NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The chemical structure of poly(ADP-ribose) suggests not only that its modification of acceptor proteins should modify the structure and function of the acceptor proteins, but also that the poly(ADP-ribose) molecule itself should possess an intrinsic structural information that can alter cellular function(s).
The localization of PARP-1 to the centrosome clearly shows that its function is not only confined to the nucleus, but plays a role also in the cytoplasm. Thus poly(ADP-ribosyl)ation should be considered an important regulatory mechanism not only in the nucleus, but in the cell at large. In this context, the interaction between nuclear and cytoplasmic events through the poly(ADP-ribosyl)ation reaction is an intriguing possibility.
Understanding poly(ADP-ribose) metabolism has an important impact for unraveling fundamental biological mechanisms ranging from chromosomal instability in cancer, the morphogenesis of the tissues and the maintenance of neuronal cell functions.
Introduction
Some 40 years have passed since poly(ADP-ribose) and the unique poly(ADP-ribosyl)ation reaction were discovered.1-4 The characterization of poly(ADP-ribose) as a huge biopolymer with branching makes poly(ADP-ribosyl)ation a unique post-translational modification of biological materials as well as a structural component of eukaryotic cell chromatin. The subcellular localization of poly(ADP-ribose) polymerase (now called PARP-1) in the nucleus and its enzymatic activation by DNA strand breaks immediately suggested that this modification should represent an important mechanism of DNA repair and other reactions involved in DNA metabolism. However the involvement of this reaction is not confined to the nucleus but might be involved in cytoplasmic events. Recent findings in our laboratory established that PARP-1 also localizes to the centrosome, and cells from PARP-1 knockout mice show several alterations linked with centrosome function. Our data also indicated an involvement of poly(ADP-ribosyl)ation in the regulation of neuronal cells. This Chapter summarizes recent findings suggesting that the metabolism of poly(ADP-ribose) is dynamic and important for the regulation of several critical cell functions, including the mechanisms of centrosome duplication, tissue morphogenesis and neurodegeneration.
Dynamic Nature of Poly(ADP-Ribose) Metabolism Due to the Interplay between PARP and PARG
The poly(ADP-ribose) chain attached to the acceptor protein possesses unique characteristics with regards to charge and structure.5 Although poly(ADP-ribosyl)ation is one out of a large number of covalent posttranslational modification of proteins, which typically alter protein structure and function, the huge size the ADP-ribose polymer can attain strongly suggests that it should have additional function as well. For example, when DNA strand breaks activate poly(ADP-ribose) synthesis on an acceptor protein on the chromatin, it might structurally hinder DNA polymerase and/or RNA polymerase to use the DNA template. It is well known that after DNA damage, cell cycle stops at S phase allowing the time for DNA repair to complete. This might be explained in one way as the physical hindrance of DNA synthesis by a huge molecule of poly(ADP-ribose) in the process of DNA polymerase traveling on the chromatin template.
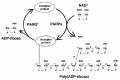
Poly(ADP-ribose) is synthesized by PARP-1 and hydrolyzed by enzymes known as poly(ADP-ribose) glycohydrolase (PARG), phosphodiesterases (PDases) and ADP-ribosyl protein lyase.6,7 Among these, PARG serves as an enzyme that hydrolyzes poly(ADP-ribose) chains quite efficiently, including the branched portion, and finally leaves the protein-proximal mono-ADP-ribose molecule, which might be removed by ADP-ribosyl protein lyase7 or released spontaneously at neutral pH. Such de-modification would enable PARP-1 to use the same acceptor protein in a new cycle of poly(ADP-ribosyl)ation (fig. 1).
Since both the modification of poly(ADP-ribose) by PARP-1 and the degradation of poly(ADP-ribose) after DNA damage occur quite rapidly,8 it follows that poly(ADP-ribose) metabolism in the cells is highly dynamic process.
Control of Cellular Function, Including Centrosome Duplication, by Poly(ADP-Ribosyl)ation
It is well known that the centrosome functions as microtubule organizing center (MTOC) and can regulate the morphology of the cell and the transport of certain proteins via motor proteins like dynein or kinesin. During the cell cycle, the centrosome starts to duplicate at the G1-S boundary, undergoes maturation in the G2 phase, and is instrumental for the equal distribution of the duplicated chromosomes to daughter cells during mitosis. Thus, the centrosome plays a vital role in maintaining chromosomal stability.9 There is a hypothesis that malignant tumors arise through centrosome defects that result in improper cell divisions and give rise to aneuploidy. This theory was proposed a century ago.10 Recently, many reports appeared showing that some posttranslational modifications including phosphorylation and ubiquitination occur in centrosome and regulate its function. Our own recent work suggests that poly(ADP-ribosyl)ation is also involved in centrosome function.
