NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Autophagy is a lysosomal degradation pathway for cytoplasmic material, which is activated during stress conditions such as amino acid starvation or viral infection. Yeast mutants defective in autophagy do not survive starvation. Mammalian cells use autophagy during short periods of starvation to degrade nonessential cellular components in order to liberate nutrients for vital biosynthetic reactions. Recent results have shown that autophagy also contributes to development, growth regulation and cancer, as well as longevity. Although the autophagic pathway was described several decades ago, the first molecular mechanisms have only been recently revealed due to the discovery of autophagy-defective yeast mutants.
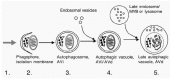
Depending on the delivery route of the cytoplasmic material to the lysosomal lumen, three different autophagic routes are known: 1) macroautophagy, or simply autophagy, 2) microautophagy (Fig. 1), and 3) chaperone-mediated autophagy. In macroautophagy, a portion of cytoplasm to be degraded is first wrapped inside a specialised organelle, the autophagosome, which then fuses with lysosomal vesicles and delivers the engulfed cytoplasm for degradation1 (Fig. 2). In microautophagy, the lysosomal membrane itself sequesters a portion of cytoplasm by a process that resembles pinching off of phagosomes or pinosomes from the plasma membrane.2 Macroautophagic uptake of cytoplasmic material appears to be a nonselective process;3 organelles are sequestered at the same frequency as they exist in the cytoplasm. In chaperone-mediated autophagy, proteins possessing a specific sequence signal are transported from the cytoplasm, through the lysosomal membrane, to the lysosomal lumen4 (Chapter 15). A fourth autophagic route, crinophagy, has also been described.5 In crinophagy, secretory vesicles directly fuse with lysosomes, which leads to degradation of the granule contents. This review concentrates on the (macro)autophagic pathway in mammalian cells.
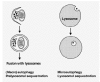
Figure 1
The different ways of sequestration of cytoplasmic material in macro- and microautophagy. See text for further details.
Macroautophagic Pathway
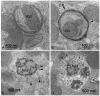
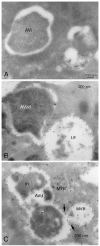
After induction by a stress signal such as amino acid starvation, the first step in autophagy is the formation of an autophagosome (Figs. 2, 3A). A flat membrane cistern elongates and wraps itself around a portion of cytoplasm, forming a double-membrane bound autophagosome. Autophagosomes next receive lysosomal constituents, such as lysosomal membrane proteins and proton pumps, from endosomal vesicles via vesicle-mediated transport. Finally the outer limiting membrane of the autophagosome fuses with the limiting membrane of a late endosome, multivesicular body (MVB), or lysosome612 (Figs. 2, 3C). In this process the cytoplasm, still engulfed by the inner limiting membrane, is delivered to the endo/lysosomal lumen (Figs. 2, 3D). Both the cytoplasm and the membrane around it are then degraded by lysosomal hydrolases, and the degradation products are transported back to cytoplasm where they can be reused for metabolism. By definition, autophagosomes, also called initial autophagic vacuoles (AVi), do not yet contain lysosomal membrane proteins or enzymes, and they are not acidic13 (Fig. 4A, 5). During the maturation process autophagosomes develop into late, or degradative autophagic vacuoles (AVd), which are acidic and contain lysosomal membrane proteins and enzymes10 (Fig. 4B, C). The latter have also been called autolysosomes . Quantitative immunoelectron microscopy has been used to demonstrate the enrichment of lysosomal membrane proteins and enzymes in late autophagic vacuoles.14,15 The lack of integral membrane proteins in autophagosomes was first revealed by freeze-fracture electron microscopy. Other cellular membranes such as lysosomal and endoplasmic reticulum membranes contain numerous integral membrane particles, considered to represent integral membrane protein molecules revealed by the freeze fracture procedure. However, the surfaces of the membranes limiting autophagosomes are almost completely smooth16,17,18 (Fig. 5).
The origin of the membrane cistern forming new autophagosomes has been the subject of numerous studies, but still this issue is unresolved in mammalian cells. Many older ultrastructural studies suggested that smooth endoplasmic reticulum (ER) cisternae are the source of autophagosome membranes,19 but evidence against this interpretation has also been published.20 Recent studies in yeast have revealed that autophagosomes originate from a unique compartment called the preautophagosomal structure (PAS).21,22 Because the autophagic pathway and genes involved in it are well conserved from yeast to mammals, it is likely that a similar unique compartment is the source of membrane in mammalian cells. Interestingly, ER cisternae are often observed on both sides of autophagosome membranes (Fig. 3B). Although in yeast membrane transport out of the ER seems to contribute to autophagosome formation,23 in mammalian cells this does not seem to be the case.24 Thus the possible function of the close ER contacts during autophagosome formation is still unknown.
