NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Biodiversity in the bacterial world is strongly influenced by “periodic selection,” in which natural selection recurrently purges diversity within a bacterial population. Owing to the extreme rarity of recombination in bacteria, selection favoring an adaptive mutation eliminates nearly all the diversity within an ecotype (defined as the set of strains using about the same ecological niche, so that an adaptive mutant or recombinant out-competes to extinction strains from the same ecotype). Diversity within an ecotype is only transient, awaiting its demise with the next periodic selection event. Ecological diversity in bacteria is governed by three kinds of mutations (or recombination events). Niche-invasion mutations found a new ecotype, such that the new genotype and its descendants escape the diversity-purging effect of periodic selection from their former ecotype. Periodic selection mutations then make the different ecotypes more distinct by purging the diversity within but not between ecotypes. Lastly, speciation-quashing mutations may occur, which can extinguish another ecotype even after it has had several private, periodic selection events. For example, an ecotype that shares all its resources with another ecotype, albeit in different proportions, may be extinguished by an extraordinarily fit adaptive mutation from the other ecotype.
Sequence clusters, as determined by a variety of criteria, are expected to correspond to ecotypes. Sequence-based approaches suggest that a typical named species contains many ecotypes.
That periodic selection occurs in nature is evidenced by the modest levels of sequence diversity observed within bacterial species, levels that are too low to be explained by genetic drift. Also, a special kind of periodic selection event, driven by “adapt globally, act locally” mutations, is inferred when strains fall into discrete sequence clusters over most of their genomes, but are aberrantly homogeneous in a small chromosomal region. Beyond establishing a history of periodic selection, this pattern can help corroborate that a set of sequence clusters correspond to ecotypes.
Introduction
One half-century ago, a simple experiment changed the way we think about the power of natural selection in bacterial populations. The classic experiment of Atwood, Schneider, and Ryan1 demonstrated the phenomenon of periodic selection, whereby diversity within a bacterial population is purged recurrently by natural selection. The principle is that in an entirely asexual population, each adaptive mutation precipitates a round of natural selection which, if successful, fixes not only the adaptive mutation but also the entire genome of the mutant cell. In the absence of recombination, the adaptive mutation is unable to enter into any other genetic background, and so selection favoring the adaptive mutation drags the entire genome associated with it to fixation.
A recent reenactment of this experiment, supported by data from modern molecular biology, illustrates the diversity-purging power of periodic selection.2 Descendants of a single Escherichia coli cell were cultured without benefit of recombination, and were allowed to evolve. Diversity within the population was monitored over time by assaying the frequency of spontaneous mutants resistant to the bacteriophage T5. The frequency of resistant cells started at zero, gradually increased due to mutation for fifty or more generations, then abruptly dropped back to zero, and this pattern was repeated several times. As in the original periodic selection paper, the crashes in frequency of the marker were interpreted as the result of periodic selection. The model is that adaptive mutations occur within the majority population of cells (in this case, marked by T5 sensitivity), and that the adaptive mutant and its clonal descendants drive to extinction all other lineages in the population (T5-resistant and T5-sensitive alike). Thus, the rise of the adaptive mutant genotype is manifested in these experiments by the disappearance of the minority marker. In Notley-McRobb and Ferenci's recent paper, the interpretation of periodic selection was supported by coincident sequence changes at mgc and mgl,loci known to play a major role in adaptation to laboratory culture.
Periodic selection clearly has the potential to sweep the diversity within a strictly asexual population, descended from a single clone, in laboratory culture. However, these laboratory experiments do not necessarily predict the effect of selection in natural populations of bacteria. While the bacteria in these periodic selection experiments were engineered to be strictly asexual, bacteria in nature undergo recombination, albeit at an extremely low rate.3-5 Thus, the diversity-purging effect of periodic selection may be diminished in natural populations. Also, while a periodic selection event can purge the diversity among the clonal descendants within a culture flask, it is not clear how broad a population would be purged of diversity in nature. For example, would all of E. coli be purged of diversity by one periodic selection event in nature?
Here I will describe the effects of periodic selection for natural populations of bacteria, and I will show how periodic selection plays a central role in the origin of ecological diversity in the bacterial world. I will demonstrate that recombination is typically too rare in bacteria to diffuse the diversity-purging effect of periodic selection. I will demonstrate that within a typical named species, there appear to be dozens of “ecotypes”— ecologically distinct populations that have their own private periodic selection events, so that each ecotype escapes the periodic selection events of all other ecotypes. I will show that the genetic changes allowing a genotype to escape periodic selection from its previous ecotype form the basis of bacterial speciation. Finally, I will show that periodic selection has actually occurred in natural populations of bacteria, and I will introduce a method for using genomic data to detect past periodic selection events.
