle RC. et al. The ski7 antiviral protein is an EF1-alpha homolog that blocks expression of non-Poly(A) mRNA in Saccharomyces cerevisiae. J Virol. 1999;73:2893–900. [PMC free article: PMC104047] [PubMed: 10074137]
- 47.
- Searfoss AM, Wickner RB. 3' poly(A) is dispensable for translation. Proc Natl Acad Sci USA. 2000;97:9133–7. [PMC free article: PMC16834] [PubMed: 10922069]
- 48.
- Wickner RB. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–65. [PMC free article: PMC239427] [PubMed: 8852903]
- 49.
- Johnson AW, Kolodner RD. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–27. [PMC free article: PMC230502] [PubMed: 7739552]
- 50.
- Jacobs JS, Anderson AR, Parker RP. The 3' to 5' degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3' to 5' exonucleases of the exosome complex. EMBO J. 1998;17:1497–506. [PMC free article: PMC1170497] [PubMed: 9482746]
- 51.
- Searfoss A, Dever TE, Wickner R. Linking the 3' poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol Cell Biol. 2001;21:4900–8. [PMC free article: PMC87206] [PubMed: 11438647]
- 52.
- Asano K, Shalev A, Phan L. et al. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–37. [PMC free article: PMC125443] [PubMed: 11331597]
- 53.
- Hentze MW. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. [PubMed: 9019810]
- 54.
- Pyronnet S, Imataka H, Gingras AC. et al. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–9. [PMC free article: PMC1171121] [PubMed: 9878069]
- 55.
- Waskiewicz AJ, Johnson JC, Penn B. et al. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–80. [PMC free article: PMC83980] [PubMed: 10022874]
- 56.
- Fortes P, Inada T, Preiss T. et al. The yeast nuclear cap-binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol Cell. 2000;6:191–6. [PubMed: 10949040]
- 57.
- McKendrick L, Thompson E, Ferreira J. et al. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21:3632–41. [PMC free article: PMC86986] [PubMed: 11340157]
- 58.
- Vilela C, Velasco C, Ptushkina M. et al. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. Embo J. 2000;19:4372–82. [PMC free article: PMC302023] [PubMed: 10944120]
- 59.
- Morino S, Imataka H, Svitkin YV. et al. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–77. [PMC free article: PMC85104] [PubMed: 10611225]
- 60.
- De Gregorio E, Preiss T, Hentze MW. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–74. [PMC free article: PMC1171558] [PubMed: 10469664]
- 61.
- Aravind L, Koonin EV. Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res. 2000;10:1172–84. [PMC free article: PMC310937] [PubMed: 10958635]
- 62.
- Ponting CP. Novel eIF4G domain homologues linking mRNA translation with nonsense- mediated mRNA decay. Trends Biochem Sci. 2000;25:423–6. [PubMed: 10973054]
- 63.
- Marcotrigiano J, Lomakin IB, Sonenberg N. et al. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7:193–203. [PubMed: 11172724]
- 64.
- Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–6. [PubMed: 7550332]
- 65.
- Deo RC, Bonanno JB, Sonenberg N. et al. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–45. [PubMed: 10499800]
- 66.
- Burd CG, Matunis EL, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–24. [PMC free article: PMC361068] [PubMed: 1675426]
- 67.
- Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. [PubMed: 8609610]
- 68.
- Sachs AB, Varani G. Eukaryotic translation initiation: there are (at least) two sides to every story. Nat Struct Biol. 2000;7:356–61. [PubMed: 10802729]
- 69.
- Marcotrigiano J, Gingras AC, Sonenberg N. et al. Cocrystal structure of the messenger RNA 5' cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–61. [PubMed: 9200613]
- 70.
- Matsuo H, Li H, McGuire AM. et al. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4:717–24. [PubMed: 9302999]
- 71.
- Varani G. Delivering messages from the 3' end. Proc Natl Acad Sci USA. 2001;98:4288–9. [PMC free article: PMC33322] [PubMed: 11296278]
- 72.
- Kozlov G, Trempe JF, Khaleghpour K. et al. Structure and function of the C-terminal PABC domain of human poly(A)- binding protein. Proc Natl Acad Sci USA. 2001;98:4409–13. [PMC free article: PMC31848] [PubMed: 11287632]
- 73.
