Intrinsically curved DNA structures are often found in or around transcriptional control regions of eukaryotic genes, and curved DNA may be common to all class I gene promoters. Although not all class II gene promoters contain curved DNA structures, both TATA-boxcontaining and TATA-box-less promoters often contain such structures. Furthermore, several studies have suggested that the TATA box itself adopts a curved DNA conformation. Curved DNA structures are likely to function in transcription in several ways. These include acting as a conformational signal for transcription factor binding; juxtaposition of the basal machinery with effector domains on upstream-bound factors; regulation of transcription in association with transcription-factor-induced bending of DNA; and organization of local chromatin structure to increase the accessibility of cis-DNA elements. This chapter presents a concise overview of studies of these functions.
Introduction
Eukaryotic cells use three different RNA polymerases, referred to as pols I, II, and III, to transcribe their genes. Pol I produces rRNAs, pol II produces mRNA molecules and most small nuclear (sn) RNAs, and pol III produces small RNA molecules such as tRNA, 5S rRNA and U6 snRNA. DNA templates that are transcribed by pols I, II and III are often called class I, II and III genes, respectively. They have their own (class-specific) promoters. Curved DNA is often found in the transcriptional control regions,1 with many reports having identified curved DNA in class II gene promoters. Both TATA-box-containing and TATA-box-less promoters often contain curved DNA. However, curved DNA is not necessarily confined to class II gene promoters. Class I gene promoters also often contain it. The role of curved DNA in eukaryotic transcription is being gradually revealed.
Here, I review what has been clarified in the field of eukaryotic transcription, and what is still unknown. Several studies have indicated that curved DNA is implicated in the packaging promoter DNA into chromatin, and this is described in Chapter 13.
Intrinsic DNA Bends in Control Regions of Transcription
Class I Gene Promoters
A large number of ribosomes are required in each cell generation. To meet the requirement, genomes have multiple copies of the rRNA gene (rDNA). For example, the human genome has about 200 copies of the gene. They are organized in tandem arrays in eukaryotes, in which the transcribed regions are separated by an intergenic spacer (fig. 1). Three of the four rRNAs (18S, 5.8S and 28S molecules) are generated from a single precursor rRNA. The nucleotide sequences of the rRNA gene promoters are highly divergent across species. As a result, in this class of genes, transcription is restricted by the cognateness between gene and transcription machinery. For example, the human rRNA gene is not transcribed by the mouse pol I machinery, or vice versa.2
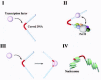
Figure 1
Schematic representation of class I and II gene promoters.
More than a decade ago, by employing a circular permutation polyacrylamide gel electrophoresis assay (Chapter 3), Schroth et al identified two intrinsically curved DNA structures near the transcription initiation site of the Physarum polycephalum rRNA gene (Table 1).3 The center of one curved DNA was 160 nucleotides upstream (-160) of the transcription start site (+1) and the center of the other was about 150 nucleotides downstream (+150). Subsequently, a curved DNA was also experimentally identified upstream of the promoter (center position: —260∼—280) for the Arabidopsis thaliana rRNA gene.4 Marilley and Pasero used several curvature prediction models to analyze rRNA gene promoters from mammals (Rattus norvegicus), amphibians (Xenopus laevis, Xenopus borealis), insects (Drosophila melanogaster), echinoderms (Paracentrotus lividus), protozoans (Tetrahymena pyriformis, Dictyostelium discoideum), dicotyledons (Arabidopsis thaliana, Pisum sativum), monocotyledons (Triticum aestivum, Zea mays), and myxomycetes (Physarum polycephalum). Each promoter had at least one curved DNA structure, although the position and extent of DNA curvature differed.5 Computational modeling also suggested that the promoter of the mouse rDNA overlaps with a curved DNA structure.6 In conclusion, a curved DNA structure may be a common feature of class I gene promoters. To the best of my knowledge, however, no study has succeeded in clarifying the function of this curved DNA.