The Nature of Recombination in Bacteria
The microcosms of periodic selection experiments were designed to be devoid of sex, but so far as we know, sexual recombination has been a part of every bacterial species' history.5,6 That a bacterial species has engaged in recombination in its past may be demonstrated through a diversity of sequence-based tests.7 For example, in the homoplasy test of Maynard Smith and Smith,8 recombination is implicated when an improbably high number of nucleotide substitutions have occurred twice or more in different parts of the phylogeny. This and similar methods have demonstrated the existence of recombination in all bacterial species investigated.
The rate at which recombination occurs in nature has been estimated through several sequence-based approaches, including the extent to which different genes or gene segments yield different phylogenetic relationships among strains; congruence of phylogenies based on different gene segments indicates rare recombination. The recombination rates may be estimated separately for recombination within populations9,10 and between populations.4,11-13 Typically, the recombination rates range from nearly an order of magnitude less than mutation in some of the most clonal of bacteria (e.g., Staphylococcus aureus),14 to about half an order of magnitude greater than mutation in some of the most frequently recombining bacteria (e.g., Neisseria meningitidis),11 with the exception so far of Helicobacter pylori, which recombines at a much higher (but not yet determined) rate.15 For example, a 450 bp segment of N. meningitidis undergoes recombination at a rate of 1.2 × 10-6 per individual per generation, which is 3.6 times the mutation rate.11,16
While recombination in bacteria is rare, it is also promiscuous; bacteria are not fastidious about their choice of sexual partners. Homologous recombination occurs even between organisms that are 25% divergent in their DNA sequences.17,18
Also, bacteria can acquire novel genes and whole operons through heterologous recombination, sometimes from extremely divergent species. Lawrence and Ochman19 have developed a method for identifying genes acquired from extremely distant sources: genes with a highly aberrant GC content are interpreted as foreign genes. Based on this principle, typically 5-15% of the genes in a bacterial genome appear to have been acquired from extremely distant relatives.6
Finally, recombination in bacteria is unidirectional, from a donor cell to a recipient cell, and usually only a small fraction of the genome is transferred.5
The Effect of Rare Recombination on Diversity within a Population
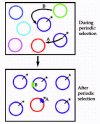
Consider next whether the rare recombination typical of bacteria should soften the diversity-purging effect of natural selection. Recombination can preserve the genetic diversity in two ways. First, if the adaptive mutation recombines into another genetic background, then the entire genome of the recipient is saved from extinction (fig. 1, example A). Alternatively, if a segment from a strain lacking the adaptive mutation should recombine into a strain with the adaptive mutation, then that segment (only) will be saved from extinction (fig. 1, example B).
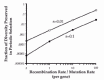
I have investigated the relationship between recombination rate and the purging of diversity using a Monte Carlo method derived from a coalescence algorithm of Braverman et al.20 In a completely asexual population, each cell after periodic selection will contain only DNA derived from the genome of the original adaptive mutation; with recombination, DNA derived from other cells existing at the time of the adaptive mutation can contribute to the genomes of cells surviving periodic selection. Figure 2 shows the diversity-purging effect of periodic selection over a range of recombination rates occurring in bacteria. When the intensity of periodic selection is strong (i.e., fitness advantage for the adaptive mutation is s = 0.1 ), each bout of periodic selection purges nearly all diversity within an ecotype. Over recombination rates observed in nature (from 0.1 to 3.6 times the mutation rate), periodic selection purges all but 0.0007% to 0.2% of the sequence diversity. Over more modest selection intensity (i.e., s = 0.01), periodic selection purges all but 0.02% to 2% of sequence diversity over naturally occurring recombination rates. It appears that recombination is ineffective at softening the diversity-purging effect of periodic selection in nature.
The Origins of Permanent Divergence
Each periodic selection event reduces the diversity within a population to little more than the clonal descendants of the original adaptive mutant. Therefore, the diversity accumulated within a bacterial population is only transient, awaiting its demise with the next periodic selection event. What, then, is the evolutionary origin of permanent diversity in bacteria?
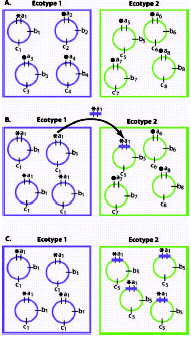
I have previously defined a bacterial “ecotype” with respect to the fate of an adaptive mutant: an ecotype is a set of strains using the same or similar ecological niches, such that an adaptive mutant from within the ecotype out-competes to extinction all other strains from the same ecotype; an adaptive mutant cannot, however, drive to extinction strains from other ecotypes (fig. 3B).18,21,22 Thus, an ecotype is the set of strains whose diversity is purged through periodic selection favoring each adaptive mutant. Periodic selection is a powerful force of cohesion within a bacterial ecotype, in that it recurrently resets the genetic diversity to near zero.
