NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Prokaryotic ribosomes, in landing at internal ribosome binding sites, have to deal with the secondary structure that is present in every RNA molecule. In previous work, we have described how the binding of ribosomes competes with the spontaneous folding of the mRNA. The strength of the mRNA structure thus determines the accessibility of the ribosome binding site and consequently the level of expression that is obtained. In this chapter, we consider a problem that was not addressed previously, namely that spontaneous unfolding of the RNA exposes the ribosome binding site only very briefly. This exposure time will often be far too short to recruit a ribosome from the cytoplasm. The fact that high expression levels are nonetheless reached suggests that a ribosome is in fact already present at or near the translational start site when the structure opens. We provide evidence from experimental data on initiation and reinitiation, from structure analysis of bacteriophage RNAs and from theoretical kinetic analysis of the initiation process, that such ribosome standby sites indeed exist and function to overcome the fast folding kinetics of mRNA.
Introduction
This is not a review in the classical sense, in that we do not claim to present a comprehensive overview of the available literature on translation (re)initiation in bacteria. Instead, we will address a paradox that arises from analysis of initiation kinetics. By examining the events taking place in translational reinitiation and extending their properties to de novo initiation, we arrive at a possible solution to this problem.
From Reinitiation to Initiation
Basic Features of de novo Initiation
Proper and efficient translational start site selection in Escherichia coli relies on at least three interactions. First, ribosomal protein S1 anchors the 30S subunit in the vicinity of the start site by its nonsequence-specific interaction with regions of single-stranded nucleotides (usually, but not necessarily, pyrimidines). Secondly, the well-known Shine-Dalgarno (SD) base pairing positions the subunit in a more precise manner close to the start codon. Finally, codon-anticodon interaction of tRNAfMet locks the ribosome in its starting position and sets the reading frame (for recent reviews on translational initiation, see refs. 1–4). This selection process, which does not consume energy, can take place in the absence of initiation factors and the initiator tRNA does not have to be charged with formyl methionine. Moreover, analysis of the 30S initiation complex by the toeprinting technique (inhibition of primer extension by mRNA-bound 30S ribosomes) showed that the bound subunit can be shifted to nearby codons by replacing tRNAfMet with their cognate tRNAs.5 This finding makes it clear that S1 and SD interactions still allow some lateral movement of the mRNA over the ribosomal surface. In vivo noninitiator tRNAs are prevented from binding by IF3 proofreading.6,7
Control of Translation by Reinitiation
Usually not all cistrons in a polycistronic mRNA are directly accessible to ribosomes from solution. Instead, translation of a downstream reading frame may depend on reading of the neighboring upstream cistron.811 In such case the terminated ribosome somehow restarts at the downstream reading frame. In most examples, stop and restart codons are only a few nucleotides apart. In Chapter 21 by R. Buckingham and M.Ehrenberg, it is described how a terminated ribosome is disassembled and we may deduce the sequence of events that lead to reinitiation. The critical step in the disassembly is probably the release of the last tRNA, a reaction catalyzed by IF3. Once this uncharged tRNA is removed, the remaining 30S ribosome is free to align with the nearby SD box and to capture an initiator tRNA. After codon-anticodon pairing, translation of the downstream cistron can begin. The essence of this coupling is that translation brings the ribosome to a region on the mRNA which it cannot reach on its own account from solution because of inhibitory RNA structure or other features disfavoring binding.
We do not understand all the details of reinitiation; in particular, it is not clear what determines its efficiency. Frequently, only a fraction of the terminated ribosomes succeed in a restart. Those that do not make it detach spontaneously from the message and before they can rebind, the mRNA refolds to its inaccessible conformation. This is of course also the path followed at stop codons not serving as a restart site. Ribosomes that do succeed in a restart probably do so by extending their weak interaction with the messenger by pairing with the SD box of the downstream reading frame12 or by interactions through protein S1. These restarts classify as kinetic escapes; at equilibrium the 30S·mRNA termination intermediate would dissociate into its components and the reverse reaction would not occur because de novo initiation from this site is by definition impossible. In other words, the terminated ribosome is rescued from release by the restart pathway.
Reinitiation at Distant Restart Signals
Not always are stop and restart sites in close proximity. One may find the restart codon rather far down or even upstream from the termination site. Reinitiation in this case is not mechanistically different from the previous one, but we treat it separately because it demonstrates an important property of the initiation process not immediately evident from the previous section.
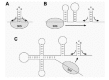
The most illuminating example is translational coupling between the coat and lysis genes of RNA phage MS2. The genetic map of the RNA phage is shown in Figure 1A. As can be seen, the restart codon of the lysis gene is located 46 nucleotides (nt) upstream of the coat termination site. When the coat gene is not translated, there is no translation of the lysis gene either, because independent ribosomal access to this gene is inhibited by the lysis hairpin (Fig. 1B).13 Activation is triggered by ribosomes terminating at the coat gene stop codon. In mutants where termination occurred downstream of the normal coat stop site, lysis expression was abolished but when termination codons were engineered more upstream, closer and closer to the lysis start, its translation became progressively enhanced.9 Activation of the lysis gene is thus clearly coupled to the nearby termination event. It was, in fact, proposed to result from the terminated but not yet released 30S ribosomes. These are thought to start a random walk along the RNA, leading some of them (˜ 5%) to the lysis start 46 nt upstream, where they reinitiate. The other 95% are released and return to the free pool. The main argument that ribosomes actually slide along the RNA is that introduction of a new start codon between termination and restart site forms a barrier for this random walk to arrive at the authentic start, causing reinitiation to occur at this new start codon instead.14 There is a close analogy here with eukaryotic scanning ribosomes that are intercepted at AUG codons introduced between the cap and the true start.15 The capacity of E. coli ribosomes to reach distant start sites after termination is confirmed by in vitro translation of very short messengers. Here the ribosome shuttles back and forth between stop and start without ever leaving the messenger.16 When the lysis hairpin is stabilized by mutations that create stronger base pairs, the scanning ribosomes are no longer able to reach the lysis start, suggesting that their “melting power” is limited.
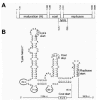
Figure 1
Translational coupling between the coat and lysis genes of RNA bacteriophage MS2. (A) Genetic map of MS2. (B) Structure model of the phage RNA around the coat stop and the lysis start.
Principles of Ribosome Sliding
The important lesson to be learned from the prokaryotic scanning model is that ribosomes bound to mRNA but not yet committed to a reading frame by codon-anticodon interaction can slide in either direction over fairly long distances. We do not know which components participate in this interaction. As continuously different sequences are encountered on the messenger, it is unlikely that the bases are involved. Moreover, these should be available to establish specific contacts such as the SD and codon-anticodon interaction. Thus, it could be either the phosphates or the riboses. On the ribosome side, an obvious stabilizing component is S1, a protein interacting with unstructured RNA,17 but also the unidentified component that allows ribosomes to bind to leaderless mRNA may be considered.18 As the interaction between the 30S subunit and the RNA probably does not involve the RNA bases, each new position has the same binding energy. Movement can be very rapid as evidenced by the lysis gene case where the distance between stop and restart site must be traversed before an elongating, coat-synthesizing ribosome blocks passage.
It is of interest to compare the speed of initiation complex formation via reinitiation and via the de novo pathway. To do so, we prepared a mutant containing a destabilized lysis hairpin. As expected, this mutant produced lysis protein even when coat translation was fully blocked. When coat translation was restored, the production of lysis protein remained unchanged. However, now there are two potential sources to feed the lysis start. There can be de novo initiation owing to the destabilized lysis hairpin and there can be restarts resulting from coat gene termination. To distinguish between the two possibilities, we transplanted the coat stop codon in the 3' direction to a position too far away to allow reinitiation. Now, the lysis gene was not translated at all. Clearly, the rapid flow of coat-translating ribosomes interfered with de novo initiation at the opened-up lysis start.19 These experiments show that lysis can only be produced by reinitiation and that reinitiation is much faster than de novo initiation, even if it includes scanning. It appears therefore that the initial binding step of free ribosomes is relatively slow.
Although the movement of 30S subunits over the mRNA is basically a heat-driven random walk, nature could conceivably introduce directionality into such movement by evolving specific RNA secondary structures that push the ribosome in one direction (Fig. 2). To our knowledge, the potential influence of RNA secondary structure in termination regions on restart events has not been studied so far.
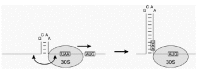
Figure 2
A possible way to confer directionality to the random walk of a terminated 30S ribosomal subunit. The subunit is not drawn to scale.
A Standby Intermediate May Solve a Kinetic Paradox in de novo Initiation
There is no obvious difference between the 30S·mRNA (IF3) termination intermediate and a 30S·mRNA (IF3) initiation intermediate. This implies that de novo initiation does not necessarily start with binding exactly at the ribosome binding site (RBS; defined as the footprint of the 30S subunit on the mRNA in a 30S initiation complex). The 30S subunits will bind with significant affinity to any piece of nonstructured RNA, like the classic example of poly(U). It is thus quite possible that the 30S subunit has the potential to land at a single-stranded region outside the RBS and then walk randomly to the true start site where it becomes fixed by additional interactions (S1, SD, tRNAfMet). This resembles eukaryotic initiation where lateral movement takes place after the initial landing, be it on the cap or on an internal ribosome entry site (IRES).20,21
In E. coli, the existence of such a landing pad or “standby site” could resolve a serious problem that exists in the kinetics of initiation. Most RBSs (like all RNAs) adopt a secondary structure of some kind. Productive ribosome binding requires the RBS to be unfolded, but this state lasts too short to allow diffusion-controlled ribosome binding (see below). Having a ribosome on standby close to its target would be a solution to this problem.