Table 1
Curved DNA-containing promoters, enhancers and regulatory sequencesa.
Class II Gene Promoters and Enhancers
The TATA box, the initiator element (INR), and the downstream promoter element (DPE), are generally regarded as the core elements of promoters of class II genes (fig. 1). The TATA box, with the consensus TATA(A/T)A(A/T), is located approximately 25-31 bp upstream of the transcription start site. The INR is centered at the transcription initiation site and has a loose consensus sequence, PyPyAN(T/A)PyPy, where Py is a pyrimidine and N is any nucleotide. Many promoters have either the TATA box (including TATA-like sequences) or some kind of INR, and some have both. Many Drosophila TATA-less promoters, and most likely many mammalian TATA-less promoters, seem to contain a DPE.7 The DPE, with the consensus PuG(A/T)CGTG, where Pu is a purine, is centered approximately 30 bp downstream of the initiation site. There is also a class of pol II promoters comprised of G/C rich sequences, and containing multiple binding sites for transcription factor Sp1 instead of the elements described above. Such promoters are often associated with housekeeping genes.
As shown in Table 1, many class II gene promoters have a curved DNA structure. Although the table does not list the reports on the conformation of TATA box per se, several groups have discussed this issue. Using models of Bolshoy et al,8 Calladine et al9 and Satchwell et al,10 Schätz and Langowski calculated helix trajectories of 504 vertebrate promoters, which suggested that the regions containing the TATA box (either canonical or non-canonical) all form curved DNA structures.11 They also predicted that the upstream half of the TATA box would be curved towards the major groove, and the remainder would be straight, or even curved towards the minor groove. de Souza and Ornstein subjected the TATA box of the adenovirus major late promoter (whose sequence is TATAAAA) to molecular dynamics simulations. Their results suggested that the direction of the curvature is toward the major groove.12 However, based on DNA cyclization kinetics, Davis et al reported that the curvature is toward the minor groove.13 Thus the issue of the TATA box conformation is not yet settled. What about the other promoter elements? Based on the consensus sequence for the INR, AAA or TTT could be formed within the box. These tracts change the direction of the helical path of DNA. However, the DNA conformation of this region has not yet been reported. DPE itself cannot possess DNA curvature as judged from its consensus sequence. To the best of my knowledge, there are no reports describing the DNA conformation of this region, though the main reason may simply be lack of studies. Concerning the G/C rich promoters, Sp1-induced DNA bending is well known.14 However, intrinsic DNA curvature has not been reported.
The DNA elements for binding transcription factors are sometimes located in curved DNA structures. Delabre et al reported that in the scorpion (Androctonus australis) AaH I_ toxin gene promoter, the region containing the putative TATA box and the CCAAT box is located in a large curved DNA structure.15 In the human cdc2 promoter,16 the binding sites for c-Myb, ATF, Sp1, E2F and CBF are located in curved DNA. In the pea rbcS-3A and rbcS-3.6 genes, both of which code for the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, each light responsive element overlaps with a curved DNA structure.17 The Xenopus A2 vitellogenin gene presents another example of a curved DNA structure that occurs between cis-DNA elements: curved DNA was found between a DNA element located around position—100, which could activate transcription in a cell-specific manner, and the estrogen-responsive element located around —300.18 Also, in the yeast GAL80 gene, a single curved DNA structure lies between the UASGAL80 (an upstream GAL80-specific activation sequence) and the more gene-proximal promoter elements.19 The GAL1 gene has two curved DNA structures between UASG (an upstream activation sequence) and the TATA box.19 The curvatures found near GAL1 and GAL80 may be involved in nucleosome formation (Chapter 13). Cis-DNA elements themselves form curved DNA structures in the following cases. The human β-actin promoter has a β-actin-specific conserved sequence (conserved among humans, rats and chickens) between the TATA and CCAAT boxes, which forms a curved DNA structure.20 In human papillomavirus type 16 (HPV-16), the E2 protein binding site in the upstream regulatory region forms a curved DNA structure.21 At present, however, we cannot find any common features shared by these regions, except that a curved DNA structure is present.