At the point that two ecologically distinct populations undergo their own private periodic selection events, they have reached a milestone toward forming new species. Such populations are now irreversibly separate, since periodic selection cannot prevent further divergence (by definition), and as has previously been shown, neither can recombination.21
Even if recombination between ecotypes were to occur at the same rate as recombination within them, natural selection against rare inter-ecotype recombinants could easily limit the frequency of recombinant genotypes to negligible levels.21 Therefore, evolution of sexual isolation is not a necessary step toward the evolution of permanent divergence in the bacterial world. The key milestone toward bacterial speciation is instead the genetic change that places a mutant (or recombinant) cell and its descendants outside the domain of periodic selection of other ecotypes.
Bacterial ecotypes, as defined by the domains of periodic selection, share the fundamental properties of species.18,22 Ecotypes are each subject to an intense force of cohesion, periodic selection, which recurrently purges diversity within an ecotype (a species attribute emphasized by the Cohesion Species Concept of Templeton).23 Once different ecotypes have diverged to the point of escaping one another's periodic selection events, there is no force that can prevent their divergence. (The irreversibility of divergence is emphasized by the Evolutionary Species Concept of Simpson24 and Wiley).25 As we shall see, ecotypes form distinct sequence clusters, owing to periodic selection purging sequence diversity within but not between ecotypes.26 (The phenotypic and molecular separateness of species is emphasized by the Phenotypic Species Concept of Sokal and Crovello27 and the Modern Synthesis Species Concept of Mallet21 , 28 Finally, bacterial ecotypes are ecologically distinct, as emphasized by the Ecological Species Concept of van Valen.29 Bacterial ecotypes are therefore evolutionary lineages that are irreversibly separate, each with its own evolutionary tendencies and historical fate. A species in the bacterial world may be understood as an evolutionary lineage bound together by ecotype-specific periodic selection.22
Effects of Periodic Selection beyond the Boundaries of the Ecotype
The ecological divergence between ecotypes allows them to coexist and to survive each other's periodic selection events. Nevertheless, if newly divergent ecotypes compete for at least some resources, they may feel the effects of periodic selection from outside the ecotype. Suppose, for example, that a parental ecotype and a nascent ecotype use the same two sugars, but the parental ecotype takes up one sugar preferentially, while the reverse is true for the nascent ecotype. Modestly adaptive mutations that increase overall efficiency in the parental ecotype will fail to extinguish the nascent ecotype, but they can decrease its population density significantly. Nevertheless, the genetic diversity of the nascent ecotype is not diminished (except minimally by increasing genetic drift). Provided that adaptive mutations are modest, these populations can coexist indefinitely, even as each adaptive mutation negatively impacts the population density of the other ecotype.
Even after newly divergent ecotypes have each undergone several rounds of their own, private periodic selection events, they may still be vulnerable to extinction caused by the other ecotype's periodic selection. This can be the case when ecotypes use entirely the same set of resources, but in different proportions. An extraordinarily fit adaptive mutant from the parental ecotype might out-compete all strains from the nascent ecotype (as well as all the other strains from its own ecotype) (fig.3c). In this case, the founding of the new ecotype would be quashed by a periodic selection event before the two incipient ecotypes had sufficiently diverged from one another.
Recombination may, in some cases, prevent a potentially speciation-quashing adaptive mutation from extinguishing another ecotype. If the adaptive mutation from one ecotype can recombine into another ecotype, the first ecotype may lose its advantage. In our “adapt globally, act locally” model of periodic selection,30 the domain of competitive superiority of an adaptive mutant (i.e., the cell) is limited to its own ecotype, as I have described, but the adaptive mutation (i.e., the allele) can be recombined into other ecotypes. Upon transfer into another ecotype, an adaptive mutation precipitates a local periodic selection event within the recipient ecotype (fig. 4). In this model, the chromosomal region near the adaptive mutation can be homogenized across different ecotypes, while the divergence elsewhere in the genome is unaffected.30
Note that a nascent ecotype is vulnerable to extinction by a parental ecotype only if its resource base is a subset of the parental ecotype's. When an ecotype utilizes at least one resource not used by the parental ecotype, it is then invulnerable to that ecotype's periodic selection events.31 If a hypothesis by Lawrence32,33 is correct, nascent ecotypes may readily acquire novel resources required to escape all periodic selection from their parental ecotype.
Lawrence32,33 has argued that nearly all ecological divergence is precipitated by horizontal transfer, in which a recipient acquires a novel (heterologous) gene locus or operon from another species. By granting an entirely new metabolic function, heterologous gene transfer has the potential to endow a strain with a new resource base that is not shared with the parental ecotype. In this case, the horizontal transfer immediately places the nascent ecotype out of range of any periodic selection emanating from the parental ecotype.