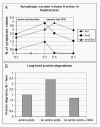
The maturation of autophagosomes to late autophagic vacuoles is a relatively quick process. Both AVis and AVds are detected soon after the beginning of amino acid starvation. The half-life of autophagosomes has been estimated to be only 8 minutes.25 When amino acid starvation is stopped and the cells are chased in full culture medium with fetal calf serum (FCS), the degradation and disappearance of the autophagic vacuoles can be followed by quantitative electron microscopy (Fig. 6A). Autophagosomes disappear first, while late autophagic vacuoles last longer. At the same time the volume fraction of endo/lysosomes undergoes a pulse-like increase reflecting the maturation of AVds to endo/lysosomes. It should be noted that the cells in Figure 6A were chased in the presence of 3-methyladenine, which may have prolonged the half-life of the AVds. When HeLa cells were first starved for amino acids for 2 h, and then chased in full medium for another 2h (without 3-methyladenine), both AVis and Avds had completely disappeared, indicating a faster consumption of AVds.
Autophagosome formation is partially inhibited by drugs that disturb actin polymerisation,26suggesting that actin filaments may have a role in this process. Microtubule inhibitors like vinblastine and nocodazole retard maturation of autophagosomes, causing either AVis or AVds, or both, to accumulate.26,27 It is possible that intact microtubules are needed for movement of autophagosomes and/or lysosomal vesicles, which in turn is necessary for fusion.
Induction and Regulation of Macroautophagy
The best known inducer of macroautophagy in mammalian cells is starvation of serum and amino acids. First autophagosomes can be detected already after 15-30 min of starvation.28 When measured by quantitative electron microscopy, the level of autophagic vacuoles reaches a steady state level in 2 - 4 h, and then it slowly starts to decrease.29 During long-term starvation, chaperone-mediated autophagy increases and autophagy decreases.30,31 This way the cell can prevent wasting too much of its cytoplasm via the nonselective macroautophagic pathway. In cultured cells, starvation of serum, but not amino acids, also increases the cytoplasmic volume fraction of autophagic vacuoles, but less than starvation of both serum and amino acids.29 Viral infections, especially Herpes simplex virus, also may induce autophagy.32
The signal transduction pathways leading to autophagy activation during amino acid starvation have received attention lately. Two amino acid sensors have been described: the protein kinase Gcn2, which is activated by uncharged transfer RNA molecules,32 and an amino acid receptor located on the plasma membrane.33 The signals from Gcn2 to autophagy are mediated by a pathway including the eukaryotic initiation factor eIF2α, which supports autophagy when phosphorylated at Ser51.32 eIF2α could thus act as a switch between protein synthesis (in the presence of amino acids) and autophagic protein degradation (in the absence of amino acids). The putative amino acid receptor at the plasma membrane seems to mediate the signals from an extracellular nontransportable leucine analogue to the autophagic pathway.33 Future studies will show whether both suggested amino acid sensors are active in the same cells, and whether different amino acids use different signal transduction pathways to regulate autophagy and protein synthesis.