Previous studies had shown that PARP-1 is mainly localized to the nucleus. However from the pattern of chromosome instability in PARP-1 knockout mouse cells, we speculated that PARP-1 might also be localized to another component related to the mitotic machinery. Indeed, we found PARP-1 localized to the centrosome in some cancer cells and mouse embryonic fibroblasts (MEF).11,12 The localization of PARP-1 at the centrosome suggested the possibility that PARP-1 at the centrosome could catalyze poly(ADP-ribosyl)ation of certain centrosomal proteins. In fact we could observe the presence of various bands in western blots of the centrosomal proteins that reacted with a monoclonal antibody to poly(ADP-ribose) (10H).13 These results showed the involvement of PARP-1-mediated poly(ADP-ribosyl)ation in centrosome regulation.
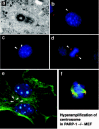
To unravel a possible cause-and-effect relationship, we used the PARP inhibitor 3-aminozenzamide (3-AB) in cell culture experiments. Surprisingly, wild type MEFs, when cultured in the presence of 3-AB for one week, showed marked centrosome hyperamplification. Further experiments using immortalized and primary cells derived from PARP-1-/- mice also showed centrosome hyperamplification (fig. 2). During cell cycle, centrosome duplication should be coupled with DNA replication and should occur only once in each cell cycle. Therefore if there is hyperamplification of the centrosome, there should be some abnormality in the coupling between the timing of initiation of the centrosome duplication and DNA replication. Indeed, PARP-1-/- MEF displayed an uncoupling of the timing between the two events, thus resulting in centrosome hyperamplification. These results showed that PARP-1-mediated poly(ADP-ribosyl)ation is necessary for the maintenance of the proper number of centrosomes.
It is important to identify each of the poly(ADP-ribosyl)ated proteins in order to clarify the role of poly(ADP-ribosyl)ation. Indeed we have identified some proteins that are poly(ADP-ribosyl)ated in the centrosome. One of these proteins is the tumor suppressor protein p53. p53 is a well known guardian of the genome, and it is essential for DNA repair and apoptosis. Interestingly, p53 also localizes to the centrosome and regulates centrosome function directly or indirectly. Our study revealed that inhibition of poly(ADP-ribosyl)ation of p53 due to administration of a PARP inhibitor or loss of PARP-1 might be involved in the defect in centrosome function and chromosomal instability.
Over the past few years a fairly large family of PARP enzymes has emerged and several PARP isoforms have been characterized to some extent.14 Especially, it was reported (i) that PARP-1 is localized to the centromere on the chromosomes and interacted with CENPA, CENPB, and Bub3;15 (ii) that vault PARP (193 kDa) is localized to the cytoplasm and also to the nucleus during interphase or the mitotic spindle during metaphase;16 and (iii) that tankyrase (142 kDa) is localized to the telomere, nuclear envelope, nuclear pore complex and pericentrimatrix.17 Together, these reports suggest that poly(ADP-ribosyl)ation might be involved in some of the chromosome functions during mitosis (fig. 3).
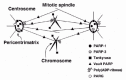
Figure 3
Localization of poly(ADP-ribose) and its metabolizing enzymes in the mitotic apparatus.
Out data show that there are many proteins that are poly(ADP-ribosyl)ated. In view of the dynamic process of poly(ADP-ribosyl)ation, a posttranslational modification undergoing rapid turnover, the recent discovery that poly(ADP-ribose) glycohydrolase (PARG) is also localized in the centrosome during the cell cycle18 is very interesting and plausible.
Identification of the poly(ADP-ribosyl)ated centrosomal proteins and clarification of the changes in subcellular localization by possible shuttling between the centrosome and the nucleus will be essential for further understanding the role of poly(ADP-ribosyl)ation. It would also be very interesting to know the trigger(s) of poly(ADP-ribosyl)ation in the centrosome, since it is believed that this structure is devoid of DNA. Such analyses would clarify a novel mechanism of the regulation of the centrosome function by posttranslational poly(ADP-ribosyl)ation.