Do We Need Standby Binding?
The possibility of standby binding of 30S subunits to mRNA as a mechanism for enhancing their binding rate to the RBS has been proposed earlier by Draper,22 but the kinetic data available at the time gave no indication as to the actual need for rate enhancement. Estimated ribosome concentrations and association rates (see below) predict that 30S binding is far more rapid than the initiation process and nothing would therefore prevent ribosomes from following each other closely on a well-translated messenger. So, if RBSs were continuously accessible, little could be gained by placing 30S subunits on standby.
While RBSs may in general be less structured than other mRNA parts,23 they are rarely structure-free. Most of the affinity of the ribosome for the messenger comes however from interactions that require the RNA to be single-stranded. The ribosome must therefore compete with the structure and capture the RBS when it happens to be in the unfolded state.1,24,25 In the following sections, we will argue that this state exists only during very brief intervals and that therefore 30S binding rates are required that are much faster than can be achieved by simple diffusion. We will show that this paradox may be solved by invoking standby binding.
Flash Exposure of a Ribosome Binding Site
We have previously used the RBS of the coat protein gene from bacteriophage MS2 to examine how simple mRNA structures control expression by limiting access to ribosomes.25,26 The MS2 coat RBS folds into a hairpin structure that encompasses both the SD region and the AUG initiation codon (Fig. 3A). Extensive mutagenesis revealed that the level of expression from this RBS is dictated directly by the stability of the hairpin structure (Fig. 3B and see below). In thermodynamic terms, the binding of free 30S subunits competes against formation of the structure at the RBS. Even though the wild-type structure is stable enough to exist more than 99% of the time, the high affinity of the 30S subunits for the unfolded RBS allows them to outcompete the structure. Expression of the wild-type gene thus reaches a maximal level that is not limited by the structure, but only by the kinetics of the initiation process and the clearance of the RBS by the elongating ribosome.
Since we proposed this quantitative thermodynamic model, data on the kinetics of spontaneous formation and disruption of hairpin structures have emerged. Not unexpectedly, simple hairpins turn out to fold extremely rapidly, with half-lives of the unfolded state in the order of microseconds. For a rather stable hairpin like the one at the RBS of the MS2 coat gene, this has important implications. Not only is it closed most of the time, but when it happens to open spontaneously, it is only for a very brief time. The question arises if a 30S subunit from the cytoplasmic pool can reach the RBS during this “flash exposure”.
The rate at which the coat hairpin refolds can be estimated in two independent ways. First, a rule of thumb has been proposed to calculate the rate of folding of a hairpin from the stability of its loop, including the first base pair.27,28 This builds on the assumption that the formation of a hairpin is most likely to commence with the formation of the topmost base pair, after which the remainder of the stem closes rapidly in a zipper-like fashion. Using the Turner parameter set 2.3 for calculating ΔG-values (see below), the rule predicts a folding rate kF at 37°C of about 105 s−1. This corresponds to an average lifetime of the open state of 1/kF = 10 μs.
An even shorter time is found when we examine the kinetics of translation of the coat gene. Analysis of polysome profiles from E. coli cells that were infected with MS2 or R17 (virtually identical bacteriophages) has shown that translated phage RNA is found mostly in dimeric and trimeric polysomes.2931 These polysomes virtually disappear when the coat protein gene contains an amber mutation, supporting the notion that they mainly represent translation of the coat protein gene.32,33 Given an elongation rate of about 16 codons per second,34 a length of the coat protein gene of 130 codons and the fact that one of the ribosomes will always occupy the RBS, two to three ribosomes per RNA would roughly correspond to one initiation event per six seconds. This is not far off from the value of one initiation per three seconds found by a different approach for another well-translated gene, lacZ.35,36
An average time span of six seconds between initiation events leaves a window of about four seconds for a new ribosome to attach to the messenger (assuming two seconds for the initiation process itself and the subsequent clearance of the RBS).37 Since the hairpin folds up immediately upon being liberated by the elongating ribosome, the observed initiation frequency prescribes that the hairpin cannot remain closed for more than four seconds.
The maximal duration of the “flash exposure” of the RBS can now be calculated from the equilibrium constant of the helix-coil transition (KF = [F]/[U]), which equals the ratio between the “open” and “closed” lifetimes. We can obtain this equilibrium constant from the free energy of formation (ΔG0f) of the hairpin (KF = e−ΔG/RT). Again using parameter set 2.3 of Turner at 37°C, one gets a value of KF = 6·107 for the MS2 coat hairpin. A “closed” time of less than four seconds thus implies an “open” time of less than 4 s /(6·107) = 0.7·10−7 s, i.e., the open RBS refolds in less than 0.1 μs. To account for the observed initiation frequency at the coat protein gene, a new ribosome must always manage to initiate within four seconds after the previous one has left the RBS. In other words, there must be a 100% probability that within 0.1 μs a 30S subunit captures the open RBS in a productive manner, resulting in the formation of a functional initiation complex. Note that this time span of 0.1 μs may represent a single exposure, or may be divided over multiple, even shorter exposures within the four-second window.
How Fast Can Free-Floating Ribosomes Be on Site?
To estimate the on-rate for 30S-mRNA association, we need to know the concentration of free 30S subunits in the cell. We have previously used an estimate of 8.5 μM, noting that this is an upper limit because not all free 30S particles may be functional.25 We assume these subunits to be saturated with initiation factors38,39 and fMet-tRNA.40 The binding rate constant kSU has been measured in vitro using a quench-flow technique (K. Andersson and M. Ehrenberg, personal communication). Its observed value of about 3·107 M−1s−1 is in good agreement with the expected diffusion rate,22,41,42 and yields an on-rate kSU· [30S] of less than 250 s−1. The RBS would thus on average take 4 ms to collide with a free 30S subunit, much longer than the available interval of less than 0.1 μs. Even if one takes into account all the uncertainties in the numbers used to arrive at this conclusion, a discrepancy of more than four orders of magnitude is difficult to ignore. It appears that ribosomes dispersed in the cytoplasm are simply too slow to jump onto the RBS within the brief instant that it is accessible.
Where then do the ribosomes that initiate at the coat start come from, if not from solution? Since expression of the coat gene is not coupled to any of the other genes present on the MS2 RNA, we can only envisage that they are sitting somewhere on the RNA, waiting for the RBS to become available. This is where we see a possible role for standby binding as proposed above.
We will now examine what standby binding sites may look like and what evidence we can find for their existence. In the final section, we will show that, given what is known about the physicochemical aspects of translation, standby binding may indeed provide a quantitatively satisfactory solution to the aforementioned kinetic paradox.
* In view of the crowded nature of the cytoplasm, in vivo binding may well be even slower. Note that crowding can also increase reaction rates, but only for reactions that are not diffusion limited.41,42
Nature and Biology of 30S Standby Binding
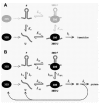
Depending on the place where the 30S subunit sits on the mRNA until it can capture an unfolded RBS, we may distinguish three fundamentally different models for standby binding. These are shown schematically in Figure 4 and will be discussed in the following subsections.
On-Site Binding: the Hammock Model
The Ribosome Can Accommodate Certain mRNA Structures
The SD interaction, the codon-anticodon interaction and the binding of S1 all require the mRNA to be locally single-stranded. Until recently, we believed that this had to be true for the entire RBS. New data have however forced us to reconsider this assumption. It now appears that the 30S subunit has the space and ability to accommodate various hairpin structures in the mRNA, as long as the interactions mentioned above are not compromised. Structures that are ignored by bound ribosomes have been found directly upstream of the SD region, between the SD region and the initiation codon and directly downstream of the initiation codon.
Noninhibitory structures upstream of the SD region are typically short, regular hairpins.1,43 The largest structure of this kind described so far is a 13 base-pair hairpin in the RBS of the E. coli thrS gene.44 It lies 5' adjacent to the SD region and forms part of the binding site for the autorepressor protein ThrRS, but by itself does not affect ribosome binding and initiation. In fact, deletion and mutation studies showed that the single-stranded RBS continues on the 5' side of the hairpin and that this upstream stretch of unpaired nucleotides is crucial for thrS translation. Moreover, nuclease and chemical protection analyses clearly showed the hairpin protruding from the bound 30S subunit. The single-stranded stretches on either side of the hairpin may thus be considered as a quasi-continuous RBS.
Structures upstream from the SD region that do function as inhibitors of translation are generally large and composite, no doubt precluding the formation of such a quasi-continuous RBS.1,24 Why very long hairpins (> 20 bp) can also inhibit ribosome binding is as yet unclear.45,46 Another mystery is the existence of long-range interactions just upstream from the SD that stimulate translation while disrupting local base pairing at the RBS.4749
Stable structures have also been discovered between the SD region and start codon of two genes from bacteriophage T4, where they are essential for fixing the correct distance between these two elements.50,51 Whether this unusual arrangement serves some regulatory purpose is not clear. The same phenomenon was observed by toeprinting on an artificial mRNA where a structure from phage T4 gene 60 was placed between an SD region and an AUG codon. In the same experiment, a long hairpin was accepted by the ribosome when placed just downstream of the AUG.52
Together, these data indicate that the 30S subunit is capable of attaching to RNAs that contain substantial base pairing in the form of stem-loop structures, as long as a quasi-continuous single-stranded stretch of sufficient length is available. Small structures are noninhibitory if they can be oriented so as to protrude from the ribosome. Indeed, various lines of evidence indicate that much of the mRNA that is protected against nuclease and chemical attack is not actually hidden inside the 30S particle, but is attached to its surface on what is known as the platform and the shoulder.5259
The Main Entry Site of RNA-Phage RNAs
Based on the above observations, we now envisage the following scenario. A free-floating 30S subunit may attach to the single-stranded stretches flanking a structured RBS and wait there for the structure to open (Fig. 4A). Once this happens, the 30S snaps into place, preventing the structure from closing again.
