NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The type 1 insulin-like growth factor receptor (IGF-1R) is widely expressed across many cell types in fetal and postnatal tissues. Signalling through the IGF-1R is the principal pathway responsible for somatic growth in fetal mammals, while somatic growth in postnatal animals is achieved through the synergistic interaction of growth hormone (GH) and the IGFs. Forced overexpression of the IGF-1R results in the malignant transformation of cultured cells and elevated levels of IGF-1R are observed in a variety of human tumor types. Downregulation of IGF-1R levels can reverse the transformed phenotype of tumor cells, and may render them sensitive to apoptosis in vivo. These discoveries have led to the emergence of IGF-1R as a therapeutic target for the development of anti-tumor agents. This Chapter will review the key developments in our understanding of the structure of the type I insulin-like growth factor receptor drawing on parallel studies with the closely related insulin receptor. These tyrosine kinase receptors are large, transmembrane proteins consisting of several structural domains. Their ectodomains have a similar arrangement of two homologous domains (L1 and L2) separated by a Cys-rich region. The L domains consist of five and a half leucine-rich repeats. The C-terminal half of their ectodomains consists of three fibronectin type 3 repeats, and an insert domain which contains the α-βcleavage site. The cytoplasmic portion of the receptor consists of a catalytic kinase domain flanked by a juxtamembrane and C-tail region, the sites of binding of various signalling molecules. Our current knowledge on the structure of these two receptors has come from a combination of multiple sequence analyses, site specific mutagenesis and chimera studies, single-molecule electron microscope images of receptor and receptor/ligand complexes, and the 3D structures of the first three domains of the IGF-1R and the tyrosine kinase domain of the insulin receptor, determined by X-ray crystallography.
Introduction
The insulin-like growth factors (IGFs) are essential for normal fetal and postnatal growth and development. The type 1 IGF receptor (IGF-IR) binds IGF-I with high affinity and initiates the physiological response to this ligand in vivo.1 The IGF-IR also binds IGF-II, albeit with lower affinity, and is in part responsible for the mitogenic effects of this polypeptide during fetal development.2 The alternately spliced form of the insulin receptor (IR) that lacks exon 11 and is expressed in many fetal tissues has recently been identified as binding IGF-II with high affinity,3 confirming an earlier genetic study implicating the insulin receptor in the growth-promoting effects of IGF-II.4
The ligands, insulin, IGF-I and IGF-II share a common three-dimensional (3D) architecture5,6,7 and can bind to both IR and IGF-IR in a competitive manner. The hIRR ligand is unknown and IRR knockouts lack a distinguishing phenotype.8
Germline deletion of both Igf-1r alleles results in severe growth retardation during the second half of gestation.9 Fibroblast cell lines established from Igf-1r knockout mice are impaired in their progression through the cell cycle in serum-rich conditions and are resistant to oncogenic transformation by a variety of viral and cellular oncogenes.9 Conversely, overexpression of IGF-IRs promoted the neoplastic transformation of cell lines in a ligand-dependent manner.10 Thus the IGF axis has an important role to play not only in normal cellular development, but also in malignant transformation.11 As a result, the IGF-IR has emerged as a candidate therapeutic target for the treatment of human cancer.
IGF-I Receptor: Discovery and Sequence
The first evidence for the presence of an IGF receptor distinct from IR came in 1974 when 25 I-labelled insulin and 25 I-labelled NSILA-s (soluble fraction of non-suppressible insulin-like activity) were used to label distinct proteins in purified rat liver plasma membranes12 and detergent solubilized fractions.13 The IGF-IR was subsequently shown, by SDS gel electrophoresis, to be a homodimer composed of two a- and two β-chains held together by disulfide bonds.14,15 The IGF-IR is synthesized as a 180 kDa precursor which is glycosylated, dimerized and proteolytically processed to yield the mature α2β2 receptor.16 The next key discovery was the demonstration that IGF-IR is a tyrosine kinase which is activated and autophosphorylated following IGF-I binding.17,18
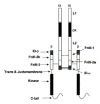
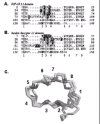
The cDNA for human (h) IGF-IR was cloned and sequenced in 1986.19 It consists of 4,989 nucleotides and codes for a 1,367 amino acid precursor (Fig. 1). The pre-proreceptor monomer includes a 30 residue signal peptide (residues -30 to -1) and an Arg.Lys.Arg.Arg furin protease cleavage site at residues 708-711, which on cleavage yields one a-chain and one α-chain. The a-chain (residues 1-707) and 195 residues of β-chain comprise the extracellular portion of the IGF-IR and contain eleven and five potential N-linked glycosylation sites, respectively.19 There is a single transmembrane sequence (residues 906-929) and a 408 residue cytoplasmic domain containing the tyrosine kinase (residues 930-1337). The cDNA for the hIR and the third member of the IR family, hIRR, have been cloned and sequenced and are similarly organized.20,21,22
The human IGF-IR gene is greater than 100 kilobasepairs in size and contains 21 exons, ten in the α-chain and eleven in the β-chain.23 An alternate human IGF-IR mRNA transcript has been reported, in which a three-basepair (CAG) deletion results in the substitution of Arg for Thr898Gly899 (Fig. 1), eight residues upstream from the start of the transmembrane region of hIGF-IR.24 The CAG- isoform shows reduced internalization and enhanced signalling properties compared to the CAG+ isoform.25
The hIGF-IR, like the hIR, is heavily glycosylated with 16 potential N-linked glycosylation sites.19,20,21 The hIR has 18 N-linked sites while hIRR, the third member of this receptor sub-family, has 11 sites.22 Most analyses have been conducted with hIR. Analytical ultracentgrifugation showed that hIR expressed in CHO-K1 cells contained 58-64 kDa of carbohydrate.26 Oligosaccharides of both the high mannose and complex type are present, the latter containing additional fucose, N-acetylglucosamine, galactose and sialic acid residues.27,28,29 O-linked glycosylation has been demonstratedonly in the β subunit of the hIR.27,30,31 Studies on the effects of removing N-linked glycosylation sites indicate that there are many redundancies in hIR glycosylation. Every site, with one exception, can be mutated individually without detriment to cell-surface expression, receptor processing and ligand binding.30-36 When combinations of sites are examined, it appears that the major domains of the receptor, particularly those closer to the N-terminus (i.e., L1, Cys-rich, L2), require at least one intact glycosylation site to ensure correct folding and processing.34
Domain Organization and Evolutionary Relationships of IGF-IR and Related Receptors
Comparative sequence analyses have revealed that many proteins, particularly eukaryotic extracellular proteins, are composed of a number of different, sometimes repeated, structural units. In the case of the IR subfamily, 11 distinct regions have been identified in each monomer (Fig. 2) The N-terminal half of the IGF-IR ectodomain contains two homologous domains (L1 and L2), separated by a Cys-rich region (Cys152 to Cys298) containing 22 cysteine residues.37,38,39 The C-terminal half of the IGF-IR ectodomain consists of three fibronectin type III (FnIII) domains, the second of which contains a large insert domain of ~120-130 residues.40,41,42,43 Intracellularly, each IGF-IR monomer contains a tyrosine kinase catalytic domain (residues 973-1229) flanked by two regulatory regions-a juxtamembrane region, residues 930-972, and an 108 residue C-tail, residues 1230-1337- that contain the phosphotyrosine binding sites for signalling molecules (Fig. 1) The C-terminal boundary of the kinase domain of IGF-IR is Phe1229 or shorter, not Ile123644 since IGF-IRs truncated at 122945 and 124346 are still catalytically active as judged by receptor autophosphorylation, the phosphorylation/activation of cellular substrates and mitogenic responses to IGF-I. The 3D structure of the hIGF-1R kinase domain has been deposited in the PDB as 1JQH.47The 3D structure of the hIR kinase domain has been described for both the inactive48 and active49 states. Phe1229, which is 10 residues downstream from the conserved CysTrp sequence located at 1218-1219 (Fig. 1), appears to be very close to the catalytic domain boundary when compared with the 3D structure of the cAMP-dependent protein kinase.48,50
