NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Introduction
Bacteriophage P22 was isolated by Zinder and Lederberg1 a half century ago, and was immediately put to work by Salmonella bacterial geneticists because of its unusual (at that time) DNA packaging properties. It was the first generalized transducing phage to be discovered — a small fraction (~2%2) of its virions carry a fragment of the host DNA instead of phage DNA, and this host DNA can be delivered into a host cell. Subsequently it has become one of the paradigm molecular genetic systems for understanding the lifecycles of temperate phages.3,4 P22 virions contain partially circularly permuted (see below for details) and 3.8% terminally redundant dsDNA molecules about 43,400 bp long,5 have very short tails6 and infect smooth (O-antigen surface polysaccharide carrying) strains of Salmonella typhimurium. The many conditional lethal mutations isolated in P22 allowed the delineation of its assembly pathway over a quarter century ago.7-10
Building a Protective Shell for Viral DNA
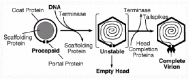
Like other large dsDNA viruses, P22 assembles a protein procapsid and then inserts the DNA chromosome into this preformed container (fig. 1). This strategy appears to be common to all dsDNA bacteriophages as well as the Herpesviridae. The genes required to build the procapsid and fill it with DNA map in a contiguous cluster on the P22 chromosome (fig. 2). The four critical protein players in this process in P22 are analogous to those in other dsDNA viruses—coat, scaffold, portal and terminase (Table 1). The coat protein shells of P22 procapsids and virions have T=7 icosahedral symmetry (each procapsid contains 420 molecules of coat, less any that are replaced by portal), and three-dimensional reconstructions of both kinds of particles have been made from cryo-electron micrographs.11-15 Procapsids contain about 250 molecules of scaffolding protein in the interior, all of which leave at or before the time of DNA packaging.8,16 Scaffolding protein has essential roles in procapsid assembly that include directing coat protein to assemble properly, recruiting portal protein into procapsids, and perhaps excluding cytoplasmic proteins from the procapsid interior.16-19 Twelve molecules of portal protein form a ring at a single capsid vertex to which tails will later attach.20 DNA is thought to pass through this ring during packaging and injection. As in other large, well-studied viruses, a terminase which contains two types of subunits is responsible for DNA recognition and cleavage and perhaps provides the force required for DNA translocation through the portal complex. Sometime after scaffolding protein exit but around the time DNA packaging, the procapsid shell expands and the conformational change to the coat protein lattice results in a physically more robust capsid that is better able to withstand chemical insult.
Table 1
Proteins required for bacteriophage P22 DNA packaging.
DNA Packaging Strategy
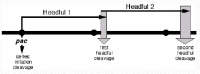
The linear P22 virion DNA is circularized by homologous recombination upon infection and replication eventually produces daughter head-to-tail concatemers that are the substrate for the DNA packaging apparatus.21 The circular permutation and terminal redundancy of the packaged DNA strongly suggests a “headful packaging” mechanism.22 Headful packaging refers to a mechanism in which the interior volume of the capsid shell measures the length of the chromosome that is encapsulated (since it is filled to a particular DNA density) and a “headful nuclease” cleaves packaged DNA from the remainder of the concatemer only when the head is “full” of DNA (this dsDNA cut is called “headful cleavage”). A more careful study of P22 virion DNA by Tye et al23 and Jackson et al24 showed elegantly that this is true, and in addition discovered that the circular permutation was not random, but that its DNA is “partially permuted”. That is, DNA ends are not randomly positioned on the sequence, but are found **largely in one region of the phage's genome sequence. The model proposed to explain partial permutation is that packaging initiates at a particular location called pac. A DNA end is thought to be created here by a DNA cleavage called the “series initiation cleavage” (fig. 3), and the DNA end on one side of this cut is inserted into the procapsid so that packaging proceeds in only one direction from the initiation cleavage point. When DNA inside this procapsid reaches sufficient density, the headful nuclease is activated to cleave the DNA (the first headful cleavage of the packaging series), and the capsid full of DNA is released from the concatemer. A second DNA headful is then inserted into a new procapsid starting at the concatemer end created by the previous headful cleavage and ending with a new headful cleavge (the second headful cleavage of the series). Subsequent packaging events then follow sequentially in the same manner (fig. 3). Such “processive” packaging series can vary from 2 to 12 packaging events long in phage P22, depending upon the infection conditions24,25—it is not known if the number of packaging events in a processive series is limited by concatemer length, or whether multiple series can initiate on a single concatemer molecule.