In summary, ecological diversity in the bacterial world appears to be determined by three kinds of genetic changes (either mutations or recombination events) (fig. 3). First, there are niche-invasion mutations (or recombination events), which allow the new genotype and its descendants to utilize a new set of resources and thereby escape periodic selection from the parental ecotype. Second, there are periodic selection mutations (or recombination events), which purge the diversity within a single ecotype; these tend to make ecotypes more distinct, since they purge the diversity within but not between ecotypes. Finally, there may be speciation-quashing mutations (or recombination events), whereby one ecotype can extinguish another. It will be interesting to quantify the rates at which these three kinds of mutations and recombination events occur, using microcosm evolution experiments that have been developed to investigate adaptive radiation in bacteria.34-36
Periodic Selection and Discovery of Bacterial Ecotypes
Discovery of Ecotypes As Sequence Clusters
By purging diversity within but not between ecotypes, periodic selection provides a rationale for discovery of bacterial diversity. Given enough time, each bacterial ecotype is expected to be identifiable as a sequence cluster, distinct from all closely related ecotypes. In addition, each ecotype is expected to be identifiable as a monophyletic group in a phylogeny based on DNA sequence data.26
A phylogenetic perspective explains the predicted correspondence between ecotypes and DNA sequence clusters.26 Suppose we begin with a single ecotype, and then one cell within the ecotype evolves new ecological properties and thereby founds a new ecotype. At this time the new ecotype appears in a phylogeny as if it is just one more lineage within the parental ecotype. However, the next adaptive mutant causing periodic selection within the parental ecotype will eliminate all other lineages within that ecotype, but will leave diversity within the nascent ecotype untouched. Likewise, periodic selection within the new ecotype will purge diversity within that ecotype but not within the parental ecotype. Recurrent selective sweeps within each lineage will result in long sequence distances on the phylogeny between each ecotype's contemporary diversity and the most recent ancestor shared by the two ecotypes (i.e., sequence distances will be much greater between than within ecotypes). Thus, each ecotype will eventually be discernible as a distinct sequence cluster and as a monophyletic group.
The Challenge of “Geotypes”
Care must be taken when using any sequence-based method to infer ecotypes. Geographically isolated populations that are members of the same ecotype could diverge into separate sequence clusters. In this case, an adaptive mutant from one geographic region is not given the chance to compete with populations from other regions, so sequence divergence between geographically isolated populations of the same ecotype could proceed indefinitely. Papke and Ward (pers. comm.) have argued that many bacterial taxa lack the means for worldwide travel, and so are expected to diverge into discrete clusters through geographic distance alone; they have termed the geographically based clusters within a single ecotype as “geotypes.” As is the case for systematics of any organism, geography-associated sequence clusters of bacteria are difficult to interpret.
It is sometimes difficult to rule out the geotype hypothesis even when bacterial sequence clusters are sympatric. This may be the case when previously allopatric geotypes have only recently become sympatric, and have not yet had time for a periodic selection event to purge diversity throughout the ecotype. We will address this issue in the final section of the paper.
Discovery of Ecotypes As Star Clades
Another issue remains. A sequence-based phylogeny from almost any named bacterial species reveals a hierarchy of clusters, subclusters, and sub-subclusters. This raises the possibility that a typical named bacterial species may contain many cryptic and uncharacterized ecotypes, each corresponding to a small subcluster. The challenge is to determine which level of subcluster corresponds to ecotypes.
The peculiar dynamics of bacteria provide a method for identifying ecotypes based on sequence data.22 Our “Star” approach assumes that the sequence diversity within an ecotype is constrained largely by periodic selection and much less by genetic drift, an assumption I will return to later.
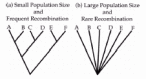
Consider the consequences of periodic selection on the phylogeny of strains from the same ecotype. Nearly all stains randomly sampled from an ecotype should trace their ancestries directly back to the adaptive mutant that caused and survived the last selective sweep. Thus, the phylogeny of an ecotype should be consistent with a star clade, with only one ancestral node, such that all members of an ecotype are equally closely related to one another (fig. 5B). In contrast, a population whose sequence diversity is limited by genetic drift will have a phylogeny with many nodes (fig. 5A).
In an asexual ecotype, a sequence-based phylogeny would yield a perfect star clade, with only minor exceptions due to homoplasy. However, in a bacterial ecotype subject to modest rates of recombination, particularly with other ecotypes, the sequence-based phylogeny can deviate significantly from a perfect star. We have developed a computer simulation to determine how closely a sequence-based phylogeny of strains from the same ecotype should resemble a star clade. Taking into account the taxon's mutation and recombination parameters, the Star simulation determines the likelihood that the phylogeny of strains from a single ecotype would have only one significant node (i.e., a perfect star), versus two, three, four, or more significant nodes (significance determined by 95% bootstrap support).22
We found that for S. aureus,which is among the most clonal of bacteria, an ecotype's phylogeny should almost never have more than one node.22 In the case of N. meningitidis, which is among the most frequently recombining bacteria, the phylogeny of an ecotype is expected to have one or two significant nodes, but almost never three or more. Accordingly, we may tentatively identify ecotypes of N.meningitidis as the largest clades that contain up to two significant nodes.