Representatives of the IGF-IR/IR receptor family have been characterized in some of the simplest multicellular animals, including cnidarians (polyps and jellyfish), nematodes, gastropods and insects.43 The IGF-IR/IRs from primitive organisms such as Caenorhabditis elegans (nematode) and Drosophila melanogaster (insect) have additional sequences at their N- and C-termini.51,52 The emergence of distinct IR and IGF-IR genes appears to coincide with the evolutionary transition from protochordates to vertebrates. The protochordate amphioxus contains only a single IR-like receptor cDNA in contrast to the hagfish, considered to be the most primitive extant vertebrate, which appears to contain two IR/IGF-IR-like cDNAs.53
Secondary Structure
The α-chain of IGF-IR has a total of 38 cysteine residues, while the β-chain has three extracellular and five intracellular cysteine residues (Fig. 1). There are disulphide bonds at each end of the L1 domain (Cys3-Cys22 and Cys120-Cys148) and L2 domain (Cys302-Cys323 and Cys425-Cys458) based on chemical analysis of hIR,54,55 sequence alignments38 and the 3D structure of the first three domains of the IGF-IR.39 The Cys-rich region consists of eight disulfide-linked modules,38,39 similar to those found in the tumor necrosis factor (TNF) receptor56 and subsequently seen in laminin.57
There are at least two a-a disulphide bonds involved in the IGF-I dimer. One involves Cys514, in the first FnIII domain, linked to Cys514 in the second monomer based on chemical analysis55 and site specific mutagenesis58,59,60 of the corresponding bond in hIR. The second involves the triplet of Cys residues at positions 669, 670 and 672 in the insert domain of IGF-IR, based on chemical analyses of the corresponding region of hIR.54 It was not possible to determine which one of the three, or whether all three Cys residues were involved in dimer disulfides. The sequence around this triplet resembles those found in the hinge region of antibodies61 where multiple disulfide bonds occur. The IGF-I receptor has an additional Cys residue (Cys662), seven residues upstream of the Cys triplet in the insert domain, and lacks the Cys residue equivalent to Cys884 in hIR (Fig. 1).DrosophilaIR62,63 also lacks the cysteine residue equivalent to Cys884 in hIR as well as the equivalent of one of Cys669 or Cys670. The two reports differ with regard to the presence or absence of the cysteine residue equivalent to the Cys514, known to form one of the a-a dimer bonds.55
There is only a single α-βdisulfide link in the hIGF-IR, between Cys633 in the first FnIII repeat and Cys849 in the second FnIII repeat,54 which is consistent with the mutagenesis data for hIR60,64 and the predictions of Ward et al,38 but not those of Schaefer et al.65 The structural implications of the disulfide bond between Cys633 and Cys849 are that the two FnIII domains are aligned side by side,38 not end to end, as is the more common configuration.66 Finally, there is a single, intra-chain disulfide linkage between the β-chain residues Cys776 and Cys785 in the predicted F and G strands of the third FnIII domain based on chemical analysis of the corresponding disulfide bond in hIR.54 Interestingly, the reported sequence for rat, but not mouse, IR67 shows a Ser residue at the position equivalent to Cys785 in human IR.
3D Structure of the L1/Cys-rich/L2 Domains of the IGF-IR
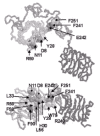
The 3D structure of the L1/Cys-rich/L2 domain fragment of the IGF-IR has been solved.39 As shown in Figure 3, the molecule adopts an extended bilobal structure (approximately 40 x 48 x 105 with the L domains at either end. The Cys-rich region runs two-thirds the length of the molecule, making contact along the length of the L1 domain but having very little contact with the L2 domain. This leaves a space at the centre of the molecule of approximately 24 diameter and of sufficient size to accommodate the ligands, IGF-I or IGF-II as shown (Fig. 3). The space is bounded on three sides by the regions of IGF-IR which are known to contribute to ligand binding, based on studies of chemical cross-linking, receptor chimeras, and natural or site-specific mutants.39
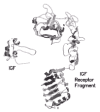
Figure 3
Polypeptide fold for residues 1-459 of the human IGF-I receptor and 1-70 of IGF-1. The L1 domain is at the top viewed from the N-terminal end. Helices are depicted as curled ribbons and β-strands as broad arrows. Based on refs. and .
The L Domains
Each L domain (residues 1-150 and 300-460) adopts a compact shape (approx. 24 x 32 x 37 being formed from a single-stranded, right-handed β-helix, capped at each end by short α-helices and a disulfide bond. The body of each L domain looks like a loaf of bread with three flat sides (β sheets) and an irregular top (Fig. 3). The two domains are superimposable.39 The repetitive nature of the β-helix is reflected in the sequence where a five-fold repeat, centered on a conserved glycine, had been identified by sequence analyses.38 The structure, however, revealed that the L domains comprise six helical turns and a fold that was quite unexpected.39
A notable difference between the L1 and L2 domains is found at their C-terminal ends. The indole ring of Trp176, from the first module in the Cys-rich region, is inserted into a pocket in the hydrophobic core of L1 formed by residues Ile98, Gly99, Leu100, and Leu103 from the fourth turn and Val125, Trp127 and Ile130 from the fifth turn of L1.39 The sequence motif of residues which form the Trp pocket in L1 does not occur in L2 of the IR/IGF-IR family. However, in EGFR, which has a second Cys-rich region after the L2 domain, the motif can be found in both the L1 and L2 domains and the Trp is conserved in the first module of both Cys-rich regions.39 It appears to contribute to the stability of the L2 domain of the EGFR since a construct that extends to the end of the first module of the second Cys-rich region (sEGFR501) binds ligand with high affinity while the shorter fragment (sEGFR476), which lacks this Cys-rich module, fails to bind ligand.68
Recently, evidence has been presented to show that the L domains are members of the leucine-rich repeat superfamily.69 Multiple sequence alignments, coupled with the 3 D structure of the L1 and L2 domains of the IGF-IR,39 enabled the residues equivalent to the eight conserved positions in the known LRR motif, LxxLxLxxNx-Lxx-Lxx-Lxx-Lxx-, to be identified (Fig. 4). Isoleucine (or valine) is preferred over leucine at some positions of the repeat and the L domains of the IR and EGFR families contain five full repeats with the 6th partially truncated.69
The Cys-rich Domain
As anticipated,38 the Cys-rich domain is composed of modules with disulfide bond connectivities resembling parts of the TNF receptor56 and laminin57 repeats. The first module sits at the end of L1 domain while the remaining seven form a curved rod running diagonally across L1 and reaching to L2 (Fig. 3). The strands in modules 2-7 run roughly perpendicular to the axis of the rod39 in a manner more akin to laminin than to the TNF receptor, where the strands run parallel to the axis (Fig. 5). The modular arrangement of the IGF-IR Cys-rich domain is different to other Cys-rich proteins for which structures are known.39 The first three modules of IGF-IR have a common core, containing a pair of disulfide bonds, but show considerable variation in the loops. These modules have been referred to as C2 (two disulfide bonds).51,52 The connectivity of the cysteines is the same as the first part of an EGF motif (Cys 1-3 and 2-4). Modules 4 to 7 have a single disulfide bond and have been referred to as C1 modules.51,52 Each C1 module is composed of three polypeptide strands, the first and third being disulfide bonded and the second and third forming a βribbon. The βribbon of each β-finger (or C1) module lines up antiparallel to form a tightly twisted eight-stranded β-sheet.39 Module 6 deviates from the common pattern with the first segment being replaced by an a-helix followed by a large loop that is implicated in ligand binding (see below). As modules 4-7 are similar, it is possible that they arose from a series of gene duplications. The final (eighth) module is a disulfide-linked bend of five residues.39
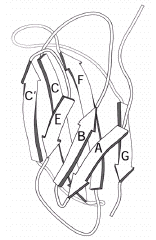
The Fibronectin Type III Domains
The FnIII domain is one of the most common structural modules found in many proteins, including membrane-anchored receptors. 3D structures have been reported for several such domains and have the fold and topology shown in Figure 6. The FnIII domain is relatively small (~100 residues) and has a fold similar to that of immunoglobulins but with a distinctive sequence motif. The domain consists of a seven-stranded β-sandwich in a three-on-four (EBA:GFCC') topology. Its main functions appear to be to mediate protein-protein interactions including ligand binding and to act as spacers to correctly position functionally important regions of extracellular proteins.