Suggestive support for the existence of this type of “on-site” standby binding comes from the secondary structure models for the RNAs of various RNA bacteriophages that were developed by phylogeny and probing.6063 These RNAs, which function both as genome and as messenger, have an intricate structure composed of large structural domains that hardly leave any single-stranded stretches longer than a few nucleotides. A remarkable feature of all these RNAs, however much they differ in sequence, is the presence of one region devoid of large-scale structures (Fig. 5A). This region invariably comprises about 120 nt and includes the RBS of the coat protein gene.
The coat gene RBS functions in these phages as the central control point for translational regulation and it is the primary entry site for ribosomes. In all cases the SD region and initiation codon are taken up in a small but fairly stable hairpin structure. The gene is nonetheless translated at a very high level.
The extent of the poorly structured region of 120 nt appears quite meaningless, until one counts only the unpaired nucleotides, ignoring the paired ones and the various loops. Figure 5B shows that in all cases these quasi-continuous single-stranded stretches add up to between 44 and 49 nt. This is only marginally larger than the 30S footprint on the mRNA backbone in the absence of initiator tRNA.64 We therefore like to picture this region as a kind of hammock, bounded on either side by the multi-branched structural domains of the remainder of the phage RNA, in which a 30S subunit can rest and wait for the structure to open.
Among the wealth of experimental data on in vitro ribosome binding to the various phage RNAs, there is one observation that may relate directly to the standby complex we propose. The 30S subunits can be bound to MS2 or Qβ RNA in the absence of initiator tRNA with an association constant of about 106 M−1.65 When prepared at 37°C, the resulting complex can be chased into translation, even in the presence of inhibitors of de novo initiation such as edeine, aurintricarboxylic acid or anti-S1 antibodies. In striking contrast, this is not possible with a similar complex formed at 0°C.66,67
A new interpretation of these observations could be as follows. At 37°C, the hairpin at the coat protein gene RBS is weak enough to allow the 30S subunit to shift rapidly from its standby position to form the true pre-initiation complex, which needs no further reorganization to enter into translation. At 0°C, on the other hand, the hairpin is stabilized to such a degree that it does not open within the time span of the experiment, leaving the 30S subunit sitting on the structured RBS as an on-site standby complex. To allow this complex to enter translation at 37°C, the 30S subunit has to resettle on the RNA in a somewhat shifted position as soon as the hairpin has opened. During this resettling, the 30S subunit is apparently sensitive to the aforementioned inhibitors. Note that S1 is involved in both the 0°C and the 37°C complex.68 In agreement with this explanation, the complex formed at 37°C remains insensitive when cooled down to 0°C, indicating that the unfolded RBS cannot refold once the true pre-initiation complex has formed.
A similar interaction of the 30S subunit with single-stranded stretches between hairpins (or with their loops) was recently proposed as an important intermediate in translational autocontrol of the E. coli rpsA gene, the gene that encodes ribosomal protein S1.69 Details of this system await further elucidation. Evidence for a standby complex with the SD interaction already in place was obtained by probing the structure of the cro mRNA in 30S·mRNA complexes with or without initiator tRNA. A hairpin structure downstream from the AUG start codon was shown to exist in the binary 30S·mRNA complex, but disappeared upon the addition of initiator tRNA.70 This confirms that the 30S subunit can indeed bind to a partially structured RBS and capture the RNA in its single-stranded form when it opens.
A Standby Site Activated by Translating Ribosomes?
The RBS of the replicase genes of the single-stranded RNA bacteriophages is by itself inactive, because it lies hidden in a heavily structured region composed of local hairpins and long-range stems (Fig. 6A). It is only activated when ribosomes translating the upstream coat-protein gene pass through the long-range components of these structures, known as the MJ and VD interactions.7174 This is believed to open the replicase start region for long enough to allow one round of initiation, yielding the expression ratio of about 1:1 as observed in expression studies with MS2 cDNA plasmids.75
Like the coat gene start, the start of the replicase gene forms a hairpin structure comprising both the SD region and the AUG initiation codon (Fig. 6A), but by itself this structure is weak enough to allow high-level translation.76 However, just like in the case of the coat gene, we are faced with the problem of the relative kinetics of 30S binding and hairpin folding. The replicase RBS is predicted to exist in its unfolded state about 0.02% of the time. This implies that a random collision with a free 30S subunit is five thousand times less likely to be productive than if the RBS would be single-stranded permanently.
If elongation takes place at a rate of 16 codons per second,34 a ribosome translating the coat gene keeps the long-range interactions open for about one second. To obtain an expression ratio of 1:1, this one second must thus be enough to achieve a successful initiation event at the replicase start. The aforementioned binding rate constant (3·107 M−1s−1) and 30S concentration (8.5 μM) predict that 30S subunits encounter the RBS at a frequency of 3·107 × 8.5·10−6 = 250 times per second. At a fraction of open RBSs of 0.02%, we can therefore expect no more than 0.05 productive collisions within the available second, yielding a coat : replicase ratio of 20 : 1. This is not consistent with the experimental observations.
A closer look at the structures proposed around the replicase RBS reveals that the passage of a coat-translating ribosome may in fact create a quasi-continuous single-stranded region, much like the one present at the coat gene start. In Figure 6B, the two long-range interactions have been opened, as they would be by a translating ribosome. As a result, a series of single-stranded stretches has become available, adding up to 49 nt. The possibility to create a region of this size is conserved within the single-stranded RNA phages (Fig. 6B),60,61 but we need to postulate that the bottom part of the hairpin following the replicase start codon is also open. The unusually asymmetric internal loop that terminates this helix in all the different RNA phages (See Fig. 6A) may function as a hinge to facilitate the opening of the standby site.77,78
Until now, it was assumed that opening of the MJ interaction activates the replicase start because it forms part of the RBS, albeit upstream from the SD region. The VD interaction was proposed to contribute to regulation because it might stabilize the MJ interaction by coaxial stacking.74 The possibility of standby binding to the surroundings of the replicase start while the local hairpins are still intact suggests a different interpretation. Movement of ribosomes through both VD and MJ is now essential to create an “on-site” standby site that is large enough to accommodate a 30S subunit. There, the subunit can wait for as long as its off-rate allows, until the hairpin at the replicase RBS opens and a true initiation complex can be formed.
The only reason why we think that an inducible standby binding site is necessary here, is the presence of a small but stable hairpin at the replicase RBS, which limits its accessibility to intervals of only a few microseconds. This hairpin however is crucial to the phage, as it is the main binding site for the MS2 coat protein. Late in the life cycle of the phage, the binding of coat protein at this hairpin blocks replicase translation and initiates packaging of the RNA into capsids. Thus, the replicase RBS has evolved under opposite selection pressures from the coat protein and the ribosome. We suggest that the creation of an inducible standby binding site has allowed the union of three otherwise incompatible functions in one stretch of RNA: a locally structured binding site for the coat protein; a long-range structure permitting translational coupling; and a single-stranded region for the 30S subunit to enter. With every new detail we learn about the RNA phages, it becomes more striking how intricately the many control mechanisms interrelate in these tiny survival machines.
Close-by Binding: Scanning, Shunting and the Landing-Pad Model
Linear diffusion is a common principle in “target location” by DNA-binding proteins.79 The relative uniformity of the DNA double helix allows these proteins to bind nonspecifically anywhere on the DNA and shift to their final position by random lateral movement. In contrast, the applicability of linear diffusion to ribosome binding has been considered limited because at first sight it would seem to require extensive stretches of structure-free RNA.22 There is however good evidence that certain secondary structures can be skipped during prokaryotic scanning, obviating the need for the region between a potential landing site and the target site (RBS) to be fully unfolded.
Analysis of reinitiation patterns in nonsense mutants of the lacI gene has indicated that the prokaryotic scanning ribosome does not necessarily stop or dissociate when it encounters structures in the mRNA. Instead, it can sometimes skip over them and continue scanning on the other side.80 Thus, large distances between termination sites and reinitiation sites can be bridged. Here again the presence of a quasi-continuous single-stranded stretch of sufficient length seems pivotal.
The same principle could apply to the binding of free subunits, if they can attach to a single-stranded region at some distance from an RBS that itself is inaccessible (Fig. 4B). By random movement over the single-stranded stretches, they could hit upon the initiation site during its “flash exposure”, in a prokaryotic equivalent of ribosome shunting.21 Prokaryotic scanning must be relatively fast, since in the coupling between the MS2 coat and lysis genes terminated ribosomes manage to reach the upstream lysis start against the current of coat-gene translating ribosomes (see above).