Curved DNA structures have also been found in enhancers, or adjacent to enhancers. In the Saccharomyces cerevisiae genome, the UAS of the mating pheromone α factor 1 (MF a 1) gene, and the UAS of the STE3 gene, adopt curved DNA structures.22 The enhancer of the human adenovirus type 2 (Ad2) E1A gene has a sharply curved DNA structure.23 As shown in Figure 2, most of the enhancer elements are within the curved DNA. In the enhancer of the human interferon-β (IFN β) gene, positive regulatory domains (PRDs) II and IV also form curved DNA structures: PRDII is curved towards the minor groove and PRDIV towards the major groove.24 The simian cytomegalovirus IE94 gene has a wide curved DNA region (about 1 kb) upstream of the enhancer.25
Class III Gene Promoters
The important promoter elements of the 5S rRNA genes and tRNA genes are located downstream of the transcription start site. On the other hand, the promoters of human and Saccharomyces cerevisiae U6 snRNA genes are similar to class II gene promoters in the sense that they adopt a TATA box at the “correct” site (-32 to -25 and -30 to -23, respectively).26 The TATA box is likely to have a curved conformation as described above. Therefore, they may be counted as promoters that contain curved DNA. To the best of my knowledge, there are no reports of curved DNA structures in or around the promoters of 5S rRNA genes and tRNA genes, but presumably this is simply due to lack of studies.
Role of Intrinsic DNA Curvature in Transcription
Does curved DNA actually function in eukaryotic transcription? The sequence and conformation of DNA are two sides of the same coin. Thus, we must always consider the possibility that it is the DNA sequence itself that is important, and the curved conformation may simply be a by-product. It is usually very difficult to experimentally prove significance of DNA conformation per se without impairing the DNA sequence. In the following two cases, however, this difficulty was overcome. Using a constrained minicircle strategy,27 Parvin et al showed that the bending of the TATA box enhances its affinity for the TATA box binding protein (TBP).28 Moreover, they suggested that the direction of bending is important: a bend towards the major groove showed 100-fold higher affinity than an unbent structure, and 300-fold higher affinity than a bend towards the minor groove. They observed similar discrimination with the holo-TFIID transcription complex. However, this study did not describe the effect of the intrinsically curved DNA conformation of the TATA box, but reported the effect of “constrained” bending of the box. If the TATA box intrinsically curves toward major groove, as suggested by Sch_ and Langowski11 and de Souza and Ornstein,12 the role of the conformation may be to facilitate TBP binding. In the adenovirus E1A gene enhancer, the bent conformation per se activated transcription.29 Run-off transcription assays using the wild-type DNA template, with a temperature-dependent conformational change of DNA curvature, showed that a moderately curved enhancer was superior to a highly curved enhancer in stimulating transcription. In the study, curved DNA of the phage λ origin of replication could substitute for the E1A enhancer, and stimulated transcription of the E1A gene to some extent, when it had an appropriate DNA conformation.