In yeast, the protein kinase TOR plays a crucial role in autophagy regulation. Active TOR kinase inhibits autophagy, and inhibition of the kinase activity by rapamycin activates autophagy. 34 In mammalian cells the role of mTOR in autophagy regulation is less clear, the induction of autophagy by rapamycin is weaker than that by amino acid starvation.35 It is possible that some regulatory pathways, such as insulin, include mTOR, while others, such as amino acids, do not.33 Phosphatidylinositol 3-kinases (PI3-kinases) also regulate autophagy. The activity of class III PI3-kinase Vps34 is absolutely required for autophagosome formation29,36 and accordingly this process is completely inhibited by PI3-kinase inhibitors including 3-methyladenine (3MA),37 wortmannin and LY294002,38 as well as microinjection of inhibitory antibodies against Vps34.29 Interestingly, class I PI3-kinases have an opposite effect, the activation of these kinases inhibits autophagy.36 Since the activities of class I PI3-kinases promote cell growth and these kinases are upregulated in many cancers, it is possible that this signalling pathway might be linked to the role of autophagy in growth control. Recent extensive reviews on the signalling pathways controlling autophagy are available.39,40
Functions of Autophagy
Autophagy is a stress response. The role of autophagy as a survival mechanism during short-term amino acid starvation was discussed above. Induction of autophagy during Herpes simplex virus infections and localisation of viral particles inside autophagic vacuoles has been proposed to indicate that autophagy acts as a host-defence mechanism in the infected cells.32 Interestingly, the Herpes virus virulence protein, ICP34.5, was shown to inhibit autophagy, suggesting that the virus has developed a way to prevent the autophagic defence of the host cell. Autophagy may also help cells to defend against some intracellular bacteria.41
In muscle and heart cells autophagy seems to have a special housekeeping role in turnover of cytoplasmic constituents including mitochondria. This is revealed by myopathy and cardiomyopathy in patients and mice possessing a defective autophagic degradation due to deficiency of the lysosomal membrane protein LAMP-215,42,43 (Chapter 4). In addition autophagy has a role in growth regulation, as suggested by decreased autophagy during growth of kidney after unilateral nephrectomy.44 Further, impaired autophagy has been proposed to contribute to cancer development. Beclin 1, a mammalian homologue of the yeast autophagy gene Apg6/ Vps30, is monoallelically deleted in a large proportion of human breast and ovarian cancers. Overexpression of beclin 1 in a breast cancer cell line increases autophagy and decreases the growth and tumorigenicity of these cells.45 Further, mice homoallelically deleted of beclin 1 have less autophagy and more tumors than control mice.46,47 In addition beclin 1-/- mouse embryos, which are completely unable to perform autophagy, die around embryonic day 8,47 suggesting that autophagy has important roles in early embryonic development. Finally, autophagy may also contribute to longevity. Long-term reduced caloric intake increases longevity in several species. Recent evidence suggests that increased autophagic turnover of cytoplasmic constituents including mitochondria may contribute to the longer life in the long-term dieting animals.48 Further evidence that autophagy contributes to longevity has come from C. elegans mutants possessing a defective insulin receptor (daf2 mutant), which live longer than control worms. The increased lifetime of these mutant worms was recently shown to depend on a functional autophagic pathway.49
Autophagy also has roles in apoptosis, or programmed cell death. Type II programmed cell death, or autophagic cell death, has been described in mammary carcinoma cells.50,51 The so-called death kinase and tumor suppressor, DAPk, may function in the signalling pathway that links autophagy to cell death.52 In this process autophagy is needed for the cell death program. It is also possible that autophagy may protect cells against another type of apoptosis, type I programmed cell death.53
Autophagy Genes
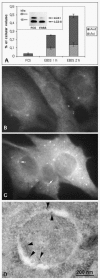
Yeast genetics has immensely enlarged our knowledge of the molecular mechanisms involved in autophagy. Today, at least 27 autophagy-related yeast genes are known, and numerous reviews have been recently written on their functions.5456 Originally these genes were called Apg, Aut, or Cvt genes, but a consensus has now been reached, with all autophagy-related genes being described by a common name, Atg.57 Several mammalian homologues of yeast autophagy genes have been identified (summarised in Table 1), and this has shown that the mechanisms of yeast autophagy are conserved in mammals. The first autophagosome marker protein, MAP LC3, was described recently.28 This protein was originally identified as a microtubule associated protein and named ‘microtubule-associated- protein-light-chain-3’. LC3 is a small 16-18 kDa protein that is soluble in nonstarved cells, but becomes peripherally membrane-associated during amino acid starvation (Fig. 7B, C). By immunoelectron microscopy LC3 has been shown to associate to the inner and outer limiting membranes of autophagosomes28 (Fig. 7D). During maturation of autophagosomes to autolysosomes, LC3 seems to dissociate from the outer limiting membrane.28 In Western blots, two forms of LC3 are seen, LC3I and LC3II (Fig. 7A). After cell homogenisation and differential centrifugation, LC3I is found in the soluble fraction, and LC3II in the pelletable membrane fraction.28 In yeast, and probably also in mammals, the membrane association is mediated by a covalent conjugation to a lipid, phosphatidylethanolamine.58 Both LC3I and LC3II are seen in nonstarved cells, but during autophagy induction the proportion of LC3II increases28 (Fig. 7A).
Table 1
Known mammalian autophagy genes.