Poly(ADP-Ribose) Metabolism in Dosophila melanogaster
In addition to the crucial function of centrosomes in chromosome segregation and cytokinesis, centrosomes coordinate all microtubule-related cellular functions, including cell shape, polarity, adhesion and motility, as well as the intracellular transport and positioning of organelles by controlling the number, polarity and distribution of microtubules.19 Thus it was interesting to see the effects of gain or loss of poly(ADP-ribosyl)ation activity in vivo.
Drosophila melanogaster is an organism with only about 5% of the genome size of humans or mice. Thus it should be much easier to analyze the significance of gain or loss of poly(ADP-ribosyl)ation function in vivo. The role of poly(ADP-ribosyl)ation in the development of an embryo or tissue, a fundamental biological process in vivo, had not been studied intensively so far. We therefore produced transgenic Drosophila strains using dPARP-I, an orthologue of mammalian PARP-1, and found that overexpression of this enzyme induced disruption of tissue polarity and disorganization of cytoskeleton in Drosophila melanogaster.20 Tulin and Spradling found that downregulation of dPARP caused some developmental defect.21,22
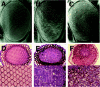
The GMR-PARP fly, which is one of the PARP-transgenic lines expressing dPARP-I specifically in developing eye, showed mild roughening of the adult compound eye, which consists of about 800 individual units of ommatidia. In this transgenic strain, the arrangement of ommatidia was disordered, with disruption of tissue polarity characterized by improper rotation and chirality of ommatidia. By contrast, in the wild type, there is a highly ordered array of an asymmetric trapezoidal pattern of seven rhabdomeres in the photoreceptors (fig. 4). The disruption of tissue polarity in the GMR-PARP mutant was already found at the initial stage of eye development and was also observed in various tissues including eye in hs-PARP, which is another PARP-transgenic line expressing dPARP-I in the whole body (data not shown).
It is known that the polarity of the Drosophila eye is regulated by the wnt/frizzled signaling pathway. The tissue polarity phenotype in GMR-PARP or hs-PARP closely resembled that of mutants or transgenic flies with alterations of wnt signaling components. In addition to tissue polarity disruption, PARP-1 overexpression induced disorganization of cytoskeletal F-actin. Furthermore, overexpression of PARP-1 neutralized the effect of overexpression of the small GTPase RhoA (fig. 5). RhoA is reported to regulate tissue polarity downstream of wnt signaling pathway in Drosophila. From these observations, it is concluded that overexpression of PARP interferes with wnt signaling or other tissue polarity signaling pathways.
While there exist at least two members of the PARP family in Drosophila, only one gene encoding PARG can be retrieved in the available databases of the Drosophila genome. Thus it was interesting to knock out the parg gene in Drosophila in order to study the importance of poly(ADP-ribosyl)ation in vivo. The Drosophila parg gene has been mapped to the X chromosome. A mutant, EP351, with a P element in the 5'UTR of the parg gene was available from the Berkeley Drosophila Genome Project (GenBank accession no. AQ025499). We made deletion mutants by imprecise excision of the P element. One of the mutants, parg27.1, lacked two-thirds of the parg ORF, including the conserved catalytic domain.23 The allele, parg27.1, was maintained with a balancer X chromosome. Almost all parg27.1/Y embryos laid by parg27.1/balancer X females that had been crossed with balancer X/Y males hatched, and two-thirds of the larvae developed to the pupal stage, but they showed lethality before eclosion at 25°C. However when parg27.1/Y males were maintained at 29°C, approximately one-fourth of the parg27.1/Y embryo developed into apparently normal adult flies. It was necessary to elevate the culture temperature from 25°C to 29°C before or just after pupation.