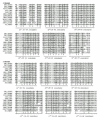
O'Bryan et al40 were the first to describe the existence of FnIII domains in members of the IR subfamily following their cloning and characterization of the tyrosine kinase axl. Their sequence alignments and descriptions covered the two C-terminal FnIII domains in the IR subfamily and these were given structural assignments by Schaffer et al65 following comparisons with the FnIII modules present in the growth hormone receptor. Recently it has been shown that members of the IR subfamily contain an additional FnIII domain in the region previously referred to as the connecting domain.41,42,43 This first FnIII domain (equivalent to residues 461-579 in IGF-IR) is 118-122 residues long, while the second FnIII domain (equivalent to 580-798 in IGF-IR) has a major insert of 120 to 130 amino acids. The third FnIII domain (equivalent to residues 799-901 in IGF-IR) is of normal size. The different authors differ in the residues assigned to the seven βstrands in each of these FnIII domains, with the regions of greatest disagreement being the locations of the C' and E strands in all three modules (Table 1).
Table 1
Caveolin-3 Mutations in Caveolinopathies.
3D Structure of the Tyrosine Kinase Domain
The crystal structure of the unphosphorylated (basal state) tyrosine kinse domain from the closely related hIR was solved by Hubbard et al48 and is shown in Figure 7. Like the protein serine kinases,50 the insulin receptor kinase is composed of two lobes with a single connection between them. The N-terminal lobe comprises a twisted β-sheet of five antiparallel βstrands (β1-β5) and one α-helix (αC). The larger C-terminal lobe comprises eight α-helices (αD, αE, αEF, αF, αG, αH, αI, αJ) and four β-strands (β7, β8, β10, β11).48 The hIR kinase lacks βstrands 6 and 9 present in cAMP protein kinase.50 In the unactivated kinase, one of the three tyrosines in the activation loop, Tyr1162, is bound in the active site but cannot be phosphorylated (in cis) because part of the A-loop interferes with the ATP binding site and the catalytic Asp1150 is improperly positioned to co-ordinate MgATP.48 On activation, autophosphorylation of Tyr1162, Tyr1158 and Tyr1163 occurs in trans by the kinase domain of the second monomer. Thus in the basal state, Tyr1162 competes with the neighboring β-chain and other protein substrates, for binding to the active site, but is not cis-phosphorylated because of steric constraints that prevent simultaneous binding of Tyr1162 and MgATP.48 The structure of the activated phosphorylated IR kinase reveals that autophosphorylation of the three tyrosines in the A-loop, leads to a dramatic change in its configuration.49 In the phosphorylated state the A-loop is displaced by approximately 30, resulting in unrestricted access to the binding sites for ATP and protein substrates.49 This movement facilitates the proper spatial arrangement of Lys1030 and Glu1047, the residues involved in MgATP coordination and Asp1150 of the highly conserved Asp-Phe-Gly triad.49 The loop A rearrangement also leads to closure of the N and C-terminal lobes, which is necessary for productive ATP binding.49 This closure involves significant rotation of the N-terminal lobe as shown in Figure 7.
The IGF-IR and IR Ectodomain Dimers
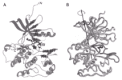
The major feature which separates the IGF-IR and other members of the IR family from most other receptor families is that they exist on the cell surface as disulfide-linked dimers and require domain re-arrangements rather than receptor oligomerization for cell signalling. There is currently no high resolution 3D structure to reveal how the various domains are organized in the dimeric, native receptor. The first clues have come from electron microscopy (EM) of single-molecule images of the hIGF-IR ectodomain and, more particularly, the hIR ectodomain and its complexes with three different monoclonal antibody-derived Fab fragments.70 These images show that the hIGF-IR and hIR ectodomains resemble a U-shaped prism of approximate dimensions 90 x 80 x 120. The images show clearly the dimeric structure of these ectodomains, with the length of the images ~80 along each bar, and the width of ~90 across the two bars. The width of the cleft (assumed membrane-distal) between the two side arms is sufficient to accommodate ligand.70
Fab molecules from the monoclonal antibody 83-7 bound hIR half-way up one end of each side arm in a diametrically opposite manner, indicating a two-fold axis of symmetry normal to the membrane surface.70 Mab 83-7 recognizes an epitope between residues 191 and 297 in the Cys-rich region of hIR.71,72 Examination of the location of the sequence differences between mouse and human IRs and the EM images suggests residues 210 in the third Cys-rich module and 236 in the fourth Cys-rich module form part of the 83-7 epitope. They are located at one corner of the fragment in line with the centre of the L1 domain (Fig. 3). This suggests that the L1/Cys-rich/L2 fragment spans the cleft between the parallel bars rather than lying within each parallel bar. Fabs 83-14 and 18-44, which have been mapped respectively to the first FnIII repeat (residues 469-592) and residues 765-770 in the insert domain,73 bound near the base of the prism at opposite corners.70
The single molecule images, together with the 3D structure of the first three domains of hIGF-IR,39 suggest that the ectodomain dimer is organized into two layers. The L1/Cys-rich/L2 domains are suggested to occupy the upper (membrane-distal) region of the U-shaped prism with the Fn III domains, the insert domains (and the disulfide bonds involved in dimer formation) located predominantly in the membrane-proximal region.70 The nature of the interaction between the two L1/Cys-rich/L2 fragments in the ectodomain dimer is not clear from these EM studies. However, some clues to these domain associations may be gained by examining the surface properties of the L1/Cys-rich/L2 fragment. Surfaces that are involved in the dimer interface would be expected to have complementary properties. In addition, the surfaces involved in either ligand binding or the interface with the bottom layer of fibronectin type III and insert modules, would be expected to be more conserved than those regions that are exposed on the surface of the dimer. The distribution of amino acid sequence conservation and the electrostatic potential of the IGF-IR L1/ Cys-rich/L2 domains have been mapped on to the molecular surface of the IGF-1R L1/Cys-rich/L2 domain structure allowing some predictions of domain association to be made.51
High-resolution data is required to establish the precise arrangement of the 14 modules that make up the ectodomain dimer and the way they interact with the respective ligands to generate signal transduction. Recently, whole receptors solubilized from human placental membranes have been examined by electron cryomicroscopy and 3D reconstruction performed using a library of 700 images.74,75,76 Gold-labeled insulin was used to locate the insulin-binding domain. The images seen were compact and globular, measuring 150 μ diameter. Some domain-like features became evident at intermediate-density thresholds, which indicated a strong two-fold vertical rotational symmetry. When this symmetry was applied to the reconstruction, some structural features became evident. The overall model showed the L1/Cys-rich/L2 domains arranged in an antiparallel manner and at an angle to each other when viewed from the side. The six FnIII domains and the two L2 domains are placed in a central band with the two tyrosine kinase domains at the base of the model.74,75,76 The images described in these two studies70,74,75,76 are substantially different from the “T”-, “X”- or “Y”- shaped objects reported for recombinant ectodomain,65 detergent-solubilized whole receptors or vesicle-reconstituted whole receptors.77,78,79