The DNA length in capsids is 43,400±750 bp, where the range represents the actual variation in length, not the uncertainty of the measurement.5 This corresponds to a DNA density within the phage head of about 0.56 bp per cubic nm which is similar to DNA densities found in crystals of of short dsDNA molecules.26 There is no evidence for any proteins bound to the DNA in the P22 virion, and there would be little room for them if they were present. The nucleotide sequence of the P22 genome is 41,725 bp long and so on average each DNA has an approximately 1800 bp direct repeat (terminal redundancy) at the ends. Homologous recombination between these repeats is responsible for circularization of the DNA in the next round of infection.
Initiation of DNA Packaging
The first step in P22 (or any virus) DNA packaging is recognition of DNA to be packaged. Current evidence points strongly toward the P22 gene 3 protein being responsible for this recognition. P22 is a generalized transducing phage in that about 1-2% of the time it “mistakenly” initiates DNA packaging on the bacterial chromosome and encapsidates a phage chromosome-sized fragment of host DNA.2 The resulting “virus” particle can deliver the host DNA it contains into another Salmonella cell as if it were a phage chromosome. Any such host DNA that is injected into such a cell can recombine with the resident chromosome to alter or “transduce” its genetic properties. Mutants originally isolated by Horst Schmieger as having higher than wild type frequencies of generalized transduction map in gene 3. Some of these do alter the target specificity of DNA recognition by gene 3 protein.27-30 Such mutational changes in specificity are considered to be strong in vivo evidence of direct interaction between DNA and a putative DNA binding protein, since it is unlikely that altering one protein would cause it to, for example, change the specificity of a partner protein. Thus gene 3 protein is thought to be responsible for recognizing DNA to be packaged by P22. Gene 3 protein is found bound to gene 2 protein in infected cells.31 Although its biochemical functions have not yet been successfully studied in the test tube, the gene 2/3 protein complex no doubt constitutes the P22 “terminase”. The gene 2 and 3 proteins are not found in procapsids or in virions,8 but these proteins are able to bind purified rings of portal protein.32 Nonsense mutations in either of these genes completely block DNA packaging.7 In addition, genes 3 and 2 (the latter encodes an ATPase motif, whereas the portal gene does not appear to encode such a motif ) occupy positions on the chromosome expected for the recognition and DNA cleavage/ATPase subunits of terminase, respectively (The gene order in nearly all tailed-phages is small terminase - large terminase - portal - scaffold - coat; fig.2; see ref. 33).
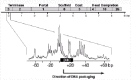
The DNA-terminase complex formation and the DNA cleavage that initiates a packaging series appear not to be as simple as might have been imagined. Both terminase subunits are required for this in vivo cleavage of P22 DNA, however series initiation cleavage occurs in the absence of procapsids (it remains unknown why all pac sites are not cleaved in the absence of procapsids). The site on P22 DNA that is recognized by gene 3 protein, the pac site, has been characterized, and it is an asymmetric site about 22 bp long that lies inside gene 3.29,34 The series initiation cleavage does not occur at a discrete site, but cleavages are scattered across an approximately 120 bp region (fig. 2). The pac site itself lies near the center of this region. The distribution of cuts within this region is not random, but cuts are (1) concentrated at 20 bp intervals, and (2) occur almost exclusively at 2 bp intervals throughout the region (fig. 2). The locations of these cleavages are a property of the terminase alone, since they do not depend upon the presence of procapsids, are modified by mutations that alter the amino acid sequence of the gene 3 protein, but are not strongly affected by the nucleotide sequence outside the pac site itself.30 The correct explanation for this complex pattern of cleavages is unknown, but it could be the result of a large repetitive protein complex being built on the DNA in which any of the units could perform the cleavage, or (2) movement of the terminase on the DNA after recognition of the pac site. P22 virion DNAs made under conditions in which the levels of terminase were lowered substantially have an unaltered pac series initiation DNA end pattern, suggesting that if model (1) is correct the complex does not enlarge along the DNA as more terminase is supplied. Recent findings with phage Sf6 suggest that (2) could be correct. Phage Sf6 is a close relative of P22 whose terminase is very different in sequence from that of P22. Sf6 appears to recognize a sequence within its gene 3 equivalent (its exact pac site has not yet been identified) but packaging series-initiating cleavages in the DNA are made over a nearly 2 kbp region.35 The simplest model for this is a ± 1 kbp-diffusion of the terminase (nuclease) after recognition but before cleavage. A terminase complex large enough to span this length of DNA seems extremely unlikely, since only small amounts of terminase are made during infection. If sliding explains Sf6 series initiation, it could explain P22 as well.