Another mammalian autophagy protein, Apg5/Atg5, was also recently identified and characterised.59 Apg5 localises to small crescent-shaped membrane structures that may be the mammalian preautophagosomal structure, the compartment that delivers membrane for newly forming autophagosomes. In developing autophagosomes, Apg5 localises mainly to the cytoplasmic side of the outer limiting membrane. Apg5 dissociates from the membranes of sealed autophagosomes. Interestingly, Apg5 is conjugated covalently to Apg12/Atg12 by a lysine located in the middle of Apg5.60 These conjugates are then linked to a polymer by a third protein, Apg16L.61,62 It has been proposed that this polymer acts as membrane coat, which assists membrane curvature during autophagosome formation.
The tumor suppressor gene beclin 1 is the mammalian homolog of Apg6/Atg6. Interestingly, like the yeast Apg6, beclin 1 forms a complex with the class III PI3-kinase Vps34.63,64 Vps34 function is needed for autophagosome formation,29,36 although the precise role of this kinase is not known.
The remainder of the known mammalian autophagy genes (Table 1) are enzymes or other factors assisting in LC3 processing or modification of LC3I to LC3II, or conjugation of Apg5 to Apg12.
Quantitation of Autophagy
The classical ways to measure autophagy are quantitative electron microscopy and the degradation rate of long-lived proteins. Although electron microscopy is work-intensive and requires dedicated personnel and expensive equipment, it is by far the most sensitive and accurate way to detect the induction of autophagy (Fig. 7A). With most continuous cultured cell lines, the cytoplasmic volume fraction of autophagic vacuoles increases up to 10-fold during a 2-h amino acid starvation32 (Fig. 7A). At the same time, the degradation rate of long-lived proteins increases up to two-fold.32 However, to find out how much of the increased proteolysis is due to autophagy, an additional sample incubated with an autophagy inhibitor (3-methyladenine or wortmannin) must be included. Autophagic protein degradation is then revealed as the difference between the amino acid starved cells without and with the inhibitor32 (Fig. 6B). Quantitative electron microscopy estimates the accumulation of autophagic vacuoles, which can be elevated either due to increased formation of autophagosomes, or decreased degradation in late autophagic vacuoles. Degradation of long-lived protein is a measure of the degradation rate of cytoplasmic constituents, i.e., delivery of cytoplasm to lysosomal compartment or the capacity of lysosomes to degrade the delivered substrates. In other words, it is possible to have increased accumulation of autophagic vacuoles but decreased degradation of long-lived proteins in the same cells, or vice versa.
Another biochemical method to estimate autophagic activity was developed by Seglen. This assay measures the sequestration of a soluble cytoplasmic marker to a membrane-bound and thus sedimentable cell fraction.65,66 The cytoplasmic markers used include lactate dehydrogenase (enzyme activity assay) or a radioactive sugar introduced by electroporation (radiactivity measurement). For the assay cells are usually incubated in the presence of lysosomal enzyme inhibitors such as leupeptin to prevent loss of the marker protein after delivery to the lysosomal compartment. This assay thus follows the rate of formation of new autophagosomes but does not tell anything about the degradative capacity in late autophagic vacuoles.
The fluorescent dye, monodansylcadaverine (MDC), was suggested to be a specific marker for autophagic structures.67,68 However, several researchers have found that this dye is not specific for autophagic vacuoles. In fact the staining is punctate irrespective of whether the cells are induced for autophagy or not, although the amount of punctae may increase during starvation. 68 Thus the increase in MDC-stained punctae may indicate increased autophagy, but as such MDC is not a specific marker of autophagic vacuoles.
LC3 was introduced as the first protein that localises specifically to autophagosome membranes. A small amount of LC3 is present also in late autophagic vacuoles, because the portion of LC3II associated with the inner limiting membrane of autophagosomes is trapped inside the vacuoles.28 Immunofluorescence staining for endogenous LC3, or LC3 Western blotting, can be used to assay autophagy (Fig. 7A-C). However, as demonstrated by Figure 7A, although the cytoplasmic volume fraction of autophagic vacuoles increases 10-fold during 2-h amino acid starvation, the amount of LC3II does not change, but instead a decrease in LC3I is observed. In other cell lines such as HeLa cells the amount of LC3II seems to better correlate with the amount of autophagic vacuoles.28 It seems however that LC3II Western blotting is not as sensitive measure for autophagic activity as quantitative electron microscopy, and that it may be better suited for a limited number of cell lines or tissues. LC3 immunofluorescence is heavily dependent on high-quality antibodies, and it does not work in all cell types, possibly due to low expression levels of the protein. GFP-tagged LC3 expression can offer a way around these problems, and this approach has been successfully used in transgenic mice.46 However, when transiently expressed in cultured cells, LC3-GFP forms punctate structures also without autophagy induction. This is possibly due to autophagy induction by LC3 overexpression. The number of punctae does however seem to increase during starvation, so combined with quantitation, the fluorescence pattern of transiently expressed LC3-GFP also might be useful as an autophagy assay. In any case, as a first specific autophagosome marker, LC3 is extremely useful for helping to identify and define autophagic structures in both fluorescence and electron microscopy. Various structures have been misidentified as autophagic vacuoles in the literature, mostly because of lack of specific marker proteins as well as morphological expertise to correctly interpret conventional plastic-embedded EM images.