While Star produces a theory-based criterion for testing whether a set of strains belong to the same ecotype, this approach does not help us choose the groups of strains to be tested for membership within an ecotype. As I have previously shown,22 the Multilocus Sequence Typing method (MLST) developed by B. Spratt and coworkers37 produces reasonable hypotheses for demarcating strains of a named species into ecotypes.
Discovery of Ecotypes through Multilocus Sequence Typing
In MLST, partial sequences (450 bp) of seven gene loci that produce housekeeping proteins are surveyed. The evolutionary distance between strains is quantified in MLST as the number of loci that are different, whether by substitution of a single nucleotide or a swath of nucleotides (possibly due to a recombination event). Strains are then classified into “clonal complexes”: all strains that are identical with a particular strain at five or more loci (in some cases, six or more loci) are deemed members of a clonal complex. E. Feil has developed the “Burst” computer algorithm for assigning strains into clonal complexes according to criteria set by the user (web site: www.mlst.net).
The clonal complexes defined by MLST correspond remarkably well to ecologically distinct groups, even in taxa such as N. meningitidis where recombination is unusually high. I have hypothesized that the clonal complexes identified by MLST are ecotypes.5,22 Because periodic selection is recurrently purging the diversity within an ecotype, ecotypes are expected to accumulate only a limited level of sequence diversity between periodic selection events, depending on the rates of mutation and recombination (which generate variation) and the time between periodic selection events. We may speculate that ecotypes typically have only enough time between selective sweeps for a given strain to accumulate divergence at one or two loci out of seven, on average, whether by mutation or recombination. This yields the 5/7 and 6/7 criteria used in MLST. In general, one would expect that frequently recombining bacteria (in which a locus is ten times more likely to be struck by a recombination than a mutation event) would diverge at more loci between periodic selection events, compared to rarely recombining bacteria, where nearly all divergence accumulates simply through mutation.
Because MLST's 5/7 and 6/7 criteria are intuitively based, we should test whether MLST's clonal complexes do indeed correspond to ecotypes. The Star algorithm can test whether the clonal complexes identified with MLST have phylogenies consistent with ecotypes, taking into account the recombination and mutation parameters estimated for the particular taxon.
I have tested whether the strains of each of the ten clonal complexes found within N. meningitidis are consistent with the Star simulation's expectations for a single ecotype.22 It turns out that the phylogenies of all but one of the ten MLST clonal complexes within N. meningitidis contain one or two nodes, as expected given N. meningitidis's recombination parameters. Similarly, all but three of the 26 clonal complexes within S. aureus are consistent with the expectation for a single ecotype (i.e., containing no more than one significant node). The three exceptional clonal complexes, when pooled together, contain only one significant node among them, suggesting that they are members of the same ecotype. Taking into account the rarity of recombination in S. aureus, perhaps the criterion for inclusion within a clonal complex in this taxon should be 6/7 instead of 5/7 identical loci. It will be interesting to use the Star approach to calibrate the Burst criterion.
In summary, Star demonstrates that the clonal complexes yielded by MLST have phylogenies consistent with ecotypes, at least within S. aureus and N. meningitidis. The clonal complexes produced by MLST do indeed yield reliable hypotheses about the membership of ecotypes, and these hypotheses can be tested using Star.
It is striking that each named species studied by MLST has so many clonal complexes.11,12 If each of these clonal complexes can be shown definitively to be a separate ecotype, each with the universal properties of species, a named bacterial species may actually be more like a genus than a species.22
We should regard the ecotypes identified by sequence-based approaches as only putative until each ecotype can be shown to be ecologically distinct. Ideally, we should also demonstrate that each group has undergone its own private periodic selection events. This is because two putative ecotypes that are only slightly different ecologically may be subject to extinction by one another's periodic selection events. To show that each putative ecotype has already undergone one or more separate periodic selection events would bolster our claim that the clusters and clonal complexes we identify are actually distinct ecotypes. In the next section, I outline a sequence-based method for demonstrating that putative ecotypes have undergone their own, private periodic selection events.
Has Periodic Selection Occurred in Nature?
Periodic Selection Is Inevitable at Bacterial Recombination Rates
We have seen that the rare recombination typical of bacteria is not sufficient to preserve a bacterial population's genetic diversity (fig. 2). Each adaptive mutation that moves to fixation will eliminate nearly all of the sequence diversity, depending on the recombination rate and the intensity of selection (and to a lesser extent, the population size). Given the inevitability of mutational improvements over time, should we not expect to see recurrent purges of diversity?
Notley-McRobb and Ferenci2 have argued that recurrent adaptive mutations will not necessarily purge diversity within a rarely sexual population of bacteria. This may be the case if, in each bout of selection, there are many independently derived adaptive mutations, each stemming from a different genetic background.38 For example, Notley-McRobb and Ferenci2 found that as many as 13 adaptive mutant alleles at the mlc locus swept simultaneously through an experimental population of E. coli. Thus, instead of one genetic background sweeping through the population, there would be 13, diminishing the diversity-purging effect of periodic selection.