- Deo RC, Sonenberg N, Burley SK. X-ray structure of the human hyperplastic discs protein: an ortholog of the C-terminal domain of poly(A)-binding protein. Proc Natl Acad Sci USA. 2001;98:4414–9. [PMC free article: PMC31849] [PubMed: 11287654]
- 74.
- Khaleghpour K, Kahvejian A, De Crescenzo G. et al. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol. 2001;21:5200–13. [PMC free article: PMC87244] [PubMed: 11438674]
- 75.
- Marcotrigiano J, Gingras AC, Sonenberg N. et al. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. [PubMed: 10394359]
- 76.
- Hershey PE, McWhirter SM, Gross JD. et al. The Cap-binding protein eIF4E promotes folding of a functional domain of yeast translation initiation factor eIF4G1. J Biol Chem. 1999;274:21297–304. [PubMed: 10409688]
- 77.
- Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–56. [PMC free article: PMC84368] [PubMed: 10409716]
- 78.
- Mitchell P, Tollervey D. mRNA turnover. Curr Opin Cell Biol. 2001;13:320–5. [PubMed: 11343902]
- 79.
- Tarun SZ Jr, Sachs AB. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol. 1997;17:6876–86. [PMC free article: PMC232544] [PubMed: 9372919]
- 80.
- Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5'-cap structure [published erratum appears in J Biol Chem 1997 Nov 14;272(46):29398] J Biol Chem. 1997;272:21677–80. [PubMed: 9268293]
- 81.
- Ptushkina M, von der Haar T, Vasilescu S. et al. Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5' cap in yeast involves a site partially shared by p20. EMBO J. 1998;17:4798–808. [PMC free article: PMC1170809] [PubMed: 9707439]
- 82.
- Ptushkina M, von der Haar T, Karim MM. et al. Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J. 1999;18:4068–4075. [PMC free article: PMC1171482] [PubMed: 10406811]
- 83.
- Wei CC, Balasta ML, Ren J. et al. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry. 1998;37:1910–6. [PubMed: 9485317]
- 84.
- Pe'ery T, Mathews MB. Viral translational strategies and host defense mechanismsIn: Sonenberg N, Hershey JBW, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000371–424.
- 85.
- Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–7. [PMC free article: PMC232551] [PubMed: 9372926]
- 86.
- Gradi A, Svitkin YV, Imataka H. et al. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–94. [PMC free article: PMC21600] [PubMed: 9736694]
- 87.
- Kerekatte V, Keiper BD, Badorff C. et al. Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol. 1999;73:709–17. [PMC free article: PMC103878] [PubMed: 9847377]
- 88.
- Joachims M, Van Breugel PC, Lloyd RE. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol. 1999;73:718–27. [PMC free article: PMC103879] [PubMed: 9847378]
- 89.
- Wickens M, Goodwin EB, Kimble J. et al. Translational control of developmental decisionsIn: Sonenberg N, Hershey JBW, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000295–370.
- 90.
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–9. [PubMed: 11433366]
- 91.
- Macdonald P. Diversity in translational regulation. Curr Opin Cell Biol. 2001;13:326–31. [PubMed: 11343903]
- 92.
- Richter J. Influence of Polyadenylation-induced Translation on Metazoan Development and Neuronal Synaptic FunctionIn: Sonenberg N, Hershey JBW, Mathews B, eds.Translational Control of gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000785–805.
- 93.
- McGrew LL, Dworkin-Rastl E, Dworkin MB. et al. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803–15. [PubMed: 2568313]
- 94.
- Fox CA, Sheets MD, Wickens MP. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–62. [PubMed: 2628165]
- 95.
- Paris J, Swenson K, Piwnica-Worms H. et al. Maturation-specific polyadenylation: in vitro activation by p34cdc2 and phosphorylation of a 58-kD CPE-binding protein. Genes Dev. 1991;5:1697–708. [PubMed: 1653174]
- 96.
- Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–27. [PubMed: 7954828]
- 97.
- Lantz V, Ambrosio L, Schedl P. The Drosophila orb gene is predicted to encode sex-specific germline RNA-binding proteins and has localized transcripts in ovaries and early embryos. Development. 1992;115:75–88. [PubMed: 1638994]
- 98.