The parg27.1/Y male and parg27.1/parg27.1 female flies showed neurological abnormalities and reduced locomotor activity, which became progressively more severe. Most mutant flies died within 10 days after eclosion. Most parg27.1 adult flies dragged their wings and could not fly. Three-fourth of them developed a black spot(s) in one or both of the base joints of the second limb. Both parg27.1/Y males and parg27.1/parg27.1 females were sterile. The above neurological disorders and sterility found in mutant flies were rescued by expression of transgene containing the parg ORF with 1 kb of its upstream sequence, or parg cDNA with a heat inducible promoter.23
Accumulation of poly(ADP-ribose) was analyzed by ELISA using an anti-poly(ADP-ribose) antibody, 10H. The parg27.1/Y male flies showed strong signals, while the wild type, parg27.1/ X balancer female and X balancer/Y males had only faint signals. Using immunohistochemistry, poly(ADP-ribose) was widely detected in the mutant, being prominent in the central nervous system including the eye and the thoracic ganglion regions. Strong signals were also detected at the surface of the brain, where the neuronal cells were clustered.
The microscopic findings indicated a remarkable neurodegeneration occurring at two weeks after eclosion. The normal structure of axons in the wild-type optic lobe was completely absent in the mutant brain, as examined by immunostaining of phosphorylated neurofilament. Electron microscopic analysis showed aggregate(s) of uniform particles adjacent to nucleolus in the mutant brain a few days after eclosion. The localization of the abnormal aggregate(s) might be poly(ADP-ribose) or poly(ADP-ribosyl)ated proteins. Most of the cells in the brain of the mutants were swollen, and the organelles and the nuclei were no longer visible clearly. There were many condensed bodies indicating cell death (fig. 6).
The observed neurodegeneration of the mutant fly indicates an indispensable role of PARG in the maintenance of neuronal function and cell survival. Interestingly, in a plant system, the period length of the circadian oscillator of Arabidopsis was recently reported to be distorted by mutation of the PARG orthologue, the tej gene.24 Although the molecular mechanism of the neurodegeneration in Drosophila is not clear, it is known that some neurological disorders are ameliorated by expression of the heat shock proteins.25 It has been suggested that the accumulation of protein aggregates as inclusion bodies will cause such neurological damage as was found in a number of neurodegenerative diseases including Alzheimer's disease, Parkinson's disease, prion disease, polyglutamine disease, the Tauopathies, and familial amyotrophic lateral sclerosis.26 Progressive neurological deterioration and renal failure due to the accumulation of glutamyl ribose-5-phosphate, which might be the degradation product of poly(ADP-ribosyl)ated protein, has also been reported.27 Understanding the metabolism and also the protein-protein interactions with poly(ADP-ribosyl)ated target proteins should yield important information to clarify the pathogenesis of the above diseases.
Acknowledgment
We would like to thank Drs. WM Tong and ZQ Wang of IARC, Dr. K Fukasawa of University of Cincinnatti College of Medicine, Drs. M Yamada and H Takahashi of Niigata University, Drs. F Uchiumi, H Maruta and S Tanuma of Tokyo University of Science, Drs. K Sawamoto and H Okano of Osaka University and Drs. S Ohashi, K Okamoto, E Sugihara and N Uematsu of our Department for their cooperation in this work. We also thank Ms. Ayako Kabayama for secretarial assistance.
References
- 1.
- Bouchard VJ, Rouleau M, Poirier GG. PARP-1, determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. [PubMed: 12829019]
- 2.
- Masutani M, Nakagama H, Sugimura T. Poly(ADP-ribose) and carcinogenesis. Genes Chromosomes Cancer. 2003;38:339–348. [PubMed: 14566854]
- 3.
- Bürkle A. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays. 2001;23:795–806. [PubMed: 11536292]
- 4.
- Shall S, de MurciaG. Poly(ADP-ribose) polymerase-1: What have we learned from the deficient mouse model? Mutat Res. 2000;460:1–15. [PubMed: 10856830]
- 5.
- Miwa M, Saikawa N, Yamaizumi Z. et al. Structure of poly(adenosine diphosphate ribose): Identification of 2'-[1"-ribosyl-2"-(or 3"-)(1"_-ribosyl)]adenosine-5',5",5"_-tris(phosphate) as a branch linkage. Proc Natl Acad Sci USA. 1979;76:595–599. [PMC free article: PMC382995] [PubMed: 218210]
- 6.
- Miwa M, Sugimura T. Phosphodiesterases and poly(ADP-ribose) glycohydrolaseIn: Hayaishi O, Ueda K, eds.ADP-ribosylation Reactions New York: Academic Press,1982263–277.
- 7.