Nevertheless, I believe we should not conclude that periodic selection is generally ineffective at purging diversity. First, even if the periodic selection events in Notley-McRobb and Ferenci's2 study are typical of nature, periodic selection should still purge population diversity, albeit at a slower rate. Note that strong selection favoring a single adaptive mutation sweeps all but 0.2% of the diversity from a population, even under high recombination rates. If there were instead ten adaptive mutations sweeping the population simultaneously, the fraction of diversity saved would be increased by only a factor of ten, in this case to 2% of the population's original diversity. So, whether one or several adaptive mutations sweep through each periodic selection event, recurrent sweeps will indeed limit population diversity to very low levels.
Moreover, we cannot expect every selective sweep to be driven by an ensemble of equally fit adaptive mutations. This will be the case only when a population is placed in a new environment and many equally good mutations can accommodate the environmental change. For example, we know that the glucose-limiting environment of Notley-McRobb and Ferenci's experiment favors changes in the regulatory loci mlc, mglD, and malT, and many mutations at these loci appear equally good at adjusting the cells to this environment.
On a global or even regional scale, environmental change is not likely to cause selective sweeps across the entire geographical range of an ecotype, unless the environmental change is global. Because different local populations are likely to see environmental changes in different directions at a given moment, the adaptive mutations that accommodate environmental change are not the ones that would sweep an entire ecotype in all its localities.
What are the adaptive mutations most likely to sweep an ecotype throughout all its diverse habitats? These are mutations (or recombination events) that bring about a novel and generally useful adaptation, which improves the fitness of the ecotype throughout its range of living situations. For example, these might be overall improvements in efficiency, which are adaptive in any circumstance. All the easily accessible mutations of this nature have already been obtained (e.g., all single-nucleotide substitutions are immediately accessible owing to the large population sizes of bacteria); those that remain are much less frequent changes that involve two or more simultaneous mutations (e.g., where none is adaptive without the others), or perhaps recombination with other species. In contrast to the case for a change in environment, where many mutations can independently and simultaneously rise to the occasion, the wait for an innovation is rewarded by only a single rare event. And when this event occurs in a rarely recombining ecotype of bacteria, it will purge the diversity, ecotype-wide, as predicted in (fig. 2).
Periodic Selection Explains the Small Effective Population Sizes of Bacteria
A named species of bacteria typically has only a modest level of DNA sequence variation in protein-coding genes, ranging from less than 0.01 to 0.05 (average pair wise sequence divergence). If sequence diversity were limited by genetic drift, a typical species-wide diversity level (e.g., 0.02 for N. meningitidis) would correspond to an effective population size (Ne) of 2.2 X 107 (assuming the whole named species to be a single ecotype).21 Given the enormous census sizes of bacteria in nature, this estimate of Ne appears absurdly low. However, periodic selection can reasonably explain the low levels of sequence diversity typical for bacteria. Periodic selection occurring once in Ne generations will yield the same amount of diversity as pure drift would in a population of size Ne. Thus, periodic selection occurring once in 2.2 X 107 generations would yield the diversity levels typical of named species (e.g.,N. meningitidis). Periodic selection need not occur often to constrain ecotype-wide diversity to modest levels.
As I have argued earlier, the range of periodic selection events is more likely to correspond to MLST's clonal complexes than to named species. In this case, a much lower frequency of ecotype-wide periodic selection would be required to explain observed diversity levels. For example, the average sequence diversity within N. meningitidis's clonal complexes (0.005) would require ecotype-wide periodic selection occurring once in 6 X 106 generations.
Identification of Periodic Selection Events
A survey of sequence diversity in E. coli39 has provided the clearest evidence for a periodic selection event in nature. Most genes in the survey corroborated the population structure already known from allozyme data: strains fell into four major sequence clusters. However, within one gene region, near gapA, all strains were anomalously homogeneous in sequence. These results were interpreted as evidence for a selective sweep throughout E. coli, driven by an adaptive mutation in the gapA region.
This interpretation would be entirely appropriate if E. coli were a highly sexual species. In the case of animals and plants, the diversity-purging effect of natural selection is limited to the chromosomal region near the adaptive mutation, where recombination with the adaptive mutation is infrequent. In bacteria, however, recombination between any two genes, regardless of their distance on the chromosome, is extremely rare. Therefore, if all of E. coli were a single ecotype, and the sequence homogeneity around gapA were caused by an ecotype-wide purging of diversity, we would expect all of E. coli to be purged of diversity over the entire chromosome.
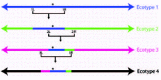
Jacek Majewski and I30 previously proposed the adapt globally, act locally model to explain anomalous homogeneity around a small chromosomal region, as seen for gapA in E. coli. Because E. coli forms four major sequence clusters, as well as many smaller subclusters, we may tentatively conclude that E. coli contains several ecotypes. We proposed that the adaptive mutation around gapA was generally useful for all of the ecotypes of E. coli, and that the allele was passed between ecotypes, precipitating a periodic selection event within each (fig. 4). Thus, for genes closely linked to the adaptive mutation, there would be nearly total purging of diversity both within and between ecotypes, but for genes not linked to the adaptive mutation, selection would purge only the diversity within ecotypes. Whenever a small chromosomal segment is homogenized across strains that otherwise form distinct clusters, a generally useful adaptive mutation is likely to have passed from ecotype to ecotype, causing local periodic selection in each.