A stretch of accessible RNA could thus function as a ribosomal landing pad for a nearby RBS. It should be noted however that we have no mechanism to give a 30S subunit on standby a push in the right direction at the right moment, viz. when the structure at the RBS opens. In contrast to the “on-site” binding discussed in the previous section, the subunit is now not necessarily in place when the RBS is exposed. Such “off-site” standby binding would therefore only function by increasing the probability of a productive encounter and we consider its potential therefore limited compared to the “on-site” model.
Intersegment Transfer and Dissociation/Association
The fast rate at which the LacI transcriptional repressor protein reaches its target on double-stranded DNA is believed to depend in part on a process termed direct intersegment transfer.81,82 It implies that the protein binds initially to a nonspecific site on the nucleic acid and from there is handed over to its site of action. A similar scenario might be imagined for the binding of ribosomes to mRNA (Fig. 4C). The LacI repressor functions as a tetramer and therefore has multiple identical DNA binding sites, which allows the existence of an intermediary situation where secondary and primary targets are bound at the same time. Although 30S subunits do not form multimers, it is not inconceivable that they may attach to a single-stranded section of the RNA through ribosomal protein S1 and then capture a distant RBS with, for example, the anti-SD region. Subsequently, S1 may follow suit and jump over to the RBS as well. As an aside, we note that the “mRNA-standby site” on the 30S subunit proposed by Gualerzi and coworkers could not be used for this kind of handing over, since it occupies roughly the same space on the subunit as the final binding site.4,65,83
Alternatively, even simple dissociation from an initial nonspecific binding site and re-association at the RBS may raise the probability of functional complex formation. Limiting the distance over which the 30S subunits diffuse (from other parts of the same RNA, rather than from anywhere in the cytoplasm) would increase the frequency with which 30S subunits collide with the RBS. In other words, it increases the “local concentration” of subunits in the vicinity of the RBS. This would parallel the way in which the movement of, among others, RNA polymerase is confined to a local DNA domain until it finds its specific binding site.84,85
If one applies the concept of local concentration to the MS2 coat gene RBS, then the apparent 10,000-fold increase in binding rate over simple diffusion (see above) would require a local 30S concentration of 10,000 × 8.5 μM. This would imply that the ribosomes be confined to a space smaller than their own dimensions. Confinement of the ribosomes to a limited volume by dissociation/association, involving secondary binding sites elsewhere on the mRNA, is apparently not sufficient to explain this particular case. A more satisfactory explanation would be that there is permanently a 30S subunit in place at the RBS. Its mRNA binding track, already oriented towards the RNA, then indeed covers only a fraction of the ribosome volume. This situation coincides with the on-site binding model proposed earlier (Fig. 4A).
Dissociation/association or intersegment transfer may nonetheless play a supportive role in the MS2 coat gene system. We have previously reported that the upstream 1300 nucleotides of native MS2 sequence strongly enhance the capability of the ribosome to compete with the hairpin structure at the RBS. Under conditions of maximal sensitivity (i.e., strong competition by the RBS structure), expression was 25-fold higher in the presence of the upstream RNA, presumably through an increase in the 30S binding rate.86 This enhancement could not be localized to a specific segment of the upstream sequence. The native structure of this highly organized piece of RNA appears to guide the 30S subunit to the coat gene RBS. By itself, such guidance would be insufficient to put the subunit in place within the 0.1 μs that the hairpin is unfolded (see above). But it is conceivable that it is needed to put and keep the 30S in its standby position on the quasi-continuous single-stranded region shown in Figure 5. Many DNA-binding proteins also appear to reach their targets by a combination of long-range three-dimensional movement and local linear sliding.87,88
Ribosome Recycling in cis
An intriguing way in which intersegment transfer could be employed for translational control, is the recycling of terminated ribosomes back to the original initiation site, where they may reinitiate for another round of translation. Recycling of ribosomes in cis has been proposed in eukaryotes, where circularized messengers were directly observed by atomic force microscopy.8991 In prokaryotes, recycling is reportedly happening on tripeptide-encoding minigene messengers, but there the ribosomes are simply slipping back over a very short distance.16 No direct evidence exists, to our knowledge, for the long-range transfer that would be needed in the case of genes of normal length. However, a very suggestive arrangement of termination and initiation codons is found in the hok-like operons for plasmid stabilization.
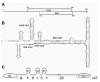
All operons of the hok family consist of two open reading frames that overlap by almost their entire lengths (Fig. 7A). 92,93 While the downstream frame encodes a killer protein that has a central function in plasmid stabilization, the polypeptide encoded by the upstream frame has no function. This frame (called mok in the prototype hok system) only exists to activate the killer gene (hok) through translational coupling. When a cell loses the plasmid during cell division, the mok start on the remaining mRNA is activated through the degradation of an inhibitory antisense RNA. This leads to expression of hok and the subsequent death of the plasmid-free cell.
The structure of the hok mRNA has been solved and is shown in Figure 7B.94,95 Both RBSs are located in stable structures, but the mok hairpin is weak enough to allow the low level of mok translation that is required to activate the hok start.95 One would expect translational coupling to function perfectly well if the mok termination codon were located next to the hok start. Nonetheless, the unusual arrangement with the mok stop at the end of the hok gene (Fig. 7A) is conserved throughout the highly divergent hok family. It is intriguing to see that the secondary structure puts the two termination codons opposite the RBS of hok. This strongly suggests that termination, rather than elongation, is the crucial event activating the hok start. While that may be related to the fact that termination consists of a rather time-consuming sequence of steps, we cannot ignore the possibility that ribosomes actually recycle in cis on this particular class of messengers, by being handed over from the mok stop codon to the hok start (compareFig. 4C). Moreover, since hok-translating ribosomes also terminate opposite the hok RBS, activation of mok would lead to a self-sustaining avalanche of hok translations, ultimately resulting in the killing of the cell. We imagine that such a highly unusual positive feedback loop would only be allowed in a situation where rapid cell death is the final aim anyway.
Whether recycling of ribosomes on the hok messenger occurs or not, we note that the terminating ribosomes may once again create an “on-site” standby site (Fig. 7C). When we assume that the asymmetric internal loop downstream of the hok start functions as a hinge, as proposed for the replicase start (Fig. 6), the stretches of unpaired nucleotides around the hok start add up to 47 nt. Standby binding would be necessary here because once the long-range interaction is disrupted, the tiny hairpin upstream from the hok start will extend to include the hok SD-region (Fig. 7C).94,95
Standby Binding and Translational Repression
Draper was the first to point out that the intuitive mechanism for the functioning of translational repressors may not be valid for thermodynamic reasons.96 Simple competition between a repressor protein and the 30S subunit would require high concentrations of repressor or high affinities for the RNA, neither of which was supported by available quantitative data. A variety of mechanisms have since been proposed that would help the repressor to do its job by taking advantage of mRNA structure, binding kinetics, mRNA degradation and even of the ribosome itself.22,9699
As an additional possibility, we propose that the existence of an obligatory standby 30S·mRNA intermediate may allow translational regulation by direct competition without the need for extreme concentrations or affinities. As argued in the following section, standby binding does not need to be very strong to be functional. But if a repressor would prevent the 30S subunit from occupying its standby site by competing with this low-affinity binding, it would effectively block initiation at the structured RBS.
Once more, we can turn to the RNA bacteriophages for a possible example. During the life cycle of the phage there comes a point when its RNA has to be replicated rather than translated. This functional switch begins with the binding of the phage-encoded replicase just upstream from the coat gene RBS (Fig. 5).100,101 This apparently suffices to block translation of the coat gene and consequently, through translational coupling, of the lysis and replicase genes. The remaining maturation gene has already switched itself off by that time.102,103 Remarkably, the replicase uses a recruited copy of ribosomal protein S1 to bind at the coat gene RBS.104,105 While the replicase may be unable to compete with formation of a 30S initiation complex on a single-stranded RNA, it may well outcompete the proposed weak standby complex, especially if its binding would negate the enhancement of 30S binding by the upstream RNA (see above). A picture now emerges in which the hairpin, the quasi-continuous single-stranded stretch and the stimulatory upstream structure have co-evolved to allow efficient regulation of the translation-replication switch that is central to the life cycle of these phages.
Physical Chemistry of 30S Standby Binding
Ribosomal Affinity and the Sensitivity to RBS Structure
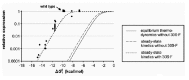
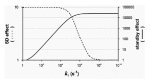
We have previously approached the mechanism by which local mRNA structures control translational initiation as an equilibrium competition scheme (Fig. 8A). For the coat-RBS of phage MS2 and for several other RBSs, we showed that the expression decreases tenfold with every −1.4 kcal/mol added to the stability of the structure (Fig. 3B).25,106 Theoretically, every -1.4 kcal/mol reduces the fraction of RNA molecules in which the hairpin is open at a given point in time by the same factor of ten. The expression data thus indicated that hairpin formation and 30S binding compete directly for the unfolded RBS. In addition, the affinity of the 30S subunit for the RBS was predicted, and subsequently shown, to determine the sensitivity to inhibition by the structure.26,107 In other words: the stronger the binding of the 30S to the unfolded mRNA is, the stronger the structure at the RBS has to be to inhibit translation. This is indicated in Figure 3B by theoretical expression curves shifting to the left as the 30S-mRNA affinity (KSU) increases. Relative expression is defined here as the fraction of the RBSs being in complex with a 30S subunit and it thus approaches the value of one as the messengers become saturated with ribosomes.