Ligand Binding by Receptor Chimeras
The results of binding studies with IR/IGF-IR chimeras are summarized in Figure 8 and indicate that the determinants of specificity for insulin and IGF-I binding reside in different regions of the two receptors. Data for whole receptor chimeras showed that residues 1-137 in the L1 domain of hIR and residues 325-524, comprising most of the L2 domain and part of the first fibronectin III domain of hIR, were important determinants of insulin binding while residues 131-315 in the IGF-IR (Cys-rich plus flanking regions from L1 and L2), were prime determinants of IGF-I binding. 80,81 Further support to the importance of the Cys-rich region in IGF-I binding is seen in the differential binding specificities of the hIR-based chimeric receptors containing residues 1-217 of IGF-IR and 1-274 of IGF-IR.82 Finally, an hIR-based chimera where residues 450-601 were replaced with the corresponding residues from hIRR showed decreased insulin binding.71
The studies with ectodomain chimeras provide similar findings to those obtained with whole-receptor chimeras (Fig. 8). The hIR-based chimera containing IGF-IR residues 1-284 resembled the IGF-IR ectodomain and showed high affinity for IGF-I and poor binding for insulin.83 The chimeras with smaller IGF-IR fragments, either 1-180 or 1-62, lack the appropriate specificity determinants for either ligand and showed poor binding of both ligands as expected.84 The ectodomain chimera with IGF-IR residues 184-279 showed high affinity for both ligands, particularly insulin. The region controlling IGF-I specificity has been further narrowed to the 14 residues (amino acids 253-266) in the variable loop of module 6 of the Cys-rich region.85 The chimeric receptor where residues 260 to 277 of hIR were replaced with residues 253 to 266 from hIGF-IR was not significantly different from wild-type hIR in terms of insulin binding affinity, but was more amenable to displacement of radiolabelled insulin by IGF-I than the parent hIR.85
The importance of the N-terminal region in insulin binding was confirmed by examining a series of IGF-IR-based chimeric ectodomains84 where the N-terminus contained decreasingly smaller proportions (191, 83 and 68 residues, respectively) of hIR-derived sequences. All showed similar binding affinities, binding insulin with comparable affinity to wild-type hIR while retaining relatively high (10-20%) binding affinity for IGF-I.84,86 The 1-68/63-1337 IR/IGF-IR whole receptor chimera displayed relative ligand affinities similar to the corresponding ectodomain construct, validating the use of ectodomain constructs in such studies86 and the IGF-IR chimera with only residues 38-50 from IR still bound insulin almost as well as the IR ectodomain.87
Residues 38-43 are predicted to lie in the second rung of the L1 β-helix domain39 at the edge of the putative binding pocket (Fig. 3). The region 223-274 in IGF-IR, implicated in IGF-I specificity, contains major sequence differences when compared to hIR (Fig. 1). It corresponds to modules 4-6 in the Cys-rich region and includes a large and somewhat mobile loop [residues 255-263, mean B(Cα atoms) = 57 A2] which extends into the central space (see Fig. 3). In hIR, this loop is four residues bigger, differs totally in sequence (Fig. 1) and is stabilized by an additional disulfide bond.38,88 The improvement in IGF-I binding by the hIR Cys-loop exchange chimera, hIR_CLX, suggests that the larger loop of hIR may serve to exclude IGF-I from the hormone binding site but allows the smaller insulin molecule to bind.85 The third region implicated in insulin binding, residues 326-524, starts in the middle of the first rung of the L2 β-helix domain39 and extends to Cys514 in the middle of the first Fn III domain (Fig. 1).
Effect of Point Mutations on Ligand Binding
As chimeras only address residues which differ between the two receptors, a more precise analysis of binding determinants can be obtained from single-site mutants. Alanine scanning mutagenesis has been carried out on three distinct regions of the receptor that have been implicated in ligand binding. The first region examined was the L1 domain where 29 residues in the second and third β-sheets of this domain were mutated.89,90 Two mutants Tyr54 and Thr93 failed to yield detectable protein. Of the other 27 Ala mutants, 10 caused a significant impairment of IGF-1 binding as summarized in Table 2. The greatest effect was seen with the Phe90 mutant, which showed a 23-fold reduction in affinity while mutations of Asp8, Asn11, Tyr28, His30, Leu33, Leu56, Phe58 and Arg59 showed reductions in affinity of 3 to 9 fold.89,90 The second region examined was the Cys-rich region where 25 residues, predicted to be accessible to ligand on the basis of the IGF-1R fragment 3D structure,39 were mutated to alanine.90 The region scanned was residues 240-284 and the results are summarized in Table 2. Only 4 of the mutants produced significant decreases in affinity for IGF-I.90 Three of these (residues 240-242) are located in Cys-rich module 5 while Phe251 is at the start of module 6.39
Table 2
Effect of Ala mutations on ligand binding by hIGF-IR.
The residues in the L1 domain (10 residues) and the Cys-rich region (four residues) implicated in IGF-I binding are located in two discontinuous regions.90 The first site includes Asp8, Asn11, Tyr28, His30, Leu33, Leu56, Phe58, Arg59 and Phe90 which are distributed across the first four repeats of the L1 domain (Fig. 9) and form a footprint on the second β-sheet which faces the central cavity of the 3D structure of IGF-1R fragment (Fig. 10). The residues implicated in insulin binding to the insulin receptor by alanine scanning mutagenesis89 occur in a similar but not identical location over the first four repeats of the hIR L1 domain (Fig. 9). The second site consists of Trp79 from the L1 domain loop and Arg240, Phe241, Glu242 and Phe251 from the Cys-rich region. These residues form a small patch on the Cys-rich domain90 adjacent to and in the same plane as the L1 footprint, providing an extended flat face as shown in Figure 10. Ala mutations of residues in the negatively charged region 255-284 (Fig. 1) had negligible effects on binding.90 This is in contrast to the effects of exchanging loop residues 260-277 of hIR with the corresponding loop residues 253-266 from hIGF-1R, which increased the capacity of IGF-1 to diplace bound insulin by more than 10 fold.85 The third region subjected to alanine mutations was the eleven residue sequence at the C-terminal end of the a-chain of hIGF-IR.90 The results are summarized in Table 2 and reveal that this region of the receptor appears to provide the majority of the free energy of the interaction between the receptor and IGF-1.90 Three mutations (Phe695, Ser699 and Val702) had no effect on IGF-I binding. Mutation at Phe701 produced a receptor with no detectable IGF-I binding, while mutation at Phe692, Glu693, Asn694, Leu696, His697, Asn698 and Ile700 resulted in decreases in affinity for IGF-I ranging from 10 to 29 fold (Table 2).90
The importance of Phe701 was reinforced in the studies of hIR- or hIGF-IR-based chimeric minireceptors. 91 Swapping the carboxy terminal domains (16 amino acids) with that from human hIRR completely abolished insulin and IGF-I binding by either hIR- or hIGF-IR-based minireceptor chimeras, while chimeras involving the carboxy terminal domains of hIR or hIGF-IR were little affected.91 Sequence comparisons of hIRR, hIR and hIGF-IR suggest the substitution of Thr for Phe at the position equivalent to 701 in IGF-IR is responsible for this loss of ligand binding by the chimeras containing the carboxy-terminal peptide from the hIRR a-chain.