After DNA is recognized by the gene 3 protein, a cleavage is made in the DNA and one of the ends created (the end to the right of the cleavage on the standard map) is inserted into the procapsid (fig. 3). Analogy with other phage suggests that a gene 2 protein/gene 3 protein/DNA complex docks with the portal containing vertex of the procapsid, and that this interaction somehow inserts the DNA end into the portal hole, thus poising it for translocation into the capsid interior.
Filling the Capsid with DNA
As in other tailed phages, the motor that drives P22 DNA into the capsid is thought to be composed of the portal protein and terminase components. Because P22 is a headful packaging phage, its head filling device must have two parts—the force generating motor itself and a “headful sensing” device that controls the cleavage of DNA when the head is full. P22 heads contain DNA molecules that are on average 43,400 bp long, but their actual lengths vary from 42650 to 44150 bp.5 The headful nuclease appears to have weak sequence specificity in vivo, since right end fragments of virion DNA are only created at a fraction of the bp in the headful cleavage region. Nonetheless, there are many potential cleavage sites in any region determined by the headful sensor to denote a headful of DNA (i.e., weak sequence specificity may determine the many exact cleavage sites that are observed within the general regions chosen for cleavage by the sensing device). Experiments with deleted P22 genomes,5,36 mutants that package a longer DNA molecule than wildtype37 and mutants with small T=4 capsids (Moore S and Prevelige P, personal communication) have demonstrated unambiguously that it is the size of the phage head that determines the length of DNA molecule encapsidated. Thus, this ±750 bp variation in chromosome length represents the ±1.8% imprecision of the headful-measuring device.
It is not known if the P22 DNA entry motor and headful sensor are separable functions or are two manifestations of the same device. Molecular genetic evidence suggests a more than simply passive role for the P22 portal protein in the DNA entry process. A mutant of P22 exists which encapsidates chromosomes that are 4.7% longer than wildtype. Although it is technically very difficult to directly prove that the mutant capsid interiors are not about 5% larger, measurements of internal DNA hydration showed that that the volume of water missing from these particles (compared to wildtype) is virtually identical to the volume of the “extra” DNA present in the mutant virions.37 It thus appears that the capsid interior volumes of the mutant and wildtype are the same, and the DNA length difference is the result of either a less sensitive headful-sensing device or of a motor that can push DNA in tighter or both. In either case, since this mutation alters a single amino acid in the portal, this finding shows that the portal ring is not a purely passive hole through which DNA is driven. In addition, experiments with procapsids that contain a mixture of mutant and wildtype portal proteins showed that the subunits of the portal ring can integrate information over the twelve subunits to determine where the headful cleavages occur, since a procapsid with about half wildtype and half mutant subunits packages a chromosome about 2.5% longer than wildtype, about halfway between procapsids with only wildtype or only mutant portal proteins.26
Although P22 DNA packaging is dependent upon ATP hydrolysis in crude extracts,38 it has not been studied with purified components and the protein that hydrolyzes the ATP remains unknown. Nonetheless, it seems likely by analogy with other phages that the large terminase (gene 2 protein) subunit is the ATPase, and the P22 gene 2 protein does carry an ATPase amino acid sequence consensus.39 Other circumstantial evidence suggests that P22 terminase may participate in DNA entry. Analysis of structures made by various mutants have shown that when gene 4, 10 or 26 protein is missing, DNA packaging occurs apparently normally, but the DNA is packaged unstably and falls out the particles before they can be isolated.40,41 These three proteins are called “head completion protein” (see fig. 1). The gene 4 and 10 proteins have been found only at the portal vertex of the mature virion,42 and have been