Beyond providing evidence for periodic selection, the sequence pattern found by Guttman and Dykhuizen39 can provide additional evidence that sequence clusters correspond to ecotypes. Recall that the clusters we discover may correspond either to ecotypes or to geotypes, which are populations of the same ecotype with a history of geographic isolation. This issue can be resolved when we find evidence of the Guttman-Dykhuizen pattern.
Different sequence clusters can correspond to geotypes within the same ecotype only if there has not been an opportunity for periodic selection to sweep through all of the clusters. If we can show that these clusters have survived as distinct groups through a periodic selection event, we can rule out the geotype hypothesis. This is indeed the case for the various clusters within E. coli. The homogenization of the gapA region across the various major clusters in E. coli shows that these clusters have maintained their distinctness, even as an adaptive mutation has caused periodic selection events within each cluster.
The Guttman-Dykhuizen pattern can provide further evidence of multiple ecotypes. We should expect that the adaptive allele driving the periodic selections in all of the ecotypes is passed to each ecotype in a separate recombination event. Therefore, the region that is homogenized should be somewhat different for each pair of ecotypes, reflecting the junctions of the recombination events that transferred the adaptive mutation across ecotypes (fig. 6). We may thus predict that if the sequence clusters correspond to ecotypes, the junctions of the homogeneous region will be unique for each pair of sequence clusters.
Guttman and Dykhuizen's39 discovery of a periodic selection event was based on a serendipitous choice of loci to survey, but today comparisons of whole genomes should provide ample opportunities for genome-wide screening of periodic selection events driven by “adapt globally, act locally” mutations. Discovery of these periodic selection events would allow us to confirm that the many sequence clusters found within named species are distinct ecotypes.
Acknowledgements
I am grateful to Michael Dehn for help in clarifying the manuscript. This work was supported by National Science Foundation grants DEB-9815576 and EF-0328698 and research funds from Wesleyan University.
References
- 1.
- Atwood KC, Schneider LK, Ryan FJ. Periodic selection in Escherichia coli. Proc Natl Acad Sci USA. 1951;37:146–155. [PMC free article: PMC1063322] [PubMed: 14808170]
- 2.
- Notley-McRobb L, Ferenci T. Experimental analysis of molecular events during mutational periodic selections in bacterial evolution. Genetics. 2000;156(4):1493–1501. [PMC free article: PMC1461358] [PubMed: 11102352]
- 3.
- Maynard SmithJ, Smith N, O'Rourke M. et al. How clonal are bacteria? Proc Natl Acad Sci USA. 1993 [PMC free article: PMC46515] [PubMed: 8506277]
- 4.
- Cohan FM. Clonal structure: An overview. In: Pagel M, ed. Encyclopedia of Evolution. New York: Oxford University Press, 2002a:158-161.
- 5.
- Cohan FM. Population structure and clonality of bacteria. In: Pagel M, ed. Encyclopedia of Evolution. New York: Oxford University Press, 2002b:161-163.
- 6.
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. [PubMed: 10830951]
- 7.
- Posada D. Evaluation of methods for detecting recombination from DNA sequences: Empirical data. Mol Biol Evol. 2002;19(5):708–717. [PubMed: 11961104]
- 8.
- Maynard SmithJ, Smith NH. Detecting recombination from gene trees. Mol Biol Evol. 1998;15(5):590–599. [PubMed: 9580989]
- 9.
- Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;145(3):833–846. [PMC free article: PMC1207867] [PubMed: 9055092]
- 10.
- Hudson RR. Estimating the recombination parameter of a finite population model without selection. Genet Res. 1987;50(3):245–250. [PubMed: 3443297]
- 11.
- Feil EJ, Maiden MC, Achtman M. et al. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16(11):1496–1502. [PubMed: 10555280]
- 12.
- Feil EJ, Smith JM, Enright MC. et al. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics. 2000;154(4):1439–1450. [PMC free article: PMC1461021] [PubMed: 10747043]
- 13.
- Guttman DS, Dykhuizen DE. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994a;266(5189):1380–1383. [PubMed: 7973728]
- 14.
- Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–590. [PubMed: 11544367]
- 15.
- Suerbaum S, Maynard SmithJ, Bapumia K. et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. [PMC free article: PMC22880] [PubMed: 9770535]
- 16.
- Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88(16):7160–7164. [PMC free article: PMC52253] [PubMed: 1831267]
- 17.