IGF Mutations
IGF-I and -II contain two extra regions compared to insulin; the C region between the B and A domains and the D region at the C-terminus (Fig. 11). Replacing residues 1-16 of IGF-I with the first 17 residues of the insulin B-chain, which involves 10 sequence differences (Fig. 11), gave only a two-fold reduction in IGF-IR binding.92 Changing just two of these residues in IGF-I, to the corresponding sequence in insulin (Gln15Tyr/Phe16Leu), increased affinity for IR by 10-fold but had no change in affinity for IGF-IR or IGF-IIR.92 A different double mutant, Glu3Gln/Thr4Ala, showed normal IGF1 binding to IR, IGF-IR and IGF-IIR.92
Residues in the IGF-I A-region are important for binding to the type 2 receptor and to the IGFBPs, but not the type 1 receptor. Replacing the A-domain residues 42-56 of IGF-I with A1-15 of insulin, together with a Thr41Ile mutation (eight differences, Fig. 11) had no effect on IGF-IR binding, reduced IGF-IIR binding 20-fold and increased IR binding by seven-fold.93 The areas of greatest divergence between the A domains of insulin and the IGFs are residues 49-51 and 55-56 (Fig. 11). Replacement of residues 49-51 with the corresponding sequence from insulin had no effect on IGF-IR or IR binding, but reduced IGF-IIR binding by 20-fold.93 The double mutant Arg55Tyr/Arg56Gln, where the residues in IGF-I are replaced by the corresponding residues in the insulin sequence, had no effect on IGF1-R or IR binding but resulted in a seven-fold increase in affinity for the type 2 receptor.93The triple exchange mutant Arg50Ser/Arg55Tyr/Arg56Gln in a recombinant construct gave a similar result, with no effects on IGF-IR or IR binding. 94
The structure of IGF-I is very sensitive to amino acid substitutions, with the loss of binding due to either direct effects on receptor interactions or indirect effects on ligand structure.95 Residues implicated in IGF-IR binding are Phe23 and Tyr24 in the B-region, Tyr31 in the C-domain and Tyr60 in the A-region of IGF-I96,97 and the corresponding residues Phe26 and Tyr27 in IGF-II.98,99 Tyr24 and Phe25 can be changed to the Phe.Tyr sequence found in insulin without affecting IGF-IR, IGF-IIR and IR binding.96 Tyr24, Tyr31 and Tyr60 are protected from iodination when bound to IGF-IR.100 Residues Phe23, Tyr24 and Tyr31 form part of a hydrophobic patch on the surface of IGF-I while Tyr60 is largely buried and unlikely to interact directly with the IGF-IR. Similarly, the reduction in IGF-IR binding by a Phe16Ala mutant is concluded to be due to the loss of structural integrity rather than direct binding interactions.95 The mutations Ala8Leu and Met59Phe, on opposite sides of the IGF-I molecule, decreased binding to IGF-IR and IR ectodomains five- to six-fold and 17- to 28-fold, respectively.101 Asp12, which is adjacent to Ala8 on the IGF-I surface, appears to contribute to IGF-IR binding, as the Asp12Ala mutant showed a four-fold reduction in affinity.95 In addition, an Ala62Leu mutant showed eight-fold and two-fold reductions in binding to the IGF-IR and IR ectodomains, respectively.101 The B-domain helix mutants, Val11Ala, Val11Thr, Gln15Ala and Gln15Glu, showed smaller (1.5 to three-fold) reductions in affinity, which have been correlated with structural defects such as the loss of a-helix content in the mutant ligand.95,102 Finally, the A-chain helix mutant of IGF-II, Val43Leu, showed a 16-fold reduction in IGF-IR binding, and a 220-fold reduction in IR binding, but normal binding to the IGF-IIR.99 The corresponding mutant in IGF-I has not been examined.
One notable feature of IGF-I and -II is the large number of charged residues and their uneven distribution over the surface. Replacement of the C-region of IGF-I by a four Gly linker reduced affinity for IGF-IR by 40-100-fold, with a 10-fold or no reduction in binding to IR.103,104 An IGF-I analogue in which residues 29-41 of the C-region have been deleted (mini IGF-I) showed no affinity for either receptor, indicating that the C-region of IGF-I contributes directly to the free energy of binding to the IGF-IR.104 These authors suggested that binding of IGF-I to the IGF-IR resembles insulin/IR binding, and involves a conformational change in which the C-terminal B-region residues are displaced from the body of the molecule to expose the underlying A-region residues. It has been shown that deletion of the C-region of IGF-I results in a substantial tertiary structural rearrangement that can account for the loss of receptor affinity.105 Truncation of the nearby D peptide in IGF-II reduced IGF-IR binding six-fold,98 while the corresponding truncation of IGF-I had no effect.99 Ala mutants have implicated Arg21, Arg36 and Arg37 in IGF-IR binding while the Arg50, Arg55 and Arg56 mutations had smaller effects,106 in agreement with the IGF-I/insulin sequence exchanges involving Arg50, Arg55 and Arg56.93,94 The region of the receptor responsible for recognizing Arg36Arg37 was shown to be the Cys-rich region 217-284.94 The putative binding site of the receptor, which incorporates these residues, has a sizeable patch of acidic residues in the corner where the Cys-rich domain departs from L1. Other acidic residues which are specific to this receptor are found along the inside face of the Cys-rich domain and the loop (residues 255-263) extending from module 6. Thus it is possible that electrostatics play an important part in IGF-I binding, with the C-region binding to the acidic patch of the Cys-rich region near L1, and the acidic patch on the other side of the hormone directed towards a small patch of basic residues (residues 307-310) on the N-terminal end of L2.39,51
Concluding Remarks
The weight of experimental and, more recently, epidemiological evidence points to deregulated signalling through the IGF-IR as a contributing factor in the pathogenesis of some cancers. With respect to the IGF-IR, a number of experimental strategies, particularly those based on antisense technology, offer clinical promise. The recent determination of the 3D structures of the IGF-IR kinase domain47 and the L1/Cys-rich/L2 domain fragment of the IGF-IR39 provide high resolution templates for the development of selective tyrosine kinase inhibitors or small molecule antagonists of receptor function. Clearly the goal is to obtain atomic resolution data for the whole IGF-IR ectodomain in complex with ligand to elucidate the precise nature of ligand/receptor interactions. Ultimately, the goal is to obtain 3D structural information of the high affinity complex, to reveal the conformational changes associated with ligand binding and signal transduction.
References
- 1.
- LeRoith D, Werner H, Beitner-Johnson D. et al. Molecular and cellular aspects of the insulin-like growth factor I receptor. Endocr Rev. 1995;16:143–163. [PubMed: 7540132]
- 2.
- Baker J, Liu JP, Robertson EJ. et al. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed: 8402902]
- 3.
- Frasc F, Pandini G, Scalia P. et al. Insulin receptor isoform A, a newly recognized, high-affinity insulinlike growth factor II receptor in fetal and cancer cells. Mol Cell Biol. 1999;19:3278–3288. [PMC free article: PMC84122] [PubMed: 10207053]
- 4.
- Louvi A, Accili D, Efstratiadis A. Growth-promoting interaction of IGF-II with the insulin receptor during mouse embryonic development. Dev Biol. 1997;189:33–48. [PubMed: 9281335]
- 5.
- Torres AM, Forbes BE, Aplin SE. et al. Solution structure of human insulin-like growth factor II: relationship to receptor and binding protein interactions. J Mol Biol. 1995;248:385–401. [PubMed: 7739048]
- 6.
- Vajdos FF, Ultsch M, Schaffer M. et al. Crystal structure of human insulin-like growth factor-1: determinant binding inhibits binding protein interactions. Biochemistry. 2001;40:11022–11029. [PubMed: 11551198]
- 7.
- Ciszak E, Smith GD. Crystallographic evidence for dual coordination around zinc in the T3R3 human insulin hexamer. Biochemistry. 1994;33:1512–1517. [PubMed: 8312271]
- 8.
- Kitimura T, Kido Y, Nef S. et al. Preserved pancreatic β-cell development and function in mice lacking the insulin receptor-related receptor. Mol Cell Biol. 2001;21:5624–5630. [PMC free article: PMC87283] [PubMed: 11463843]
- 9.
- Sell C, Dumenil G, Deveaud C. et al. Effect of a null mutation of the insulin-like growth factor I receptor gene on growth and transformation of mouse embryo fibroblasts. Mol Cell Biol. 1994;14:3604–3612. [PMC free article: PMC358728] [PubMed: 8196606]
- 10.
- Kaleko M, Rutter WJ, Miller AD. Overexpression of the human insulin-like growth factor I receptor promotes ligand-dependent neoplastic transformation. Mol Cell Biol. 1990;10:464–473. [PMC free article: PMC360815] [PubMed: 2153917]
- 11.
- Baserga R, Hongo A, Rubini M. et al. The IGF-I receptor in cell growth, transformation and apoptosis. Biochim Biophys Acta. 1997;1332:F105–F126. [PubMed: 9196021]
- 12.
- Megyesi K, Kahn CR, Roth J. et al. The NSILA-s receptor in liver plasma membranes. Characterization and comparison with the insulin receptor. J Biol Chem. 1975;250:8990–8996. [PubMed: 391]
- 13.