proposed to plug the portal hole so that DNA does not “leak” back out after packaging. Virion-like “empty heads” that have lost their DNA can be isolated from viruses containing mutations in genes 4, 10 or 26, and these empty heads have 5 to 10 molecules of both terminase subunits tightly bound to them10 (Adams M, Casjens S, unpublished). Terminase subunits are also present in the “empty head” fraction of particles isolated from wildtype infections, suggesting that this is not an aberrant association seen only under the mutant conditions (Adams M, Casjens S, unpublished). Since it is required for packaging initiation and present in the particles after packaging is complete, it seems that terminase molecules are most likely also present and bound to the procapsid during the DNA entry process. It is quite possible, therefore, that they could participate directly in that process. Furthermore, the stoichometry of the bound terminase suggests that DNA packaging is carried out by a complex consisting of the procapsid and multiple terminase subunits. How the structure and organization of such a packaging complex might affect the packaging strategy is not known. Since we and others have failed to observe any ATPase activity by portal protein in any virus system, our current model for P22 DNA entry is thus that the DNA translocase motor consists of the portal ring and 5-15 molecules of each of the terminase subunits. We imagine, currently without proof (largely by analogy to other phage systems that are better studied in this regard), that the P22 large terminase subunit is both the ATPase that provides the packaging energy as well as the nuclease that acts during initiation of packaging series and headful DNA cleavage. The portal appears to be more than a passive hole, and is likely part of the headful-sensing device.
Termination of DNA Packaging
After the headful DNA cleavage releases the capsid containing the newly packaged chromosome from the concatemer, the three head completion proteins (the products of genes 4, 10 and 26) add to the particle and stabilize the packaged DNA (fig. 1). After the head completion proteins add to the stucture, 18 molecules of one more protein, called “tailspike” protein, add to the portal vertex. Tailspike protein is responsible for binding virions to the outer surface of cells to initate the next round of infection.3,4 Tailspike defective virions and wildtype virions contain no terminase subunits, but particles made by mutants defective in any of the three head completion genes do contain them (above), suggesting that addition of the head completion proteins causes the release of terminase. The head completion proteins are known to add in the following obligate order (each in small numbers and to the portal vertex portion of the virion): gene 4 protein, gene 10 protein, and finally gene 26 protein.41,42 Since 26 defective particles, which have bound 4 and 10 proteins, contain terminase, we have proposed that 26 protein addition is responsible for removing terminase from the particle to complete each DNA packaging event.43
It is not known what causes processive packaging series to end. Do packaging series terminate with an abortive attempt to package a too short DNA and simply run off the end of the DNA being packaged? Or might they collide with a replication apparatus? Or might the packaging machine have some way of only allowing a packaging event to start if enough DNA is available for a productive event? Many questions remain about headful packaging and the processive packaging series.
Summary
P22 virion assembly is one of the prototypic virus nucleic acid packaging systems. Its terminase, portal, scaffold, coat and head completion proteins have little sequence similarity to the analogous proteins of other well-studied dsDNA virus types, yet a perfect parallel exists between these P22 general functions and those of their analogs in other systems. This relationship suggests that either the large dsDNA viruses (perhaps including herpesviruses, and even iridoviruses and adenoviruses?44,45) have an extremely distant common ancestor from which all their DNA packaging machines are derived or that the procapsid/DNA packaging strategy represents an optimal solution arrived at more than once during the evolution of bacterial and eukaryotic viruses.