- Gebauer F, Richter JD. Mouse cytoplasmic polyadenylylation element binding protein: an evolutionarily conserved protein that interacts with the cytoplasmic polyadenylylation elements of c-mos mRNA. Proc Natl Acad Sci USA. 1996;93:14602–7. [PMC free article: PMC26180] [PubMed: 8962099]
- 99.
- Luitjens C, Gallegos M, Kraemer B. et al. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000;14:2596–609. [PMC free article: PMC316992] [PubMed: 11040214]
- 100.
- Stebbins-Boaz B, Cao Q, de Moor CH. et al. Maskin is a CPEB-associated factor that transiently interacts with elF- 4E [published erratum appears in Mol Cell 2000 Apr;5(4):following 766] Mol Cell. 1999;4:1017–27. [PubMed: 10635326]
- 101.
- Mendez R, Hake LE, Andresson T. et al. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–7. [PubMed: 10749216]
- 102.
- Mendez R, Murthy KG, Ryan K. et al. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000;6:1253–9. [PubMed: 11106762]
- 103.
- Kuge H, Richter JD. Cytoplasmic 3' poly(A) addition induces 5' cap ribose methylation: implications for translational control of maternal mRNA. EMBO J. 1995;14:6301–10. [PMC free article: PMC394754] [PubMed: 8557049]
- 104.
- Gillian-Daniel DL, Gray NK, Astrom J. et al. Modifications of the 5' cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol Cell Biol. 1998;18:6152–63. [PMC free article: PMC109201] [PubMed: 9742132]
- 105.
- Zelus BD, Giebelhaus DH, Eib DW. et al. Expression of the poly(A)-binding protein during development of Xenopus laevis. Mol Cell Biol. 1989;9:2756–60. [PMC free article: PMC362352] [PubMed: 2761544]
- 106.
- Voeltz GK, Ongkasuwan J, Standart N. et al. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–88. [PMC free article: PMC312653] [PubMed: 11274061]
- 107.
- Gray NK, Coller JM, Dickson KS. et al. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–33. [PMC free article: PMC302064] [PubMed: 10970864]
- 108.
- Wakiyama M, Imataka H, Sonenberg N. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol. 2000;10:1147–50. [PubMed: 10996799]
- 109.
- Gebauer F, Xu W, Cooper GM. et al. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J. 1994;13:5712. [PMC free article: PMC395537] [PubMed: 7988567]
- 110.
- Stutz A, Conne B, Huarte J. et al. Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes Dev. 1998;12:2535–48. [PMC free article: PMC317088] [PubMed: 9716406]
- 111.
- Oh B, Hwang S, McLaughlin J. et al. Timely translation during the mouse oocyte-to-embryo transition. Development. 2000;127:3795–803. [PubMed: 10934024]
- 112.
- Tay J, Hodgman R, Richter JD. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev Biol. 2000;221:1–9. [PubMed: 10772787]
- 113.
- Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1:201–213. [PubMed: 11702780]
- 114.
- Wu L, Wells D, Tay J. et al. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–39. [PubMed: 9856468]
- 115.
- Richter JD. Think globally, translate locally: what mitotic spindles and neuronal synapses have in common. Proc Natl Acad Sci USA. 2001;98:7069–71. [PMC free article: PMC34624] [PubMed: 11416189]
- 116.
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. [PubMed: 8811193]
- 117.
- Schwartz DC, Parker R. Interaction of mRNA translation and mRNA degradation in Saccharomyces cerevisiaeIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000807–825.
- 118.
- Lykke-Andersen J. mRNA quality control: Marking the message for life or death. Curr Biol. 2001;11:R88–91. [PubMed: 11231165]
- 119.
- Culbertson MR. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. [PubMed: 10098411]
- 120.
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–10. [PubMed: 10025395]
- 121.
- Jacobson A, Peltz SW. Destabilization of nonsense-containing transcripts in Saccharomyces cerevisiaeIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000827–847.
- 122.
- Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104:173–6. [PubMed: 11207359]
- 123.
- Schwartz DC, Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol. 2000;20:7933–42. [PMC free article: PMC86404] [PubMed: 11027264]
- 124.
- von Der Haar T, Ball PD, McCarthy JE. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5'- Cap by domains of eIF4G. J Biol Chem. 2000;275:30551–5. [PubMed: 10887196]
- 125.
- Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–32. [PubMed: 7557393]
- 126.
- Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–35. [PMC free article: PMC317214] [PubMed: 9784497]
- 127.
- Mitchell P, Tollervey D. Musing on the structural organization of the exosome complex. Nat Struct Biol. 2000;7:843–6. [PubMed: 11017189]
- 128.
- Beelman CA, Stevens A, Caponigro G. et al. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–6. [PubMed: 8757137]
- 129.
- LaGrandeur TE, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. Embo J. 1998;17:1487–96. [PMC free article: PMC1170496] [PubMed: 9482745]
- 130.
- Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–22. [PMC free article: PMC1171610] [PubMed: 10508173]
- 131.
- Dunckley T, Tucker M, Parker R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics. 2001;157:27–37. [PMC free article: PMC1461477] [PubMed: 11139489]
- 132.
- Hsu CL, Stevens A. Yeast cells lacking 5'->3' exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5' cap structure. Mol Cell Biol. 1993;13:4826–35. [PMC free article: PMC360109] [PubMed: 8336719]
- 133.
- Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'->3' digestion of the transcript. Genes Dev. 1994;8:855–66. [PubMed: 7926773]
- 134.
- Mitchell P, Petfalski E, Shevchenko A. et al. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3'->5' exoribonucleases. Cell. 1997;91:457–66. [PubMed: 9390555]
- 135.
- Allmang C, Petfalski E, Podtelejnikov A. et al. The yeast exosome and human PM-Scl are related complexes of 3' -> 5' exonucleases. Genes Dev. 1999;13:2148–58. [PMC free article: PMC316947] [PubMed: 10465791]
- 136.
- Tucker M, Valencia-Sanchez MA, Staples RR. et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–86. [PubMed: 11239395]
- 137.
- Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–55. [PMC free article: PMC55743] [PubMed: 11410650]
- 138.
- Dehlin E, Wormington M, Korner CG. et al. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–86. [PMC free article: PMC305646] [PubMed: 10698948]
- 139.
- Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3'- exoribonuclease. J Biol Chem. 1997;272:10448–56. [PubMed: 9099687]
- 140.
- Hatfield L, Beelman CA, Stevens A. et al. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–8. [PMC free article: PMC231584] [PubMed: 8816497]
- 141.
- Boeck R, Lapeyre B, Brown CE. et al. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–72. [PMC free article: PMC109091] [PubMed: 9710590]
- 142.
- Bouveret E, Rigaut G, Shevchenko A. et al. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–71. [PMC free article: PMC310234] [PubMed: 10747033]
- 143.
- Tharun S, He W, Mayes AE. et al. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–8. [PubMed: 10761922]
- 144.
- Kambach C, Walke S, Young R. et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–87. [PubMed: 10025403]
- 145.
- Fromont-Racine M, Mayes AE, Brunet-Simon A. et al. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast. 2000;17:95–110. [PMC free article: PMC2448332] [PubMed: 10900456]
- 146.
- Gradi A, Imataka H, Svitkin YV. et al. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–42. [PMC free article: PMC121501] [PubMed: 9418880]
- Introduction
- The Mechanistic Role of the Poly(A) Tail During Initiation of Translation
- Structural Information on the Building Blocks of the Bridge between Cap Structure and Poly(A) Tail
- Molecular Concepts to Explain Translational Synergy
- Mechanisms of Translational Control Involving the Poly(A) Tail
- Translation and mRNA Degradation
- Components of the mRNA Degradation Machinery
- Perspectives
- Acknowledgements
- References
- The End in Sight: Poly(A), Translation and mRNA Stability in Eukaryotes - Madame...The End in Sight: Poly(A), Translation and mRNA Stability in Eukaryotes - Madame Curie Bioscience Database
- Origin and Evolution of DNA and DNA Replication Machineries - Madame Curie Biosc...Origin and Evolution of DNA and DNA Replication Machineries - Madame Curie Bioscience Database
- Polyomavirus Allograft Nephropathy: Clinico-Pathological Correlations - Madame C...Polyomavirus Allograft Nephropathy: Clinico-Pathological Correlations - Madame Curie Bioscience Database
- STATs and Infection - Madame Curie Bioscience DatabaseSTATs and Infection - Madame Curie Bioscience Database
- Regulation of Medical Devices - Madame Curie Bioscience DatabaseRegulation of Medical Devices - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...