- Marshall RN, Underwood LE, Voina SJ. et al. Characterization of the insulin and somatomedin-C receptors in human placental cell membranes. J Clin Endocrinol Metab. 1974;39:283–292. [PubMed: 4370626]
- 14.
- Bhaumick B, Bala RM, Hollenberg MD. Somatomedin receptor of human placenta: solubilization, photolabeling, partial purification, and comparison with insulin receptor. Proc Natl Acad Sci USA. 1981;78:4279–4283. [PMC free article: PMC319773] [PubMed: 6270667]
- 15.
- Chernausek SD, Jacobs S, Van Wyk JJ. Structural similarities between human receptors for somatomedin C and insulin: analysis by affinity labeling. Biochemistry. 1981;20:7345–7350. [PubMed: 6275879]
- 16.
- Jacobs S, Kull Jr FC, Cuatrecasas P. Monensin blocks the maturation of receptors for insulin and somatomedin C: identification of receptor precursors. Proc Natl Acad Sci USA. 1983;80:1228–1231. [PMC free article: PMC393568] [PubMed: 6298786]
- 17.
- Jacobs S, Kull Jr FC, Earp HS. et al. Somatomedin-C stimulates the phosphorylation of the beta-subunit of its own receptor. J Biol Chem. 1983;258:9581–9584. [PubMed: 6309774]
- 18.
- Rubin JB, Shia MA, Pilch PF. Stimulation of tyrosine-specific phosphorylation in vitro by insulin-like growth factor. Nature. 1983;305:438–440. [PubMed: 6312321]
- 19.
- Ullrich A, Gray A, Tam AW. et al. Insulin-like growth factor 1 receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986;5:2503–2512. [PMC free article: PMC1167146] [PubMed: 2877871]
- 20.
- Ullrich A, Bell JR, Chen EY. et al. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature. 1985;313:756–761. [PubMed: 2983222]
- 21.
- Ebina Y, Ellis L, Jarnagin K. et al. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell. 1985;40:747–758. [PubMed: 2859121]
- 22.
- Shier P, Watt VM. Primary structure of a putative receptor for a ligand of the insulin family. J Biol Chem. 1989;264:14605–14608. [PubMed: 2768234]
- 23.
- Abbott AM, Bueno R, Pedrini MT. et al. Insulin-like growth factor I receptor gene structure. J Biol Chem. 1992;267:10759–10763. [PubMed: 1316909]
- 24.
- Yee D, Lebovic GS, Marcus RR. et al. Identification of an alternate type I insulin-like growth factor receptor beta subunit mRNA transcript. J Biol Chem. 1989;264:21439–21441. [PubMed: 2557327]
- 25.
- Condorelli G, Bueno R, Smith RJ. Two alternatively spliced forms of the human insulin-like growth factor I receptor have distinct biological activities and internalization kinetics. J Biol Chem. 1994;269:8510–8516. [PubMed: 7510688]
- 26.
- Cosgrove L, Lovrecz GO, Verkuylen A. et al. Purification and properties of insulin receptor ectodomain from large-scale mammalian cell culture. Protein Express Purif. 1995;6:789–798. [PubMed: 8746631]
- 27.
- Hedo JA, Kasuga M, Van ObberghenE. et al. Direct demonstration of glycosylation of insulin receptor subunits by biosynthetic and external labeling: evidence for heterogeneity. Proc Natl Acad Sci USA. 1981;78:4791–4795. [PMC free article: PMC320249] [PubMed: 6946427]
- 28.
- Hedo JA, Gorden P. Biosynthesis of the insulin receptor. Horm Metab Res. 1985;17:487–490. [PubMed: 3905553]
- 29.
- Herzberg VL, Grigorescu F, Edge AS. et al. Characterization of insulin receptor carbohydrate by comparison of chemical and enzymatic deglycosylation. Biochem Biophys Res Commun. 1985;129:789–796. [PubMed: 2990467]
- 30.
- Collier E, Gorden P. O-Linked oligosaccharides on insulin receptor. Diabetes. 1991;40:197–203. [PubMed: 1991570]
- 31.
- Collier E, Carpentier JL, Beitz L. et al. Specific glycosylation site mutations of the insulin receptor alpha-subunit impair intracellular transport. Biochemistry. 1993;32:7818–7823. [PubMed: 8347587]
- 32.
- Leconte I, Carpentier JL, Clauser E. The functions of the human insulin receptor are affected in different ways by mutation of each of the four N-glycosylation sites in the beta subunit. J Biol Chem. 1994;269:18062–18071. [PubMed: 8027066]
- 33.
- Wiese RJ, Herrera R, Lockwood DH. Glycosylation sites encoded by exon 2 of the human insulin receptor gene are not required for the oligomerization, ligand binding, or kinase activity of the insulin receptor. Receptor. 1995;5:71–80. [PubMed: 7613486]
- 34.
- Elleman TC, Frenkel MJ, Hoyne PA. et al. Mutational analysis of the N-linked glycosylation sites of the human insulin receptor. Biochem J. 2000;347:771–779. [PMC free article: PMC1221015] [PubMed: 10769182]
- 35.
- Caro LHP, Ohali A, Gorden P. et al. Mutational analysis of the NH2-terminal gycosylation sites of the insulin receptor alpha-subunit. Diabetes. 1994;43:240–246. [PubMed: 8288048]
- 36.
- Bastian W, Zhu J, Way B. et al. Glycosylation of Asn397 or Asn418 is required for normal insulin receptor biosynthesis and processing. Diabetes. 1993;42:966–974. [PubMed: 8513978]
- 37.
- Bajaj M, Waterfield MD, Schlessinger J. et al. On the tertiary structure of the extracellular domains of the epidermal growth factor and insulin receptors. Biochim Biophys Acta. 1987;916:220–226. [PubMed: 3676333]
- 38.
- Ward CW, Hoyne PA, Flegg RH. Insulin and epidermal growth factor receptors contain the cysteine repeat motif found in the tumor necrosis factor receptor. Proteins Struct Funct Genet. 1995;22:141–153. [PubMed: 7567962]
- 39.
- Garrett TPJ, Mckern NM, Lou MZ. et al. Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor . Nature. 1998;394:395–399. [PubMed: 9690478]
- 40.
- O'Bryan JP, Frye RA, Cogswell PC. et al. axl, a transforming gene isolated from primary human myeloidb leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol. 1991;11:5016–5031. [PMC free article: PMC361494] [PubMed: 1656220]
- 41.
- Marino-Buslje C, Mizuguchi K, Siddle K. et al. A third fibronectin type III domain in the extracellular region of the insulin receptor family. FEBS Lett. 1998;441:331–336. [PubMed: 9883910]
- 42.
- Mulhern TD, Booker GW, Cosgrove L. A third fibronectin type-III domain in the insulin-family receptors. Trends Biochem Sci. 1998;23:465–466. [PubMed: 9868364]
- 43.
- Ward CW. Members of the insulin receptor family contain three fibronectin type III domains. Growth Factors. 1999;16:315–322. [PubMed: 10427505]
- 44.
- Hanks SK. Eukaryotic protein kinases. Curr Opin Struct Biol. 1991;1:369–383.
- 45.
- Surmacz E, Sell C, Swantek J. et al. Dissociation of mitogenesis and transforming activity by C-terminal truncation of the insulin-like growth factor-I receptor. Exp Cell Res. 1995;218:370–380. [PubMed: 7737373]
- 46.
- Hongo A, Dambrosio C, Miura M. et al. Mutational analysis of the mitogenic and transforming activities of the insulin-like growth factor I receptor. Oncogene. 1996;12:1231–1238. [PubMed: 8649825]
- 47.
- Pautsh A, Zoephel A, Ahorn H. et al. IGF-1 receptor kinase domain. PDB. 2001 [PubMed: 11591350]
- 48.
- Hubbard SR, Wei L, Elis L. et al. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372:746–754. [PubMed: 7997262]
- 49.
- Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. [PMC free article: PMC1170189] [PubMed: 9312016]
- 50.
- Knighton DR, Zeng J, Eyck LFT. et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. [PubMed: 1862342]
- 51.
- Adams TE, Epa VC, Garrett TPJ. et al. Structure and function of the type-I insulin-like growth factor receptor. Cell Mol Life Sci. 2000;57:1050–1093. [PubMed: 10961344]
- 52.
- Ward CW, Garrett TPJ, McKern NM. et al. Structure of the insulin receptor family: unexpected relationships with other proteins. Today's Life Sciences. 1999;11:26–32.
- 53.
- Pashmforoush M, Chan SJ, Steiner DF. Structure and expression of the insulin-like peptide receptor from amphioxus. Mol Endocrinol. 1996;10:857–866. [PubMed: 8813726]
- 54.
- Sparrow LG, Mckern NM, Gorman JJ. et al. The disulfide bonds in the C-terminal domains of the human insulin receptor ectodomain. J Biol Chem. 1997;272:29460–29467. [PubMed: 9368005]
- 55.
- Schaffer L, Ljungqvist L. Identification of a disulfide bridge connecting the alpha-subunits of the extracellular domain of the insulin receptor. Biochem Biophys Res Commun. 1992;189:650–653. [PubMed: 1472036]
- 56.
- Banner DW, D'Arcy A, Janes W. et al. Crystal structure of the soluble human 55 kd TNF receptor-human TNF-βcomplex: implications for TNF receptor activation. Cell. 1993;73:431–445. [PubMed: 8387891]
- 57.
- Stetefeld J, Mayer U, Timpl R. et al. Crystal structure of three consecutive laminin-type epidermal growthb factor-like (LE) modules of laminin gamma-1 chain harboring the nidogen binding site. J Mol Biol. 1996;257:644–657. [PubMed: 8648630]
- 58.
- Macaulay SL, Polites M, Hewish DR. et al. Cysteine-524 is not the only residue involved in the formation of disulfide-bonded dimers of the insulin receptor. Biochem J. 1994;303:575–581. [PMC free article: PMC1137366] [PubMed: 7980420]
- 59.
- Bilan PJ, Yip CCQ. Unusual insulin binding to cells expressing an insulin receptor mutated at cysteine 524. Biochem Biophys Res Commun. 1994;205:1891–1898. [PubMed: 7811279]
- 60.
- Lu K, Guidotti G. Identification of the cysteine residues involved in the class I disulfide bonds of the human insulin receptor: properties of insulin receptor monomers. Mol Biol Cell. 1996;7:679–691. [PMC free article: PMC275922] [PubMed: 8744943]
- 61.
- Kabat EA, Wu TT, Perry HM. et al. Sequences of Proteins of Immunological Interest. 5th ed. Bethesda: US Department of Health and Human Services, 1991.
- 62.
- Fernandez R, Tabarini D, Azpiazu N. et al. The Drosophila insulin receptor homolog—a gene essential for embryonic development encodes two receptor isoforms with different sigaling potential. EMBO J. 1995;14:3373–3384. [PMC free article: PMC394404] [PubMed: 7628438]
- 63.
- Ruan YM, Chen C, Cao YX. et al. The Drosophila insulin receptor contains a novel carboxy-terminal extension likely to play an important role in signal transduction. J Biol Chem. 1995;270:4236–4243. [PubMed: 7876183]
- 64.
- Cheatham B, Kahn CR. Cysteine 647 in the insulin receptor is required for normal covalent interaction between α- and β-subunits and signal transduction. J Biol Chem. 1992;267:7108–7115. [PubMed: 1551916]
- 65.
- Schaefer EM, Erickson HP, Federwisch M. et al. Structural organization of the human insulin receptor ectodomain. J Biol Chem. 1992;267:23393–23402. [PubMed: 1385419]
- 66.
- Campbell ID, Spitzfaden C. Building proteins with fibronectin type III modules. Structure. 1994;2:333–337. [PubMed: 8081748]
- 67.
- Goldstein BJ, Dudley AL. The rat insulin receptor: primary structure and conservation of tissue-specific alternative mRNA splicing. Mol Endocrinol. 1990;4:235–244. [PubMed: 2330003]
- 68.
- Elleman TC, Domagala T, Mckern NM. et al. Identification of a determinant of epidermal growth factor receptor ligand-binding specificity using a truncated, high-affinity form of the ectodomain. Biochemistry. 2001;40:8930–8939. [PubMed: 11467954]
- 69.
- Ward CW, Garrett TPJ. The relationship between the L1 and L2 domains of the insulin and epidermal growth factor receptors and leucine-rich repeat modules. BMC Bioinformatics. 2001;2:4. [PMC free article: PMC37351] [PubMed: 11504559]
- 70.
- Tulloch PA, Lawrence LJ, McKern NM. et al. Single-molecule imaging of human insulin receptor ectodomain and its Fab complexes. J Struct Biol. 1999;125:11–18. [PubMed: 10196112]
- 71.
- Zhang B, Roth RA. A region of the insulin receptor important for ligand bimnding (residues 450-601) is recognized by patient's autoimmune antibodies and inhibitory monoclonal antibodies. Proc Natl Acad Sci USA. 1991;88:9858–9862. [PMC free article: PMC52820] [PubMed: 1719540]
- 72.
- Schaefer EM, Siddle K, Ellis L. Deletion analysis of the human insulin receptor ectodomain reveals independently folded soluble subdomains and insulin binding by a monomeric a-subunit. J Biol Chem. 1990;265:13248–13253. [PubMed: 2198288]
- 73.
- Prigent SA, Stanley KK, Siddle K. Identification of epitopes on the human insulin receptor reacting with rabbit polyclonal antisera and mouse monoclonal antibodies. J Biol Chem. 1990;265:9970–9977. [PubMed: 1693619]
- 74.
- Luo RZT, Beniac DR, Fernandes A. et al. Quaternary structure of the insulin-insulin receptor complex. Science. 1999;285:1077–1080. [PubMed: 10446056]
- 75.
- Ottensmeyer FP, Beniac DR, Luo RZT. et al. Mechanism of transmembrane signaling: insulin binding and the insulin receptor. Biochemistry. 2000;39:12103–12112. [PubMed: 11015187]
- 76.
- Ottensmeyer FP, Beniac DR, Luo RZT. et al. Mechanism of transmembrane signaling: insulin binding and the insulin receptor-Correction. Biochemistry. 2001;40:12103–12112. [PubMed: 11015187]
- 77.
- Christiansen K, Tranum-Jensen J, Carlsen J. et al. A model for the quaternary structure of human placental insulin receptor deduced from electron microscopy. Proc Natl Acad Sci USA. 1991;88:249–252. [PMC free article: PMC50787] [PubMed: 1986371]
- 78.
- Tranum-Jensen J, Christiansen K, Carlsen J. et al. Membrane topology of insulin receptors reconstituted into lipid vesicles. J Membrane Biol. 1994;140:215–223. [PubMed: 7932656]
- 79.
- Woldin CN, Hing FS, Lee J. et al. Structural studies of the detergent-solubilized and vesicle-reconstituted insulin receptor. J Biol Chem. 1999;274:34981–34992. [PubMed: 10574975]
- 80.
- Schumacher R, Mosthaf L, Schlessinger J. et al. Insulin and insulin-like growth factor-1 binding specificity is determined by distinct regions of their cognate receptors. J Biol Chem. 1991;266:19288–19295. [PubMed: 1655782]
- 81.
- Schumacher R, Soos MA, Schlessinger J. et al. Signaling-competent receptor chimeras allow mapping of major insulin receptor binding domain determinants. J Biol Chem. 1993;268:1087–1094. [PubMed: 7678247]
- 82.
- Gustafson TA, Rutter WJ. The cysteine-rich domains of the insulin and insulin-like growth factor I receptors are primary determinants of hormone binding specificity. J Biol Chem. 1990;265:18663–18667. [PubMed: 2170418]
- 83.