Standby Binding Relies on Kinetics
Standby binding of ribosomal subunits to the messenger can be introduced into this scheme as shown in light gray in Figure 8A. However, the equilibrium approach now turns out to be too limited. In fact, standby binding would be predicted to decrease translational efficiency, as the complex between the folded RNA and the 30S subunit (30S·F) would form at the expense of the pool that drives translation (30S·U). Moreover, we predicted standby binding only to be effective with fast RNA folding kinetics, an aspect that is completely ignored in thermodynamics (note that equilibrium thermodynamics, in only dealing with equilibrium constants, by definition ignores the time factor).
Figure 8B shows how we have converted the equilibrium scheme into a kinetic model, mainly by splitting the equilibrium constants into their component rate constants and by adding a route for the RBSs and 30S subunits to return to the free pool after taking part in translation. We have also split the initiation process in two steps, so that we can examine the consequences of varying the rate of formation of the true 30S initiation complex (ki) without affecting the clearance of the RBS (kc; see ref. 2). Apart from ignoring the time factor, the original equilibrium scheme represents nothing but a special case of the kinetic scheme, namely when ki is negligible compared to k-SU and k-SFU.
The binding of 30S subunits to the mRNA is now described by no less than eight rate constants, but these are related through the first law of thermodynamics. If ki is made zero (i.e., formation of the 30S initiation complex is blocked), the circular chain of reactions should be in equilibrium, meaning that the multiplied rate constants clockwise must equal those counterclockwise. With this restriction, the model allows us to freely enter established values for the different parameters, or to vary them one by one and examine how each parameter affects the sensitivity of expression to the RBS structure. We did this by solving the corresponding steady-state equations, entering these into a spreadsheet and plotting the predicted outcome in the same format as Figure 3B. Table 1 summarizes the justifications for the various parameter values that we used as default.
Table 1
Parameter values entered into the model of Figure 8B to obtain Figures 9 to 11.
Figure 9 shows the predictions using the values of Table 1. It is clear that without involvement of standby binding, the predicted expression for the wild-type coat RBS (ΔG0f = −9.9 kcal/mol at 42°C; see below) would be practically zero (dashed curve). Standby binding, even with a binding constant as low as 105 M−1, is predicted to yield a dramatic increase in the ability of ribosomes to initiate at the coat start (dotted curve). In fact, the model now predicts maximal expression with the wild-type hairpin, as we observed in our experiments.
Standby Binding May Explain Extreme Apparent 30S-mRNA Affinities in vivo
When we first reported on our mutational analysis of the MS2 coat gene start in 1990, we noted that the obtained expression curve suggested a 30S-mRNA affinity of about −13.5 kcal/mol, i.e., a binding constant of 3·109 M−1 (Fig. 3B).25 The fact that this was about two orders of magnitude higher than the values usually measured in vitro96,108113 was blamed, at least in part, on inaccuracies in the parameters used for calculating the hairpin stability.
Since 1990, the parameters for ΔG-calculation have been updated several times, partly on the basis of new experimental data and theoretical considerations, but also by optimizing them for more accurate structure prediction.114 Presently, the Turner group proposes two alternative sets of parameters designated versions 2.3 and 3.0, where only version 2.3 can be adapted for different temperatures (see http://bioinfo.math.rpi.edu/˜zukerm/rna/energy/ for parameters and references). We have a preference for version 2.3, because it is more directly based on experimental data. Moreover, our experimental expression curves show considerably less scattering with set 2.3 than with set 3.0, suggesting that the former better approximates free-energy values in vivo. Recalculated with parameter set 2.3, the ΔG0f of the wild-type coat hairpin at 42°C is now −9.9 kcal/mol rather than the earlier −5.6 kcal/mol and the values for the mutants decrease by similar amounts. Shockingly, the updated expression curve would now suggest a 30S-mRNA affinity of −17.3 kcal/mol or KSU = 1012 M−1 (compare Figs. 3B and 9), which is five orders of magnitude beyond binding constants measured in vitro.
The kinetic scheme of Figure 8B and its predictions in Figure 9 may provide an adequate explanation for this discrepancy. The gain achieved by always having a 30S subunit on standby graphically mimics a large addition to the affinity between the 30S subunit and the RBS, yielding an apparent KSU of almost 1012 M−1. Even though this was not a priori introduced with our choice of rate-constant values, we thus obtain an expression curve that fits well with the experimental data without requiring ribosome-mRNA affinities that are unreasonably high (Fig. 9; dotted curve).
Standby binding thus solves two problems that are physically related but were encountered independently. Firstly, it answers the theoretical question of how ribosomes can bind to an RBS that is only exposed for a very brief instant. Secondly, it explains the extreme apparent 30S-mRNA affinities observed in in vivo translation from a structured RBS as compared to in vitro ribosome binding.
Further experimentation with the parameters in the standby model indicates that the leftward shift of the predicted curve as shown in Figure 9 is close to the maximum that can be obtained with reasonable parameter values. In Figure 10, we summarize the effects of moderate changes in the various parameter values and thus show that the predictions are not highly dependent on particular estimates (see the legend for more detailed information).
Comparing the experimental data in Figure 9 with the various theoretical curves on Figure 10, it seems that the MS2 coat gene system has evolved to achieve the maximum profit that can physically be gained from standby binding. Only its full exploitation would have allowed the hairpin to evolve to its present high stability without a concomitant reduction in translation. This is not so surprising when one considers the tendency of these phages to develop stable RNA structures on the one hand and their need for high-level synthesis of coat protein on the other. In addition, a hairpin that is only just noninhibitory may sensitize the coat gene RBS to translational repression by the phage replicase (see below). Evolutionary experiments in our laboratory have provided ample evidence for extremely rapid and precise optimization of translational control mechanisms in these organisms.107,115122
Kinetics of Initiation in vivo and in vitro
An important difference between the old thermodynamic model and the present kinetic one is the necessary introduction of the fast step indicated with ki in Figure 8B. When we approached ribosome binding as an equilibrium reaction, we had to assume that the drain of 30S·U complexes into translation was slow compared to the preceding reactions. This assumption was supported by in vitro translation data indicating a slow conversion of the 30S·U complex into a true 30S initiation complex.108,111 Gualerzi and coworkers have suggested that this slow step may reflect a conformational change in the ribosome, necessary for codon-anticodon base pairing at the initiation codon.108,123
Now that the existence of a fast but essentially irreversible step turns out to be necessary to make standby binding work, we need to re-examine these assumptions. In fact, one may wonder about the exact significance of 30S-mRNA affinities as measured in vitro. If a fast and essentially irreversible reaction would follow the binding of 30S subunits in such experiments, this would produce an infinitely high binding constant, but instead values of about 107 M−1 are usually found. We must therefore assume that the hypothetical fast reaction does not occur in simple binding experiments with purified compounds.
We know of only one report where ribosome binding parameters were derived from a complete in vitro translation reaction.111 The authors estimated the Michaelis constant for the binding of 30S subunits to the R17 coat-gene start (identical to MS2) as 0.07 μM. This corresponds to an apparent binding constant of 1.4·107 M%1 if one assumes, as they did, that the step following the association is negligibly slow. While at first sight this value seems in good agreement with the in vitro binding data mentioned above, it was in fact not corrected for the presence of the hairpin at the RBS, implying a serious underestimation of the affinity for the unfolded RBS. It can be calculated that the binding constant to the unfolded RBS (KSU) relates to the apparent binding constant to the structured one (KSU,app) and the equilibrium constant of hairpin formation (KF) as KSU = KSU,app·(KF + 1). Using the calculated stability of the coat hairpin, this gives a value for KSU of 8·1014 M−1, far higher than the value of 107 M−1 found in in vitro binding experiments with poly(U) and even higher than the value of 1012 M−1 derived from our in vivo data. We think that this extreme value may reflect on the one hand the existence of standby binding and on the other hand an overestimation due to a nonnegligible forward rate (ki), in agreement with our kinetic model. Although inadequate as true binding constants, these high numbers vividly illustrate the efficiency with which ribosomes are recruited for translation.
We have previously found that the strength of the SD-interaction is one of the factors determining the capability of the ribosome to compete against mRNA structure.26 The fact that expression can thus be modulated by varying the strength of the SD interaction must result from altering the dissociation rate of the 30S subunit, implying that this rate (k-SU·[30S·U]) is significant compared to the irreversible entry of the ribosome into translation (ki·[30S·U]). So while the forward rate must be fast for standby binding to be effective, it must be relatively slow for the SD interaction to modulate expression. While no experimental data are available to assess the true value of this parameter, Figure 11 provides a suggestion for how this paradox may be solved. Intrinsic to the reaction scheme of Figure 8B, there exists a value for ki where expression is almost maximally sensitive to both standby binding and the strength of the SD-interaction. In our numerical example, based upon the coat gene RBS of MS2, this point is at ki = 105 s−1.