- Okayama H, Honda M, Hayaishi O. Novel enzyme from rat liver that cleaves an ADP-ribosyl histone linkage. Proc Natl Acad Sci USA. 1978;75:2254–2257. [PMC free article: PMC392530] [PubMed: 276865]
- 8.
- Juarez-Salinas H, Sims JL, Jacobson MK. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979;282:740–741. [PubMed: 229416]
- 9.
- Doxsey S. Reevaluating centrosome function. Nat Rev Mol Cell Biol. 2001;2:688–689. [PubMed: 11533726]
- 10.
- Duesberg P, Li R, Rasnick D. et al. Aneuploidy precedes and segregates with chemical carcinogenesis. Cancer Genet Cytogenet. 2000;119:83–93. [PubMed: 10867141]
- 11.
- Kanai M, Uchida M, Hanai S. et al. Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem Biophys Res Commun. 2000;278:385–389. [PubMed: 11097846]
- 12.
- Kanai M, Tong WM, Sugihara E. et al. Involvement of poly(ADP-ribose) polymerase 1 and poly(ADP-ribosyl)ation in regulation of centrosome function. Mol Cell Biol. 2003;23:2451–2462. [PMC free article: PMC150716] [PubMed: 12640128]
- 13.
- Kawamitsu H, Hoshino H, Okada H. et al. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. [PubMed: 6206890]
- 14.
- Smith S. The world according to PARP. Trends Biochem Sci. 2001;26:174–179. [PubMed: 11246023]
- 15.
- Saxena A, Saffery R, Wong LH. et al. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J Biol Chem. 2002;277:26921–26926. [PubMed: 12011073]
- 16.
- Kickhoefer VA, Siva AC, Kedersha NL. et al. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146:917–928. [PMC free article: PMC2169495] [PubMed: 10477748]
- 17.
- Smith S, de LangeT. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112:3649–3656. [PubMed: 10523501]
- 18.
- Ohashi S, Kanai M, Hanai S. et al. Subcellular localization of poly(ADP-ribose) glycohydrolase in mammalian cells. Biochem Biophys Res Commun. 2003;307:915–921. [PubMed: 12878198]
- 19.
- Nigg EA. Centrosome aberrations: Cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. [PubMed: 12415252]
- 20.
- Uchida M, Hanai S, Uematsu N. et al. Overexpression of poly(ADP-ribose) polymerase disrupts organization of cytoskeletal F-actin and tissue polarity in Drosophila. J Biol Chem. 2002;277:6696–6702. [PubMed: 11744702]
- 21.
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. [PubMed: 12543974]
- 22.
- Tulin A, Spradling A. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. [PMC free article: PMC186441] [PubMed: 12183365]
- 23.
- Hanai S, Kanai M, Ohashi S. et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:82–86. [PMC free article: PMC314142] [PubMed: 14676324]
- 24.
- Panda S, Poirier GG, Kay SA. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev Cell. 2002;3:51–61. [PubMed: 12110167]
- 25.
- Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. [PubMed: 12065827]
- 26.
- Bahr BA, Bendiske J. The neuropathogenic contributions of lysosomal dysfunction. J Neurochem. 2002;83:481–489. [PubMed: 12390510]
- 27.
- Williams JC, Butler IJ, Rosenberg HS. et al. Progressive neurologic deterioration and renal failure due to storage of glutamyl ribose-5-phosphate. N Engl J Med. 1984;311(3):152–155. [PubMed: 6738601]
- Roles of Poly(ADP-Ribose) Metabolism in the Regulation of Centrosome Duplication...Roles of Poly(ADP-Ribose) Metabolism in the Regulation of Centrosome Duplication and in the Maintenance of Neuronal Integrity - Madame Curie Bioscience Database
- Enzyme-RNA Substrate Recognition in RNA-Modifying Enzymes - Madame Curie Bioscie...Enzyme-RNA Substrate Recognition in RNA-Modifying Enzymes - Madame Curie Bioscience Database
- DNA Demethylation - Madame Curie Bioscience DatabaseDNA Demethylation - Madame Curie Bioscience Database
- Role of Mammalian Coronin 7 in the Biosynthetic Pathway - Madame Curie Bioscienc...Role of Mammalian Coronin 7 in the Biosynthetic Pathway - Madame Curie Bioscience Database
- Lysosomal Storage Disorders - Madame Curie Bioscience DatabaseLysosomal Storage Disorders - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...