Acknowledgements
Research on P22 DNA packaging has been supported by the National Science Foundation (grant number MCB990526 to S.C.).
References
- 1.
- Zinder N, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64:679–699. [PMC free article: PMC169409] [PubMed: 12999698]
- 2.
- Ebel-Tsipis J, Botstein D, Fox MS. Generalized transduction by phage P22 in Salmonella typhimurium. I. Molecular origin of transducing DNA. J Mol Biol. 1972;71:433–448. [PubMed: 4564486]
- 3.
- Poteete AR. Bacteriophage P22 In: Calendar R, ed.The BacteriophagesNew York: Plenum Press,1988647–682.
- 4.
- Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42:385–413. [PMC free article: PMC281435] [PubMed: 353481]
- 5.
- Casjens S, Hayden M. Analysis in vivo of the bacteriophage P22 headful nuclease. J Mol Biol. 1988;199:467–474. [PubMed: 3280806]
- 6.
- Anderson T, Yamamoto N. Genomic masking and recombination between serologically unrelated phages P22 and P221. Virology. 1961;14:430–437. [PubMed: 13787142]
- 7.
- Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J Mol Biol. 1973;80:669–695. [PubMed: 4773026]
- 8.
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2:202–224. [PubMed: 4612247]
- 9.
- King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J Mol Biol. 1973;80:697–731. [PubMed: 4773027]
- 10.
- Poteete AR, King J. Functions of two new genes in Salmonella phage P22 assembly. Virology. 1977;76:725–739. [PubMed: 320755]
- 11.
- Casjens S. Molecular organization of the bacteriophage P22 coat protein shell. J Mol Biol. 1979;131:1–14. [PubMed: 385887]
- 12.
- Prasad BV, Prevelige PE, Marietta E. et al. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. [PubMed: 8496966]
- 13.
- Thuman-Commike PA, Greene B, Jakana J. et al. Three-dimensional structure of scaffolding-containing phage p22 procapsids by electron cryo-microscopy. J Mol Biol. 1996;260:85–98. [PubMed: 8676394]
- 14.
- Thuman-Commike PA, Greene B, Malinski JA. et al. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys J. 1999;76:3267–3277. [PMC free article: PMC1300296] [PubMed: 10354452]
- 15.
- Thuman-Commike PA, Greene B, Jakana J. et al. Identification of additional coat-scaffolding interactions in a bacteriophage P22 mutant defective in maturation. J Virol. 2000;74:3871–3873. [PMC free article: PMC111895] [PubMed: 10729161]
- 16.
- King J, Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974;251:112–119. [PubMed: 4421992]
- 17.
- Bazinet C, King J. Initiation of P22 procapsid assembly in vivo. J Mol Biol. 1988;202:77–86. [PubMed: 3262766]
- 18.
- Earnshaw W, Casjens S. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. [PubMed: 6447542]
- 19.
- Greene B, King J. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology. 1996;225:82–96. [PubMed: 8918536]
- 20.
- Bazinet C, Benbasat J, King J. et al. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27:1849–1856. [PubMed: 3288279]
- 21.
- Botstein D, Levine M. Intermediates in the synthesis of phage P22 DNA. Cold Spring Harb Symp Quant Biol. 1968;33:659–667. [PubMed: 4892003]
- 22.
- Streisinger G, Enrich J, Stahl M. Chromosome structure in T4. III. Terminal redundancy and length determination. Proc Natl Acad Sci USA. 1967;57:292–295. [PMC free article: PMC335503] [PubMed: 16591467]
- 23.
- Tye BK, Botstein D. P22 morphogenesis. II: Mechanism of DNA encapsulation. J Supramol Struct. 1974;2:225–238.
- 24.
- Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol. 1978;118:365–388. [PubMed: 344888]
- 25.
- Adams MB, Hayden M, Casjens S. On the sequential packaging of bacteriophage P22 DNA. J Virol. 1983;46:673–677. [PMC free article: PMC255176] [PubMed: 6842684]
- 26.
- Casjens S, Wyckoff E, Hayden M. et al. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–1074. [PubMed: 1569567]
- 27.