All in all, LC3 immunofluorescence and Western blotting may offer a rapid method for screening autophagic activity, but it is recommendable to confirm the result using one or two of the established autophagy assays, quantitative electron microscopy, long-lived protein degradation, or enzyme sequestration assay.
Guidelines for Identification of Autophagic Vacuoles in Transmission Electron Microscopy
By definition, autophagic vacuoles are membrane-bound structures that contain cytoplas m, ribosomes, endoplasmic reticulum (ER) membranes, mitochondria, or occasionally peroxisomes. Structures that do not fulfil the latter criterion should not be classified as autophagosomes or autophagic vacuoles. In other words, vesicles containing exclusively membrane whorls, small vesicles, or undefined amorphous debris should not be called autophagic vacuoles and cannot be used as an evidence of autophagic activity. When looking at plastic-embedded specimens, it is relatively easy to identify autophagic vacuoles because ribosomes have a good contrast after staining with uranyl acetate and lead citrate. Structures surrounded by a single, double, or multiple membrane, containing clearly identifiable cytoplasmic constituents, are autophagosomes or autophagic vacuoles (Fig. 3A-D). The morphology of cytoplasmic constituents is not so clear in late autophagic vacuoles, but after careful examination it is possible to recognise partially degraded rough ER remnants (Fig. 3D, asterisks) or mitochondria. Mitochondria can be used also for identification in cryosections, where the ribosomal contrast is very low (Fig. 4C, m). Immunogold labelling of a cytoplasmic protein6 to demonstrate the cytoplasmic contents, or of LC3 (Fig. 7D), can also be used for identification of autophagic vacuoles in cryosections.
Future of Autophagy
More than thirty years after the initial description of autophagy in mammalian cells by De Duve,69 we finally have the first specific marker proteins and genetic tools to elucidate the mechanisms and functions of this fascinating cellular process. Many other mammalian homologs of the yeast autophagy genes are likely to be identified in the near future. This will further assist in identifying autophagic structures and in correctly estimating the autophagic activity in cell lines and tissues. The known physiological roles and functions of autophagy are thus likely to increase in number and diversity.
Acknowledgements
I thank Paul Saftig and Paul Steven Hartley for helpful comments on the manuscript, Isei Tanida and Takashi Ueno for LC3 antibodies, and Stefenie Jäger for providing material for Figure 7. The work presented in this review was supported by Jenny and Antti Wihuri Foundation and The Royal Society.
References
- 1.
- Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53:687–733. [PMC free article: PMC2013521] [PubMed: 4300890]
- 2.
- Ahlberg J, Marzella L, Glaumann H. Uptake and degradation of proteins by isolated rat liver lysosomes. Suggestion of a microautophagic pathway of proteolysis. Lab Invest. 1982:523–32. [PubMed: 6755063]
- 3.
- Kopitz J, Kisen GO, Gordon PB. et al. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol. 1990;111:941–53. [PMC free article: PMC2116292] [PubMed: 2391370]
- 4.
- Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–03. [PubMed: 8662539]
- 5.
- Glaumann H. Crinophagy as a means for degrading excess secretory proteins in rat liver. Revis Biol Celular. 1989;20:97–110. [PubMed: 2700099]
- 6.
- Liou W, Geuze HJ, Geelen MJH. et al. The autophagic and endocytic pathways converge at the nascent autophagic vacuole. J Cell Biol. 1997;136:61–70. [PMC free article: PMC2132457] [PubMed: 9008703]
- 7.
- Berg TO, Fengsrud M, Stromhaug PE. et al. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–92. [PubMed: 9705327]
- 8.
- Gordon PB, Hoyvik H, Seglen PO. Prelysosomal and lysosomal connections between autophagy and endocytosis. Biochem J. 1992;283:361–69. [PMC free article: PMC1131042] [PubMed: 1575680]
- 9.