Synthetic curved DNA fragments sometimes help the analysis of the structure-function relationships. An early study reported that (CAAAAATGCC)19 and (CAAAAATGCC)35 activated transcription driven by the promoter element CGTATTTATTTG by two- and three-fold, respectively.30 Using synthetic curved DNA segments, Kim et al also showed that transcription could be activated by curved DNA.31 As described in Chapter 3, curved DNA with a right-handed superhelical writhe activates transcription in Escherichia coli. However, a recent study showed that it was left-handed curved DNA that activated transcription in eukaryotes.32 In the study, synthetic DNA fragments with different geometries were constructed by changing the spacing between T5 tracts, which was based on the principle described in Chapter 3. When left-handed curved DNA was linked to the herpes simplex virus thymidine kinase promoter at a specific rotational orientation and distance, the curved DNA activated transcription. Neither planar curving, nor right-handedly curved DNA, nor straight DNA had this effect (Chapter 13). Thus, in studying the role of DNA curvatures, it seems important to consider the shape of DNA curvature. It has been suggested that many eukaryotic promoters can adopt superhelical conformations.33
How does curved DNA function in transcription? The most likely role is that curved DNA structures function as recognition signals for transcription factors (fig. 3). In the study of the human β-actin promoter described above, a protein in the HeLa cell extracts bound to the curved DNA.20 The above report by Parvin et al28 seems to support this role as well. However, the TATA box may curve toward the minor groove as suggested by Davis et al.13 Their study leads us to another important concept: the binding of a transcription factor can change the direction of DNA curvature. They used the TATA box of the adenovirus major late promoter and showed, by cyclization kinetics, that the direction of bend induced by TBP (toward the major groove of the TATA box) was opposite to the direction of bend of the TATA box per se (toward the minor groove).13 They speculated that the intrinsic bend of the TATA box could repress transcription complex assembly in the absence of TBP. A similar phenomenon has been reported for the interaction between the E2F transcription factor and the E2F site.34 In the human E2F1 promoter, a consensus E2F site adopts a curved DNA structure oriented toward the major groove relative to the center of the E2F site (magnitude of the bend; ∼40°). When E2F binds, the bend is reversed and oriented toward the minor groove (net magnitude of the resulting bend; ∼25°). Furthermore, in the human IFN β enhancer, the directions of the two intrinsic bends are also counteracted or reversed by the binding of NF-κB, ATF-2/c-Jun, and HMGI(Y).24 Alteration of DNA architecture may facilitate protein-protein interactions between the transcription factors that constitute the enhanceosome.
As another possibility, curved DNA-mediated nucleosome positioning may be used to juxtapose two distant sites, and to allow interaction between transcription factors. The regulatory region of the Xenopus A2 vitellogenin gene may be such an example. In this case, curved DNA is located between the estrogen-responsive element and the cell type specific activator sequence.18 These elements may be juxtaposed by nucleosome positioning directed by the curved DNA. In fact, in the corresponding region of the Xenopus vitellogenin B1 gene, the DNA sequence strongly directs the positioning of a nucleosome. This nucleosome creates a static loop. As the result, the distal estrogen receptor binding site is brought close to the proximal promoter elements, which presumably allows interaction between transcription factors, and facilitates transcription.35 Without the help of a nucleosome, curved DNA conformation could play the same role. An experiment using an artificial promoter showed that a curved DNA juxtaposed bound factors, and activated transcription.36 Thus, there are several lines of direct or indirect evidence for the function of curved DNAs. In some cases, they may play a single role in transcription, but in others they may play multiple roles simultaneously.
Some of the putative roles of DNA curvature described above seem to be the same, in essence, as those for prokaryotic transcription. The important question we must consider next is the relationship between curved DNA, chromatin structure and transcription. This issue is described in Chapter 13.
Conclusion
The presence of a curved DNA structure may be a common feature shared by class I gene promoters. In transcription of some class II genes, curved DNA conformations per se may also play an important role. Curved DNA sequences seem to be used in several ways; some roles are presumably common between prokaryotes and eukaryotes, while others are used in transcription in the chromatin environment, and are thus specific to eukaryotes.
Acknowledgements
The author would like to acknowledge the contributions of Yoshiro Fukue, Jun-ichi Nishikawa, and Junko Ohyama. The studies reported from my laboratory were supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
References
- 1.
- Ohyama T. Intrinsic DNA bends: an organizer of local chromatin structure for transcription. Bioessays. 2001;23:708–715. [PubMed: 11494319]
- 2.
- Learned RM, Cordes S, Tjian R. Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol Cell Biol. 1985;5:1358–1369. [PMC free article: PMC366865] [PubMed: 3929071]
- 3.