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. [PubMed: 4564719]
- 28.
- Schmieger H, Backhaus H. The origin of DNA in transducing particles in P22-mutants with increased transduction-frequencies (HT-mutants). Mol Gen Genet. 1973;120:181–190. [PubMed: 4568531]
- 29.
- Casjens S, Huang WM, Hayden M. et al. Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J Mol Biol. 1987;194:411–422. [PubMed: 3041006]
- 30.
- Casjens S, Sampson L, Randall S. et al. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J Mol Biol. 1992;227:1086–1099. [PubMed: 1433288]
- 31.
- Poteete AR, Botstein D. Purification and properties of proteins essential to DNA encapsulation by phage P22. Virology. 1979;95:565–573. [PubMed: 380140]
- 32.
- Moore SD, Prevelidge P. Bacteriophage P22 portal vertex formation in vivo. J Mol Biol. 2002;315:975–994. [PubMed: 11827470]
- 33.
- Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly In: Calendar R, ed.The BacteriophagesNew York: Plenum Press,198815–91.
- 34.
- Wu H, Sampson L, Parr R. et al. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Molec Micro. 2002;45:1631–1646. [PubMed: 12354230]
- 35.
- Casjens S, Winn D, Gilcrease L. et al. The chromosome of Shigella flexneri bacteriophage Sf6: Complete nucleotide sequence, genetic mosaicism, and DNA packaging. J Mol Biol. 2004;339:379–394. [PubMed: 15136040]
- 36.
- Tye BK, Huberman JA, Botstein D. Nonrandom circular permutation of phage P22 DNA. J Mol Biol. 1974;85:501–528. [PubMed: 4853363]
- 37.
- Casjens S, Wyckoff E, Hayden M. et al. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–74. [PubMed: 1569567]
- 38.
- Poteete AR, Jarvik V, Botstein D. Encapsulation of phage P22 DNA in vitro. Virology. 1979;95:550–564. [PubMed: 380139]
- 39.
- Eppler K, Wyckoff E, Goates J. et al. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. [PubMed: 1853558]
- 40.
- Lenk E, Casjens S, Weeks J. et al. Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology. 1975;68:182–199. [PubMed: 1103445]
- 41.
- Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–543. [PubMed: 6363718]
- 42.
- Hartweig E, Bazinet C, King J. Injection apparatus of phage P22. Biophys J. 1986;49:24–27. [PMC free article: PMC1329563] [PubMed: 19431631]
- 43.
- Casjens S. Bacteriophage P22 DNA packagingIn: Adolph K, ed.Chromosomes, Eukaryotic, Prokaryotic and ViralBoca Raton: CRC Press,1989241–261.
- 44.
- Casjens S. Principles of virion structure, function and assembly In: Chiu W, Burnett R, Garcea R, eds.Structural Biology of Viruses Oxford University Press: Oxford,19973–37.
- 45.
- Newcomb WW, Juhas RM, Thomsen DR. et al. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–10932. [PMC free article: PMC114672] [PubMed: 11602732]
- DNA Packaging by Bacteriophage P22 - Madame Curie Bioscience DatabaseDNA Packaging by Bacteriophage P22 - Madame Curie Bioscience Database
- Regulation of Insulin Action and Insulin Secretion by SNARE-Mediated Vesicle Exo...Regulation of Insulin Action and Insulin Secretion by SNARE-Mediated Vesicle Exocytosis - Madame Curie Bioscience Database
- Structure, Function and Assembly of Flagellar Axial Proteins - Madame Curie Bios...Structure, Function and Assembly of Flagellar Axial Proteins - Madame Curie Bioscience Database
- The ø29 DNA Packaging Motor: Seeking the Mechanism - Madame Curie Bioscience Dat...The ø29 DNA Packaging Motor: Seeking the Mechanism - Madame Curie Bioscience Database
- Molecular Phylogeny and Evolution of the Coronin Gene Family - Madame Curie Bios...Molecular Phylogeny and Evolution of the Coronin Gene Family - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...