Conclusion: Target Location in Translation
The problem of target location in molecular biology has fascinated biologists, chemists and physicists alike for decades. Theories on the various routes that a molecule or complex can take to reach its target in a fast and efficient manner, despite the overcrowded nature of the cytoplasm, are manifold.79,88 Applied to transcription regulators and restriction enzymes in particular, they have helped in understanding quantitative aspects of their binding kinetics that would otherwise have been incomprehensible. Remarkably little of these developments has permeated into the world of translation. Presumably the complexity of the initiation process, especially in eukaryotes, has kept most people from asking this type of quantitative questions on how ribosomes reach their target in and among the tangled clews of mRNA in the cell. Moreover, it has been argued that due to precisely this crowded nature of the cytoplasm standard thermodynamic and kinetic theory, based on ideal solutions, may break down in vivo.42,124
While the values for kinetic and thermodynamic constants and concentrations in vivo that we have used may be imprecise, we believe that this “naive” modeling of the reactions involved is nonetheless a useful exercise. Ten thousand-fold discrepancies between theoretical predictions and observed reality are a strong stimulus to further thinking. In the end, this will hopefully lead to the design of targeted experiments to test possible mechanisms, preferably in vivo. Let us not forget that in a world of genomes and proteomes, it is the dynamics of their interactions that defines life.
Acknowledgements
We thank Dr Måns Ehrenberg and his coworkers for stimulating discussions and Dr Kerstin Andersson for her unpublished results. M.S. was paid by the Netherlands Organization for Scientific Research, NWO.
References
- 1.
- de Smit MH. Translational control by mRNA structure in eubacteria: molecular biology and physical chemistryIn: Simons RW, Grunberg-Manago M, eds.RNA structure and FunctionCold Spring Harbor: Cold Spring Harbor Laboratory Press,1998495–540.
- 2.
- Draper DE, Gluick TC, Schlax PJ. Pseudoknots, RNA folding, and translational regulationIn: Simons RW, Grunberg-Manago M, eds.RNA structure and FunctionCold Spring Harbor: Cold Spring Harbor Laboratory Press,1998415–436.
- 3.
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. [PubMed: 10395892]
- 4.
- Gualerzi CO, Brandi L, Caserta E. et al. Translation initiation in bacteriaIn: Garrett R, Douthwaite SR, Liljas A et al, eds.The Ribosome: Structure, Function, Antibiotics, and Cellular InteractionsWashington: ASM Press,2000477–494.
- 5.
- Hartz D, McPheeters DS, Traut R. et al. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. [PubMed: 2468068]
- 6.
- Risuleo G, Gualerzi C, Pon C. Specificity and properties of the destabilization, induced by initiation factor IF-3, of ternary complexes of the 30-S ribosomal subunit, aminoacyl-tRNA and polynucleotides. Eur J Biochem. 1976;67:603–613. [PubMed: 9282]
- 7.
- Hartz D, Binkley J, Hollingsworth T. et al. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 1990;4:1790–1800. [PubMed: 1701151]
- 8.
- Oppenheim DS, Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. [PMC free article: PMC1214269] [PubMed: 6162715]
- 9.
- Berkhout B, Schmidt BF, van Strien A. et al. Lysis gene of bacteriophage MS2 is activated by translation termination at the overlapping coat gene. J Mol Biol. 1987;195:517–524. [PubMed: 3656424]
- 10.
- Schümperli D, McKenney K, Sobieski DA. et al. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982;30:865–871. [PubMed: 6754091]
- 11.
- Ivey-Hoyle M, Steege DA. Mutational analysis of an inherently defective translation initiation site. J Mol Biol. 1992;224:1039–1054. [PubMed: 1569566]
- 12.
- Spanjaard RA, van Duin J. Translational reinitiation in the presence and absence of a Shine and Dalgarno sequence. Nucleic Acids Res. 1989;17:5501–5507. [PMC free article: PMC318173] [PubMed: 2668889]
- 13.
- Schmidt BF, Berkhout B, Overbeek G. et al. Determination of the RNA secondary structure that regulates lysis gene expression in bacteriophage MS2. J Mol Biol. 1987;195:505–516. [PubMed: 3656423]
- 14.
- Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J Mol Biol. 1990;213:811–818. [PubMed: 2193163]
- 15.
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984;12:3873–3893. [PMC free article: PMC318796] [PubMed: 6328442]
- 16.
- Pavlov MY, Freistroffer DV, MacDougall J. et al. Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J. 1997;16:4134–4141. [PMC free article: PMC1170036] [PubMed: 9233822]
- 17.
- Subramanian AR. Structure and functions of ribosomal protein S1. Prog Nucleic Acid Res Mol Biol. 1983;28:101–142. [PubMed: 6348874]
- 18.
- Moll I, Resch A, Bläsi U. Discrimination of 5'-terminal start codons by translation initiation factor 3 is mediated by ribosomal protein S1. FEBS Lett. 1998;436:213–217. [PubMed: 9781681]
- 19.
- Berkhout B, Kastelein RA, van Duin J. Translational interference at overlapping reading frames in prokaryotic messenger RNA. Gene. 1985;37:171–179. [PubMed: 3840447]
- 20.
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed: 1939050]
- 21.
- Jackson RJ. A comparative view of initiation site selection mechanisms In: Hershey JWB, Mathews MB, Sonenberg N, eds.Translational control Cold Spring Harbor: Cold Spring Harbor Laboratory Press,199671–112.
- 22.
- Draper D. Mechanisms of translational initiation and repression in prokaryotesIn: Nierhaus KH, ed.The Translational ApparatusNew York: Plenum Press,1993197–207.
- 23.
- Ganoza MC, Kofoid EC, Marlière P. et al. Potential secondary structure at translation-initiation sites. Nucleic Acids Res. 1987;15:345–360. [PMC free article: PMC340414] [PubMed: 3484332]
- 24.
- de Smit MH, van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. [PubMed: 2183291]
- 25.
- de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA. 1990;87:7668–7672. [PMC free article: PMC54809] [PubMed: 2217199]
- 26.
- de Smit MH, van Duin J. Translational initiation on structured messengers.Another role for the Shine-Dalgarno interaction. J Mol Biol. 1994;235:173–184. [PubMed: 8289239]
- 27.
- Tacker M, Fontana W, Stadler PF. et al. Statistics of RNA melting kinetics. Eur Biophys J. 1994;23:29–38. [PubMed: 7515805]
- 28.
- Gultyaev AP, van Batenburg FHD, Pleij CWA. The influence of a metastable structure in plasmid primer RNA on antisense RNA binding kinetics. Nucleic Acids Res. 1995;23:3718–3725. [PMC free article: PMC307271] [PubMed: 7479002]
- 29.
- Hotham-Iglewski B, Franklin RM. Replication of bacteriophage ribonucleic acid: Alterations in polyribosome patterns and association of double-stranded RNA with polyribosomes in Escherichia coli infected with bacteriophage R17. Proc Natl Acad Sci USA. 1967;56:743–749. [PMC free article: PMC335696] [PubMed: 4860757]
- 30.
- Koontz SW, Jakubowski H, Goldman E. Control of RNA and protein synthesis by the concentration of Trp-tRNATrp in Escherichia coliinfected with bacteriophage MS2. J Mol Biol. 1983;168:747–763. [PubMed: 6350608]
- 31.
- Truden JL, Franklin RM. Polysomal localization of R17 bacteriophage-specific protein synthesis. J Virol. 1972;9:75–84. [PMC free article: PMC356264] [PubMed: 4550780]
- 32.
- Phillips LA, Truden JL, Iglewski WJ. et al. Replication of bacteriophage ribonucleic acid: Alterations in polyribosome patterns in Escherichia coli infected with amber mutants of bacteriophage R17. Virology. 1969;39:781–790. [PubMed: 4902256]
- 33.
- Beremand MN, Blumenthal T. Overlapping genes in RNA phage: a new protein implicated in lysis. Cell. 1979;18:257–266. [PubMed: 387256]
- 34.
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rateIn: Neidhardt FC, ed.Cellular and Molecular Biology of Escherichia coli and Salmonella typhimuriumWashington DC: Am. Soc. Microbiol,19871527–1542.
- 35.
- Kennell D, Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J Mol Biol. 1977;114:1–21. [PubMed: 409848]
- 36.
- Petersen C. Translation and mRNA stability in bacteria: a complex relationshipIn: Belasco J, Brawerman G, eds.Control of Messenger RNA StabilitySan Diego: Academic Press,1993117–145.
- 37.
- Tomsic J, Vitali LA, Daviter T. et al. Late events of translation initiation in bacteria: a kinetic analysis. EMBO J. 2000;19:2127–2136. [PMC free article: PMC305682] [PubMed: 10790378]
- 38.
- Grunberg-Manago M. Initiation of protein synthesis as seen in 1979In: Chambliss G, ed.Ribosomes: Structure, Function and GeneticsBaltimore: University Park Press,1979445–477.
- 39.
- Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. [PubMed: 2200518]
- 40.
- de Smit MH. Regulation of translation by mRNA structure PhD Thesis Leiden: Leiden University1994 .
- 41.
- Goodsell DS. Inside a living cell. Trends Biochem Sci. 1991;16:203–206. [PubMed: 1891800]
- 42.
- Zimmerman SB, Minton AP. Macromolecular crowding: Biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. [PubMed: 7688609]
- 43.
- Shinedling S, Gayle M, Pribnow D. et al. Mutations affecting translation of the bacteriophage T4 rIIB gene cloned in Escherichia coli. Mol Gen Genet. 1987;207:224–232. [PubMed: 3112515]
- 44.
- Sacerdot C, Caillet J, Graffe M. et al. The Escherichia coli threonyl-tRNA synthetase gene contains a split ribosomal binding site interrupted by a hairpin structure that is essential for autoregulation. Mol Microbiol. 1998;29:1077–1090. [PubMed: 9767575]
- 45.