- Andersen AS, Kjeldsen T, Wiberg FC. et al. Changing the insulin receptor to possess insulin-like growth factor 1 ligand specificity. Biochemistry. 1990;29:7363–7366. [PubMed: 2223767]
- 84.
- Kjeldsen T, Andersen AS, Wiberg FC. et al. The ligand specificities of the insulin receptor and the insulin-like growth factor I receptor reside in different regions of a common binding site. Proc Natl Acad Sci USA. 1991;88:4404–4408. [PMC free article: PMC51668] [PubMed: 1852007]
- 85.
- Hoyne PA, Elleman TC, Adams TE. et al. Properties of an insulin receptor with an IGF-1 receptor loop exchange in the cysteine-rich region. FEBS Lett. 2000;469:57–60. [PubMed: 10708756]
- 86.
- Andersen AS, Kjeldsen T, Wiberg FC. et al. Identification of determinants that confer ligand specificity on the insulin receptor. J BiolChem. 1992;267:13681–13686. [PubMed: 1320025]
- 87.
- Kjeldsen T, Wiberg FC, Andersen AS. Chimeric receptors indicate that phenylalanine 29 is a major contributor to insulin specificity of the insulin receptor. J Biol Chem. 1994;269:32942–32946. [PubMed: 7806523]
- 88.
- Schäer L, Hansen PH. Partial characterization of the disulfide bridges of the soluble insulin receptor. Exp Clin Endocrinol Diabetes. 1996:104–89.
- 89.
- Mynarcik DC, Williams PF, Schaffer L. et al. Identification of common ligand binding determinants of the insulin and insulin-like growth factor receptors-insights into mechanisms of ligand binding. J Biol Chem. 1997;272:18650–18655. [PubMed: 9228034]
- 90.
- Whittaker J, Groth AV, Mynarcik D. et al. Alanine scanning mutagenesis of a type-I insulin-like growth factor receptor ligand binding site. J Biol Chem. 2001;276:43980–43986. [PubMed: 11500492]
- 91.
- Kristensen C, Wiberg FC, Andersen AS. Specificity of insulin and insulin-like growth factor receptors investigated using chimeric minireceptors. Role of carboxy terminal of receptor a-subunit. J Biol Chem. 1999;274:37351–37356. [PubMed: 10601304]
- 92.
- Bayne ML, Applebaum J, Chicchi GG. et al. Structural analogs of human insulin-like growth factor 1 with reduced affinity for serum binding proteins and the type 2 insulin-like growth factor receptor. J Biol Chem. 1988;263:6233–6239. [PubMed: 2966152]
- 93.
- Cascieri MA, Chicci GG, Applebaum J. et al. Structural analogues of human insulin-like growth factor (IGF) I with altered affinity for type 2 IGF receptors. J Biol Chem. 1989;264:2190–2202. [PubMed: 2536701]
- 94.
- Zhang WG, Gustafson TA, Rutter WJ. et al. Positively charged side chains in the insulin-like growth factor-1 C- and D-regions determine receptor binding specificity. J Biol Chem. 1994;269:10609–10613. [PubMed: 8144650]
- 95.
- Jansson M, Uhlen M, Nilsson B. Structural changes in insulin-like growth factor (IGF) I mutant proteins affecting binding kinetics to IGF binding protein 1 and IGF-I receptor. Biochemistry. 1997;36:4108–4117. [PubMed: 9100004]
- 96.
- Cascieri MA, Chicci GG, Applebaum J. et al. Mutants of human insulin-like growth factor I with reduced affinity for the type I insulin-like growth factor receptor. Biochemistry. 1988;27:3229–3233. [PubMed: 2839228]
- 97.
- Bayne ML, Applebaum J, Chicchi GG. et al. Role of tyrosines 24, 31, and 60 in the high affinity binding of insulin-like growth factor-1 to the type 1 insulin-like growth factor receptor. J Biol Chem. 1990;265:15648–15652. [PubMed: 2168421]
- 98.
- Roth BV, Burgisser DM, Luthi C. et al. Mutants of human IGF2: expression and characterization of analogues with a substitution of Tyr27 and/or a deletion of residues 62-67. Biochem Biophys Res Commun. 1991;181:907–914. [PubMed: 1721812]
- 99.
- Sakano K, Enjoh T, Numata F. et al. The design, expression, and characterization of human insulin-like growth factor II (IGF-II) mutants specific for either the IGF-II/cation-independent mannose 6-phosphate receptor or IGF-I receptor. J Biol Chem. 1991;266:20626–20635. [PubMed: 1657932]
- 100.
- Maly P, Luthi C. The binding sites of insulin-like growth factor I (IGF I) to type I IGF receptor and to a monoclonal antibody. Mapping by chemical modification of tyrosine residues. J Biol Chem. 1988;263:7068–7072. [PubMed: 2966799]
- 101.
- Shooter GK, Magee B, Soos MA. et al. Insulin-like growth factor (IGF)-I A- and B-domain analogues with altered type 1 IGF and insulin receptor binding specificities. J Mol Endocrinol. 1996;17:237–246. [PubMed: 8981230]
- 102.
- Hodgson DR, May FEB, Westley BR. Mutations at positions 11 and 60 of insulin-like growth factor 1 reveal differences between its interactions with the type I insulin-like-growth-factor receptor and the insulin receptor. Eur J Biochem. 1995;233:299–309. [PubMed: 7588759]
- 103.
- Bayne ML, Applebaum J, Underwood D. et al. The C-region of human insulin-like growth factor (IGF) I is required for high affinity binding to the type I IGF receptor. J Biol Chem. 1988;264:11004–11008. [PubMed: 2472386]
- 104.
- Gill R, Wallach B, Verma C. et al. Engineering the C-region of human insulin-like growth-factor-1- implications for receptor binding. Protein Eng. 1996;9:1011–1019. [PubMed: 8961354]
- 105.
- Dewolf E, Gill R, Geddes S. et al. Solution structure of a mini IGF-1. Protein Sci. 1996;5:2193–2202. [PMC free article: PMC2143290] [PubMed: 8931138]
- 106.
- Jansson M, Andersson G, Uhlen M. et al. The insulin-like growth factor (IGF) binding protein 1 binding epitope on IGF-1 probed by heteronuclear NMB spectroscopy and mutational analysis. J Biol Chem. 1998;273:24701–24707. [PubMed: 9733769]
- 107.
- Dickinson CD, Veerapandian B, Dai X-P. et al. Crystal structure of the tenth type III cell adhesion module of human fibronectin. J Mol Biol. 1992;236:1079–1092. [PubMed: 8120888]
- Introduction
- IGF-I Receptor: Discovery and Sequence
- Domain Organization and Evolutionary Relationships of IGF-IR and Related Receptors
- 3D Structure of the L1/Cys-rich/L2 Domains of the IGF-IR
- 3D Structure of the Tyrosine Kinase Domain
- The IGF-IR and IR Ectodomain Dimers
- Ligand Binding by Receptor Chimeras
- Effect of Point Mutations on Ligand Binding
- Concluding Remarks
- References
- The Structure of the Type 1 Insulin-Like Growth Factor Receptor - Madame Curie B...The Structure of the Type 1 Insulin-Like Growth Factor Receptor - Madame Curie Bioscience Database
- Phylogeny of Major Intrinsic Proteins - Madame Curie Bioscience DatabasePhylogeny of Major Intrinsic Proteins - Madame Curie Bioscience Database
- Molecular Properties of Voltage-Gated Calcium Channels - Madame Curie Bioscience...Molecular Properties of Voltage-Gated Calcium Channels - Madame Curie Bioscience Database
- Matrix Metalloproteinases, Tissue Inhibitors of Metalloproteinase and Matrix Tur...Matrix Metalloproteinases, Tissue Inhibitors of Metalloproteinase and Matrix Turnover and the Fate of Hepatic Stellate Cells - Madame Curie Bioscience Database
- MSK1 and Nuclear Receptors Signaling - Madame Curie Bioscience DatabaseMSK1 and Nuclear Receptors Signaling - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...