- Lawrence BP, Brown WJ. Autophagic vacuoles rapidly fuse with preexisting lysosomes in cultured hepatocytes. J Cell Sci. 1992;102:515–26. [PubMed: 1324248]
- 10.
- Dunn WA. Studies on the mechanisms of autophagy: Maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–45. [PMC free article: PMC2116125] [PubMed: 2161853]
- 11.
- Punnonen EL, Autio S, Kaija H. et al. Autophagic vacuoles fuse with the prelysosomal compartment in cultured rat fibroblasts. Eur J Cell Biol. 1993;61:54–66. [PubMed: 8223708]
- 12.
- Tooze J, Hollinshead M, Ludwig T. et al. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J Cell Biol. 1990;111:329–45. [PMC free article: PMC2116176] [PubMed: 2166050]
- 13.
- Dunn WA. Studies on the mechanisms of autophagy: Formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–33. [PMC free article: PMC2116114] [PubMed: 2351689]
- 14.
- Eskelinen EL, Illert AL, Tanaka Y. et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13:3355–68. [PMC free article: PMC124165] [PubMed: 12221139]
- 15.
- Tanaka Y, Guhde G, Suter A. et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2 -deficient mice. Nature. 2000;406:902–06. [PubMed: 10972293]
- 16.
- Rez G, Meldolesi J. Freeze-fracture of drug-induced autophagocytosis in the mouse exocrine pancreas. Lab Invest. 1980;43:269–77. [PubMed: 7401637]
- 17.
- Fengsrud M, Erichsen ES, Berg TO. et al. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur J Cell Biol. 2000;79:871–82. [PubMed: 11152279]
- 18.
- Punnonen EL, Pihakaski K, Mattila K. et al. Intramembrane particles and filipin labelling on the membranes of autophagic vacuoles and lysosomes in mouse liver. Cell Tissue Res. 1989;258:269–76. [PubMed: 2582478]
- 19.
- Dunn WA. Autophagy and related mechanisms of lysosomal-mediated protein degradation. Trends Cell Biol. 1994;4:139–43. [PubMed: 14731737]
- 20.
- Yamamoto A, Masaki R, Fukui Y. et al. Absence of cytochrome P-450 and presence of autolysosomal membrane antigens on the isolation membranes and autophagosomal membranes in rat hepatocytes. J Histochem Cytochem. 1990;38:1571–81. [PubMed: 2212617]
- 21.
- Kim J, Huang WP, Stromhaug PE. et al. Convergence of multiple autophagy and cytoplasm to vacuole components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–73. [PMC free article: PMC2754695] [PubMed: 11675395]
- 22.
- Suzuki K, Kirisako T, Kamada Y. et al. The preautophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. [PMC free article: PMC125692] [PubMed: 11689437]
- 23.
- Hamasaki M, Noda T, Ohsumi Y. The early secretory pathway contributes to autophagy in yeast. Cell Struct Funct. 2003;28:49–54. [PubMed: 12655150]
- 24.
- Purhonen P, Pursiainen K, Reunanen H. Effects of brefeldin A on autophagy in cultured rat fibroblasts. Eur J Cell Biol. 1997;74:63–67. [PubMed: 9309391]
- 25.
- Papadopoulos T, Pfeifer U. Regression of rat liver autophagic vacuoles by locally applied cycloheximide. Lab Invest. 1986;54:100–07. [PubMed: 3941538]
- 26.
- Aplin A, Jasionowski T, Tuttle DL. et al. Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol. 1992;152:458–66. [PubMed: 1506410]
- 27.
- Arstila AU, Nuuja IJ, Trump BF. Studies on cellular autophagocytosis. Vinblastine-induced autophagy in the rat liver. Exp Cell Res. 1974;87:249–52. [PubMed: 4415937]
- 28.
- Kabeya Y, Mizushima N, Ueno T. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–28. [PMC free article: PMC305793] [PubMed: 11060023]
- 29.
- Eskelinen EL, Prescott AR, Cooper J. et al. Inhibition of autophagy in mitotic animal cells. Traffic. 2002;3:878–893. [PubMed: 12453151]
- 30.
- Cuervo AM, Dice JF. Lysosomes, a meeting point of proteins, chaperones, and proteases. J Mol Med. 1998;76:6–12. [PubMed: 9462863]
- 31.