- Wilson IW, Praszkier J, Pittard AJ. Molecular analysis of RNAI control of repB translation in IncB plasmids. J Bacteriol. 1994;176:6497–6508. [PMC free article: PMC197003] [PubMed: 7525535]
- 46.
- Malmgren C, Engdahl HM, Romby P. et al. An antisense/target RNA duplex or a strong intramolecular RNA structure 5' of a translation initiation signal blocks ribosome binding: The case of plasmid R1. RNA. 1996;2:1022–1032. [PMC free article: PMC1369434] [PubMed: 8849778]
- 47.
- Wilson IW, Praszkier J, Pittard AJ. Mutations affecting pseudoknot control of the replication of B group plasmids. J Bacteriol. 1993;175:6476–6483. [PMC free article: PMC206756] [PubMed: 7691796]
- 48.
- Asano K, Mizobuchi K. An RNA pseudoknot as the molecular switch for translation of the repZ gene encoding the replication initiator of IncIα plasmid ColIb-P9. J Biol Chem. 1998;273:11815–11825. [PubMed: 9565606]
- 49.
- Ravnum S, Andersson DI. An adenosyl-cobalamin (coenzyme-B12)-repressed translational enhancer in the cob mRNA of Salmonella typhimurium. Mol Microbiol. 2001;39:1585–1594. [PubMed: 11260475]
- 50.
- Hartz D, McPheeters DS, Gold L. Influence of mRNA determinants on translation initiation in Escherichia coli. J Mol Biol. 1991;218:83–97. [PubMed: 1705985]
- 51.
- Nivinskas R, Malys N, Klausa V. et al. Post-transcriptional control of bacteriophage T4 gene 25 expression: mRNA secondary structure that enhances translational initiation. J Mol Biol. 1999;288:291–304. [PubMed: 10329143]
- 52.
- Ringquist S, MacDonald M, Gibson T. et al. Nature of the ribosomal mRNA track: analysis of ribosome-binding sites containing different sequences and secondary structures. Biochemistry. 1993;32:10254–10262. [PubMed: 7691171]
- 53.
- Murakawa GJ, Nierlich DP. Mapping the lacZribosome binding site by RNA footprinting. Biochemistry. 1989;28:8067–8072. [PubMed: 2481494]
- 54.
- Montessano-Roditis L, Glitz DG. Tracing the path of messenger RNA on the Escherichia coli small ribosomal subunit. J Biol Chem. 1994;269:6458–6470. [PubMed: 8119997]
- 55.
- Bhangu R, Wollenzien P. The mRNA track in the Escherichia coli ribosome for mRNAs of different sequences. Biochemistry. 1992;31:5937–5944. [PubMed: 1610836]
- 56.
- Gabashvili IS, Agrawal RK, Grassucci R. et al. Structure and structural variations of the Escherichia coli 30 S ribosomal subunit as revealed by three-dimensional cryo-electron microscopy. J Mol Biol. 1999;286:1285–1291. [PubMed: 10064696]
- 57.
- Schluenzen F, Tocilj A, Zarivach R. et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. [PubMed: 11007480]
- 58.
- Ogle JM, Brodersen DE, Clemons WM. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. [PubMed: 11340196]
- 59.
- Yusupova GZ, Yusupov MM, Cate JDH. et al. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. [PubMed: 11511350]
- 60.
- Groeneveld H. Secondary Structure of Bacteriophage MS2 RNA: Translational Control by Kinetics of RNA Folding PhD thesis Leiden: Leiden University,1997 .
- 61.
- Beekwilder J. Secondary structure of the RNA genome of bacteriophage Qβ PhD thesisLeiden: Leiden University,1996 .
- 62.
- Olsthoorn RCL. Structure and evolution of RNA phages PhD thesis, Leiden: Leiden University,1996 .
- 63.
- Skripkin EA, Jacobson AB. A two-dimensional model at the nucleotide level for the central hairpin of coliphage Qβ RNA. J Mol Biol. 1993;233:245–260. [PubMed: 8377201]
- 64.
- Hüttenhofer A, Noller HF. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 1994;13:3892–3901. [PMC free article: PMC395302] [PubMed: 8070416]
- 65.
- Canonaco MA, Gualerzi CO, Pon CL. Alternative occupancy of a dual ribosome binding site by mRNA affected by translation initiation factors. Eur J Biochem. 1989;182:501–506. [PubMed: 2666129]
- 66.
- van Dieijen G, Zipori P, van Prooijen W. et al. Involvement of ribosomal protein S1 in the assembly of the initiation complex. Eur J Biochem. 1978;90:571–580. [PubMed: 361407]
- 67.
- van Duin J, Overbeek GP, Backendorf C. Functional recognition of phage RNA by 30-S ribosomal subunits in the absence of initiator tRNA. Eur J Biochem. 1980;110:593–597. [PubMed: 7002554]
- 68.
- Szer W, Leffler S. Interaction of Escherichia coli 30S ribosomal subunits with MS2 phage RNA in the absence of initiation factors. Proc Natl Acad Sci USA. 1974;71:3611–3615. [PMC free article: PMC433825] [PubMed: 4610580]
- 69.
- Boni IV, Artamonova VS, Tzareva NV. et al. Noncanonical mechanism for translational control in bacteria: synthesis of ribosomal protein S1. EMBO J. 2001;20:4222–4232. [PMC free article: PMC149162] [PubMed: 11483525]
- 70.
- Balakin A, Skripkin E, Shatsky I. et al. Transition of the mRNA sequence downstream from the initiation codon into a single-stranded conformation is strongly promoted by binding of the initiator tRNA. Biochim Biophys Acta. 1990;1050:119–123. [PMC free article: PMC7148821] [PubMed: 2207137]
- 71.
- Min Jou W, Haegeman G, Ysebaert M. et al. Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein. Nature. 1972;237:82–88. [PubMed: 4555447]
- 72.
- Berkhout B, van Duin J. Mechanism of translational coupling between coat protein and replicase genes of RNA bacteriophage MS2. Nucleic Acids Res. 1985;13:6955–6967. [PMC free article: PMC322015] [PubMed: 3840590]
- 73.
- van Himbergen J, van Geffen B, van Duin J. Translational control by a long range RNA-RNA interaction; a basepair substitution analysis. Nucleic Acids Res. 1993;21:1713–1717. [PMC free article: PMC309405] [PubMed: 8493088]
- 74.
- Licis N, van Duin J, Balklava Z. et al. Long-range translational coupling in single-stranded RNA bacteriophages: an evolutionary analysis. Nucleic Acids Res. 1998;26:3242–3246. [PMC free article: PMC147662] [PubMed: 9628925]
- 75.
- Berkhout B, van Duin J. Mechanism of translational coupling between coat protein and replicase genes of RNA bacteriophage MS2. Nucleic Acids Res. 1985;13:6955–6967. [PMC free article: PMC322015] [PubMed: 3840590]
- 76.
- Berkhout B. Translational Control Mechanisms PhD thesis Leiden: Leiden University,1986 .
- 77.
- Papanicolaou C, Gouy M, Ninio J. An energy model that predicts the correct folding of both the tRNA and the 5S RNA molecules. Nucleic Acids Res. 1984;12:31–44. [PMC free article: PMC320981] [PubMed: 6694903]
- 78.
- Peritz AE, Kierzek R, Sugimoto N. et al. Thermodynamic study of internal loops in oligoribonucleotides: symmetric loops are more stable than asymmetric loops. Biochemistry. 1991;30:6428–6436. [PubMed: 1711369]
- 79.
- Shimamoto N. One-dimensional diffusion of proteins along DNA. Its biological and chemical significance revealed by single-molecule measurements. J Biol Chem. 1999;274:15293–15296. [PubMed: 10336412]
- 80.
- Matteson RJ, Biswas SJ, Steege DA. Distinctive patterns of translational reinitiation in the lac repressor mRNA: bridging of long distances by out-of-frame translation and RNA secondary structure, effects of primary sequence. Nucleic Acids Res. 1991;19:3499–3506. [PMC free article: PMC328371] [PubMed: 1906601]
- 81.
- Fickert R, Müller-Hill B. How Lac repressor finds lac operator in vivo. J Mol Biol. 1992;226:59–68. [PubMed: 1535665]
- 82.
- Ruusala T, Crothers DM. Sliding and intermolecular transfer of the lac repressor: kinetic perturbation of a reaction intermediate by a distant DNA sequence. Proc Natl Acad Sci USA. 1992;89:4903–4907. [PMC free article: PMC49196] [PubMed: 1594591]
- 83.
- La Teana A, Gualerzi CO, Brimacombe R. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA. 1995;1:772–782. [PMC free article: PMC1369318] [PubMed: 7493323]
- 84.
- Park CS, Hillel Z, Wu CW. Molecular mechanism of promoter selection in gene transcription. I. Development of a rapid mixing-photocrosslinking technique to study the kinetics of Escherichia coli RNA polymerase binding to T7 DNA. J Biol Chem. 1982;257:6944–6949. [PubMed: 7045098]
- 85.
- Singer P, Wu CW. Promoter search by Escherichia coli RNA polymerase on a circular DNA template. J Biol Chem. 1987;262:14178–14189. [PubMed: 3308887]
- 86.
- de Smit MH, van Duin J. Translational initiation at the coat-protein gene of phage MS2: native upstream RNA relieves inhibition by local secondary structure. Mol Microbiol. 1993;9:1079–1088. [PubMed: 7934914]
- 87.