- Duncan KE, Istock CA, Graham JB. et al. Genetic exchange between Bacillus subtilis and Bacillus licheniformis: Variable hybrid stability and the nature of species. Evolution. 1989;43:1585–1609. [PubMed: 28564334]
- 18.
- Cohan FM. Bacterial species and speciation. Syst Biol. 2001;50:513–524. [PubMed: 12116650]
- 19.
- Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95(16):9413–9417. [PMC free article: PMC21352] [PubMed: 9689094]
- 20.
- Braverman JM, Hudson RR, Kaplan NL. et al. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics. 1995;140(2):783–796. [PMC free article: PMC1206652] [PubMed: 7498754]
- 21.
- Cohan FM. The effects of rare but promiscuous genetic exchange on evolutionary divergence in prokaryotes. Am Naturalist. 1994;143:965–986.
- 22.
- Cohan FM. What are bacterial species? Annual Review of Microbiology 2002c; 56:457-487. [PMC free article: PMC94378] [PubMed: 12142474]
- 23.
- Templeton A. The meaning of species and speciation: A genetic perspectiveIn: Otte D, Endler J, eds.Speciation and its Consequences Sunderland MA: Sinauer Assoc, 1989.
- 24.
- Simpson G. Principles of Animal Taxonomy. New York: Columbia Univ Press, 1961.
- 25.
- Wiley E. The evolutionary species concept reconsidered. Syst Zool. 1978;27:17–26.
- 26.
- Palys T, Nakamura LK, Cohan FM. Discovery and classification of ecological diversity in the bacterial world: The role of DNA sequence data. Int J Syst Bacteriol. 1997;47(4):1145–1156. [PubMed: 9336922]
- 27.
- Sokal R, Crovello T. The biological species concept: A critical evaluation. Am Nat. 1970;104:127–153.
- 28.
- Mallet J. A species definition for the modern synthesis. Trends in Ecology & Evolution. 1995;10:294–299. [PubMed: 21237047]
- 29.
- VanValen L. Ecological species, multispecies, and oaks. Taxon. 1976;25:233–239.
- 30.
- Majewski J, Cohan FM. Adapt globally, act locally: The effect of selective sweeps on bacterial sequence diversity. Genetics. 1999;152(4):1459–1474. [PMC free article: PMC1460694] [PubMed: 10430576]
- 31.
- Holt RD. On the relationship between niche overlap and competition: The effect of incommensurable niche dimensions. Oikos. 1987;48:110–114.
- 32.
- Lawrence J. Gene Transfer in Bacteria: Speciation without Species? Theor Popul Biol. 2002;61(4):449. [PubMed: 12167364]
- 33.
- Lawrence J. Catalyzing Bacterial Speciation: Correlating Lateral Transfer with Genetic Headroom. Syst Biol. 2001;50(4):479–496. [PubMed: 12116648]
- 34.
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394(6688):69–72. [PubMed: 9665128]
- 35.
- Treves DS, Manning S, Adams J. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol. 1998;15(7):789–797. [PubMed: 9656481]
- 36.
- Imhof M, Schlotterer C. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc Natl Acad Sci USA. 2001;98(3):1113–1117. [PMC free article: PMC14717] [PubMed: 11158603]
- 37.
- Maiden MC, Bygraves JA, Feil E. et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95(6):3140–3145. [PMC free article: PMC19708] [PubMed: 9501229]
- 38.
- Korona R. Genetic divergence and fitness convergence under uniform selection in experimental populations of bacteria. Genetics. 1996;143(2):637–644. [PMC free article: PMC1207325] [PubMed: 8725215]
- 39.
- Guttman DS, Dykhuizen DE. Detecting selective sweeps in naturally occurring Escherichia coli. Genetics. 1994b;138(4):993–1003. [PMC free article: PMC1206276] [PubMed: 7896119]
- Introduction
- The Nature of Recombination in Bacteria
- The Effect of Rare Recombination on Diversity within a Population
- The Origins of Permanent Divergence
- Effects of Periodic Selection beyond the Boundaries of the Ecotype
- Periodic Selection and Discovery of Bacterial Ecotypes
- Has Periodic Selection Occurred in Nature?
- Acknowledgements
- References
- Periodic Selection and Ecological Diversity in Bacteria - Madame Curie Bioscienc...Periodic Selection and Ecological Diversity in Bacteria - Madame Curie Bioscience Database
- Brain Metastasis - Madame Curie Bioscience DatabaseBrain Metastasis - Madame Curie Bioscience Database
- Theory of Organelle Biogenesis: A Historical Perspective - Madame Curie Bioscien...Theory of Organelle Biogenesis: A Historical Perspective - Madame Curie Bioscience Database
- Pseudouridine Formation, the Most Common Transglycosylation in RNA - Madame Curi...Pseudouridine Formation, the Most Common Transglycosylation in RNA - Madame Curie Bioscience Database
- Modeling Metastasis In Vivo - Madame Curie Bioscience DatabaseModeling Metastasis In Vivo - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...