- Fuertes G, Martin De Llano JJ, Villarroya A. et al. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. [PMC free article: PMC1223664] [PubMed: 12841850]
- 32.
- Talloczy Z, Jiang W, Virgin HW IV. et al. Regulation of starvation and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99:190–95. [PMC free article: PMC117537] [PubMed: 11756670]
- 33.
- Kanazawa T, Taneike I, Akaishi R. et al. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes J Biol Chem Nov 10 2003[Epub ahead of print]. [PubMed: 14610086]
- 34.
- Kamada Y, Funakoshi T, Shintani T. et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. [PMC free article: PMC2150712] [PubMed: 10995454]
- 35.
- Blommaart EFC, Luiken JJFP, Blommaart PJE. et al. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–26. [PubMed: 7836465]
- 36.
- Petiot A, Ogier-Denis E, Blommaart EFC. et al. Distinct classes of phosphatidyl-inositol 3'-kinase are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–98. [PubMed: 10625637]
- 37.
- Seglen PO, Gordon PB. 3-methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–92. [PMC free article: PMC346086] [PubMed: 6952238]
- 38.
- Blommaart EFC, Krause U, Schellens JPM. et al. The phosphatidyl-inositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–46. [PubMed: 9030745]
- 39.
- Ogier-Denis E, Codogno P. Autophagy: A barrier or an adaptive response to cancer. Biochim Biophys Acta. 2003;1603:113–28. [PubMed: 12618311]
- 40.
- Petiot A, Pattingre S, Arico S. et al. Diversity of signaling controls of macroautophagy in mammalian cells. Cell Struct Funct. 2002;27:431–41. [PubMed: 12576636]
- 41.
- Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–68. [PubMed: 12814436]
- 42.
- Nishino I, Fu J, Tanji K. et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature. 2000;406:906–10. [PubMed: 10972294]
- 43.
- Nishino I. Autophagic vacuolar myopathies. Curr Neurol Neurosci Rep. 2003;3:64–69. [PubMed: 12507414]
- 44.
- Jurilj N, Pfeifer U. Inhibition of cellular autophagy in kidney tubular cells stimulated to grow by unilateral nephrectomy. Virchovs Arch B Cell Pathol Incl Mol Pathol. 1990;59:32–37.
- 45.
- Liang XH, Jackson S, Seaman M. et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–76. [PubMed: 10604474]
- 46.
- Qu X, Yu J, Bhagat G. et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. [PMC free article: PMC297002] [PubMed: 14638851]
- 47.
- Yue Z, Jin S, Yang C. et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–82. [PMC free article: PMC299911] [PubMed: 14657337]
- 48.
- Bergamini E, Cavallini G, Donati A. et al. The anti-ageing effects of caloric restriction may involve stimulation of macroautophagy and lysosomal degradation, and can be intensified pharmacologically. Biomedicine and pharmacotherapy. 2003;57:203–08. [PubMed: 12888255]
- 49.
- Melendez A, Talloczy Z, Seaman M. et al. Autophagy genes are essential for dauer development and lifespan extension in C. elegans. Science. 2003;301:1387–91. [PubMed: 12958363]
- 50.
- Bursch W, Ellinger A, Kienzl H. et al. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: The role of autophagy. Carcinogenesis. 1996;17:1595–607. [PubMed: 8761415]
- 51.
- Bursch W. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ. 2001;8:569–81. [PubMed: 11536007]
- 52.
- Inbal B, Bialik S, Sabanay I. et al. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157:455–68. [PMC free article: PMC2173279] [PubMed: 11980920]
- 53.
- Bauvy C, Gane P, Arico S. et al. Autophagy delays sulindac sulfide-induced apoptosis in the human intestinal colon cancer cell line HT-29. Exp Cell Res. 2001;15:139–49. [PubMed: 11478840]
- 54.
- Huang WP, Klionsky DJ. Autophagy in yeast: A review of the molecular machinery. Cell Struct Funct. 2002;27:409–20. [PubMed: 12576634]
- 55.
- Noda T, Suzuki K, Ohsumi Y. Yeast autophagosomes: De novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–35. [PubMed: 12062171]
- 56.
- Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article: PMC1430730] [PubMed: 12865942]
- 57.
- Klionsky DJ, Cregg JM, Dunn WAJ. et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. [PubMed: 14536056]
- 58.
- Ichimura Y, Kirisako T, Takao T. et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. [PubMed: 11100732]
- 59.
- Mizushima N, Yamamoto A, Hatano M. et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–67. [PMC free article: PMC2195787] [PubMed: 11266458]
- 60.