- Ruusala T, Crothers DM. Sliding and intermolecular transfer of the lac repressor: kinetic perturbation of a reaction intermediate by a distant DNA sequence. Proc Natl Acad Sci USA. 1992;89:4903–4907. [PMC free article: PMC49196] [PubMed: 1594591]
- 88.
- Stanford NP, Szczelkun MD, Marko JF, Halford SE. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19:6546–6557. [PMC free article: PMC305861] [PubMed: 11101527]
- 89.
- Novoa I, Carrasco L. Cleavage of eukaryotic translation initiation factor 4G by exogenously added hybrid proteins containing poliovirus 2Apro in HeLa cells: effects on gene expression. Mol Cell Biol. 1999;19:2445–2454. [PMC free article: PMC84037] [PubMed: 10082510]
- 90.
- Wells SE, Hillner PE, Vale RD. et al. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. [PubMed: 9702200]
- 91.
- Preiss T, Hentze MW. From factors to mechanisms: translation and translational control in eukaryotes. Curr Opin Genet Dev. 1999;9:515–521. [PubMed: 10508691]
- 92.
- Gerdes K, Poulsen LK, Thisted T. et al. The hok killer gene family in gram-negative bacteria. New Biol. 1990;2:946–956. [PubMed: 2101633]
- 93.
- Gerdes K, Gultyaev AP, Franch T. et al. Antisense RNA-regulated programmed cell death. Annu Rev Genet. 1997;31:1–31. [PubMed: 9442888]
- 94.
- Gultyaev AP, Franch T, Gerdes K. Programmed cell death by hok/sok of plasmid R1: coupled nucleotide covariations reveal a phylogenetically conserved folding pathway in the hok family of mRNAs. J Mol Biol. 1997;273:26–37. [PubMed: 9367743]
- 95.
- Franch T, Gultyaev AP, Gerdes K. Programmed cell death by hok/sok of plasmid R1: processing at the hok mRNA 3'-end triggers structural rearrangements that allow translation and antisense RNA binding. J Mol Biol. 1997;273:38–51. [PubMed: 9367744]
- 96.
- Draper D. Translational regulation of ribosomal proteins in Escherichia coli In: Ilan J, ed.Translational Regulation of Gene Expression New York: Plenum,19871–26.
- 97.
- Brunel C, Romby P, Sacerdot C. et al. Stabilised secondary structure at a ribosomal binding site enhances translational repression in E. coli. J Mol Biol. 1995;253:277–290. [PubMed: 7563089]
- 98.
- Ehresmann C, Philippe C, Westhof E. et al. A pseudoknot is required for efficient translational initiation and regulation of the Escherichia coli rpsO gene coding for ribosomal protein S15. Biochem Cell Biol. 1995;73:1131–1140. [PubMed: 8722030]
- 99.
- Nogueira T, de Smit M, Graffe M. et al. The relationship between translational control and mRNA degradation for the Escherichia coli threonyl-tRNA synthetase gene. J Mol Biol. 2001;310:709–722. [PubMed: 11453682]
- 100.
- Kolakofsky D, Weissmann C. Qβ replicase as repressor of Qβ RNA-directed protein synthesis. Biochim Biophys Acta. 1971;246:596–599. [PubMed: 5142076]
- 101.
- Kolakofsky D, Weissmann C. Possible mechanism for transition of viral RNA from polysome to replication complex. Nat New Biol. 1971;231:42–46. [PubMed: 5283386]
- 102.
- Poot RA, Tsareva NV, Boni IV. et al. RNA folding kinetics regulates translation of phage MS2 maturation gene. Proc Natl Acad Sci USA. 1997;94:10110–10115. [PMC free article: PMC23320] [PubMed: 9294171]
- 103.
- van Meerten D, Girard G, van Duin J. Translational control by delayed RNA folding: identification of the kinetic trap. RNA. 2001;7:483–494. [PMC free article: PMC1370103] [PubMed: 11333027]
- 104.
- Goelz S, Steitz JA. Escherichia coli ribosomal protein S1 recognizes two sites in bacteriophage Qbeta RNA. J Biol Chem. 1977;252:5177–5179. [PubMed: 328498]
- 105.
- Boni IV, Isaeva DM, Musychenko ML. et al. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. [PMC free article: PMC333546] [PubMed: 2011495]
- 106.
- de Smit MH, van Duin J. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J Mol Biol. 1994;244:144–150. [PubMed: 7966326]
- 107.
- Olsthoorn RC, Zoog S, van Duin J. Coevolution of RNA helix stability and Shine-Dalgarno complementarity in a translational start region. Mol Microbiol. 1995;15:333–339. [PubMed: 7746154]
- 108.
- Gualerzi C, Risuleo G, Pon CL. Initial rate kinetic analysis of the mechanism of initiation complex formation and the role of initiation factor IF-3. Biochemistry. 1977;16:1684–1689. [PubMed: 322704]
- 109.
- Katunin VI, Semenkov YP, Makhno VI. et al. Comparative study of the interaction of polyuridylic acid with 30S subunits and 70S ribosomes of Escherichia coli. Nucleic Acids Res. 1980;8:403–421. [PMC free article: PMC327275] [PubMed: 6999461]
- 110.
- Pon CL, Gualerzi CO. Mechanism of protein biosynthesis in prokaryotic cells: Effect of initiation factor IF1 on the initial rate of 30S initiation complex formation. FEBS Lett. 1984;175:203–207. [PubMed: 6383865]
- 111.
- Ellis S, Conway TW. Initial velocity kinetic analysis of 30S initiation complex formation in an in vitro translation system derived from Escherichia coli. J Biol Chem. 1984;259:7607–7614. [PubMed: 6376491]
- 112.
- Calogero RA, Pon CL, Canonaco MA. et al. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc Natl Acad Sci USA. 1988;85:6427–6431. [PMC free article: PMC281985] [PubMed: 3045816]
- 113.
- Czworkowski J, Odom OW, Hardesty B. Fluorescence study of the topology of messenger RNA bound to the 30S ribosomal subunit of Escherichia coli. Biochemistry. 1991;30:4821–4830. [PubMed: 2029524]
- 114.
- Mathews DH, Sabina J, Zuker M. et al. Expanded Sequence Dependence of Thermodynamic Parameters Improves Prediction of RNA Secondary Structure. J Mol Biol. 1999;288:911–940. [PubMed: 10329189]
- 115.
- Olsthoorn RC, Licis N, van Duin J. Leeway and constraints in the forced evolution of a regulatory RNA helix. EMBO J. 1994;13:2660–2668. [PMC free article: PMC395140] [PubMed: 8013465]
- 116.
- Olsthoorn RC, van Duin J. Random removal of inserts from an RNA genome: selection against single-stranded RNA. J Virol. 1996;70:729–736. [PMC free article: PMC189873] [PubMed: 8551609]
- 117.
- Olsthoorn RC, van Duin J. Evolutionary reconstruction of a hairpin deleted from the genome of an RNA virus. Proc Natl Acad Sci USA. 1996;93:12256–12261. [PMC free article: PMC37977] [PubMed: 8901567]
- 118.
- Klovins J, van Duin J, Olsthoorn RC. Rescue of the RNA phage genome from RNase III cleavage. Nucleic Acids Res. 1997;25:4201–4208. [PMC free article: PMC147046] [PubMed: 9336447]
- 119.
- Klovins J, Tsareva NA, de Smit MH. et al. Rapid evolution of translational control mechanisms in RNA genomes. J Mol Biol. 1997;265:372–384. [PubMed: 9034357]
- 120.
- Licis N, van Duin J, Balklava Z. et al. Long-range translational coupling in single-stranded RNA bacteriophages: an evolutionary analysis. Nucleic Acids Res. 1998;26:3242–3246. [PMC free article: PMC147662] [PubMed: 9628925]
- 121.
- Licis N, Balklava Z, Van Duin J. Forced retroevolution of an RNA bacteriophage. Virology. 2000;271:298–306. [PubMed: 10860884]
- 122.
- van Meerten D, Olsthoorn RC, van Duin J. et al. Peptide display on live MS2 phage: restrictions at the RNA genome level. J Gen Virol. 2001;82:1797–1805. [PubMed: 11413393]
- 123.
- Wintermeyer W, Gualerzi C. Effect of Escherichia coli initiation factors on the kinetics of N-AcPhe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry. 1983;22:690–694. [PubMed: 6340723]
- 124.
- Kuthan H. Self-organisation and orderly processes by individual protein complexes in the bacterial cell. Prog Biophys Mol Biol. 2001;75:1–17. [PubMed: 11311713]
- A Prelude to Translational (Re)Initiation - Madame Curie Bioscience DatabaseA Prelude to Translational (Re)Initiation - Madame Curie Bioscience Database
- Adding Complexity to Phagocytic Signaling: Phagocytosis-Associated Cell Response...Adding Complexity to Phagocytic Signaling: Phagocytosis-Associated Cell Responses and Phagocytic Efficiency - Madame Curie Bioscience Database
- Asparaginyl-tRNA Synthetases - Madame Curie Bioscience DatabaseAsparaginyl-tRNA Synthetases - Madame Curie Bioscience Database
- Therapies of Nonsense-Associated Diseases - Madame Curie Bioscience DatabaseTherapies of Nonsense-Associated Diseases - Madame Curie Bioscience Database
- Coronin: The Double-Edged Sword of Actin Dynamics - Madame Curie Bioscience Data...Coronin: The Double-Edged Sword of Actin Dynamics - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...