- Mizushima N, Sugita H, Yoshimori T. et al. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autopagy. J Biol Chem. 1998;273:33889–92. [PubMed: 9852036]
- 61.
- Mizushima N, Kuma A, Kobayashi Y. et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. [PubMed: 12665549]
- 62.
- Mizushima N, Yoshimori T, Ohsumi Y. Role of the Apg12 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2003;35:553–61. [PubMed: 12672448]
- 63.
- Kihara A, Kabeya Y, Ohsumi Y. et al. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Reports. 2001;2:330–35. [PMC free article: PMC1083858] [PubMed: 11306555]
- 64.
- Tassa A, Roux MP, Attaix D. et al. Class III phosphoinositide 3-kinase-beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J Sep 11. 2003;376:577–586. [PMC free article: PMC1223813] [PubMed: 12967324]
- 65.
- Hoyvik H, Gordon PB, Berg TO. et al. Inhibition of autophagic-lysosomal delivery and autophagic lactolysis by asparagine. J Cell Biol. 1991;113:1305–12. [PMC free article: PMC2289037] [PubMed: 1904444]
- 66.
- Seglen PO, Gordon PB, Tolleshaug H. et al. Use of [3H]raffinose as a specific probe of autophagic sequestration. Exp Cell Res. 1986;162(273-277) [PubMed: 3940229]
- 67.
- Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995:3–14. [PubMed: 7750517]
- 68.
- Munafo DB, Colombo MI. A novel assay to study autophagy: Regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114:3619–29. [PubMed: 11707514]
- 69.
- De Duve C. The lysosome in retrospectIn: Dingle JT, Fell HB, eds.Lysosomes in Biology and PathologyAmsterdam: North Holland Publ. Co.,19693–40.
- 70.
- Tanida I, Tanida-Miyake E, Komatsu M. et al. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GAPARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–44. [PubMed: 11825910]
- 71.
- Marino G, Uria JA, Puente XS. et al. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–78. [PubMed: 12446702]
- 72.
- Hemelaar J, Lelyveld VS, Kessler BM. et al. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP and Apg8L. J Biol Chem Oct 6. 2003;278:51841–51850. [PubMed: 14530254]
- 73.
- Liang XH, Yu J, Brown K. et al. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–49. [PubMed: 11309306]
- 74.
- Tanida I, Tanida-Miyake E, Ueno T. et al. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–06. [PubMed: 11096062]
- 75.
- Tanida I, Tanida-Miyake E, Nishitani T. et al. Murine Apg12p has a substrate preference for murine Apg7p over three Apg8p homologs. Biochem Biophys Res Commun. 2002;292:256–62. [PubMed: 11890701]
- 76.
- Mizushima N, Yoshimori T, Ohsumi Y. Mouse Apg10 as an Apg12-conjugating enzyme: Analysis by the conjugation-mediated yeast two-hybrid method. FEBS Lett. 2002;532:450–54. [PubMed: 12482611]
- 77.
- Nemoto T, Tanida I, Tanida-Miyake E. et al. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J Biol Chem. 2003;278:39517–26. [PubMed: 12890687]
- 78.
- Tanida I, Nishitani T, Nemoto T. et al. Mammalian Apg12p, but not the Amp12p-Apg5p conjugate, facilitates LC3 processing. Biochem Biophys Res Commun. 2002;296:1164–70. [PubMed: 12207896]
- Macroautophagy in Mammalian Cells - Madame Curie Bioscience DatabaseMacroautophagy in Mammalian Cells - Madame Curie Bioscience Database
- Regulation of Phagocytosis by FcγRIIb and Phosphatases - Madame Curie Bioscience...Regulation of Phagocytosis by FcγRIIb and Phosphatases - Madame Curie Bioscience Database
- Inflammatory Myopathies: Dermatomyositis, Polymyositis and Inclusion Body Myosit...Inflammatory Myopathies: Dermatomyositis, Polymyositis and Inclusion Body Myositis - Madame Curie Bioscience Database
- Caspases as Targets for Drug Development - Madame Curie Bioscience DatabaseCaspases as Targets for Drug Development - Madame Curie Bioscience Database
- Insights into the Modulation of Ceramide Metabolism by Naturally Occurring and S...Insights into the Modulation of Ceramide Metabolism by Naturally Occurring and Synthetic Sphingolipid Analogs as Monitored by Electrospray Tandem Mass Spectrometry - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...