- Schroth GP, Siino JS, Cooney CA. et al. Intrinsically bent DNA flanks both sides of an RNA polymerase I transcription start site. Both regions display novel electrophoretic mobility. J Biol Chem. 1992;267:9958–9964. [PubMed: 1577826]
- 4.
- Kneidl C, Dinkl E, Grummt F. An intrinsically bent region upstream of the transcription start site of the rRNA genes of Arabidopsis thaliana interacts with an HMG-related protein. Plant Mol Biol. 1995;27:705–713. [PubMed: 7727748]
- 5.
- Marilley M, Pasero P. Common DNA structural features exhibited by eukaryotic ribosomal gene promoters. Nucleic Acids Res. 1996;24:2204–2211. [PMC free article: PMC145945] [PubMed: 8710487]
- 6.
- Längst G, Schätz T, Langowski J. et al. Structural analysis of mouse rDNA: coincidence between nuclease hypersensitive sites, DNA curvature and regulatory elements in the intergenic spacer. Nucleic Acids Res. 1997;25:511–517. [PMC free article: PMC146485] [PubMed: 9016589]
- 7.
- Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. [PubMed: 8598298]
- 8.
- Bolshoy A, McNamara P, Harrington RE. et al. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci USA. 1991;88:2312–2316. [PMC free article: PMC51221] [PubMed: 2006170]
- 9.
- Calladine CR, Drew HR, McCall MJ. The intrinsic curvature of DNA in solution. J Mol Biol. 1988;201:127–137. [PubMed: 3418695]
- 10.
- Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–675. [PubMed: 3806678]
- 11.
- Schätz T, Langowski J. Curvature and sequence analysis of eukaryotic promoters. J Biomol Struct Dyn. 1997;15:265–275. [PubMed: 9399154]
- 12.
- de SouzaON, Ornstein RL. Inherent DNA curvature and flexibility correlate with TATA box functionality. Biopolymers. 1998;46:403–415. [PubMed: 9798428]
- 13.
- Davis NA, Majee SS, Kahn JD. TATA box DNA deformation with and without the TATA box-binding protein. J Mol Biol. 1999;291:249–265. [PubMed: 10438619]
- 14.
- Sjottem E, Andersen C, Johansen T. Structural and functional analyses of DNA bending induced by Sp1 family transcription factors. J Mol Biol. 1997;267:490–504. [PubMed: 9126833]
- 15.
- Delabre ML, Pasero P, Marilley M. et al. Promoter structure and intron-exon organization of a scorpion α-toxin gene. Biochemistry. 1995;34:6729–6736. [PubMed: 7756304]
- 16.
- Nair TM. Evidence for intrinsic DNA bends within the human cdc2 promoter. FEBS Lett. 1998;422:94–98. [PubMed: 9475177]
- 17.
- Cacchione S, Savino M, Tufillaro A. Different superstructural features of the light responsive elements of the pea genes rbcS-3A and rbcS-3.6. FEBS Lett. 1991;289:244–248. [PubMed: 1915854]
- 18.
- Döbbeling U, Roß K, Klein-Hitpaß L. et al. A cell-specific activator in the Xenopus A2 vitellogenin gene: promoter elements functioning with rat liver nuclear extracts. EMBO J. 1988;7:2495–2501. [PMC free article: PMC457119] [PubMed: 2847920]
- 19.
- Bash RC, Vargason JM, Cornejo S. et al. Intrinsically bent DNA in the promoter regions of the yeast GAL1-10 and GAL80 genes. J Biol Chem. 2001;276:861–866. [PubMed: 11013248]
- 20.
- Kawamoto T, Makino K, Orita S. et al. DNA bending and binding factors of the human β-actin promoter. Nucleic Acids Res. 1989;17:523–537. [PMC free article: PMC331601] [PubMed: 2915921]
- 21.
- Bedrosian CL, Bastia D. The DNA-binding domain of HPV-16 E2 protein interaction with the viral enhancer: protein-induced DNA bending and role of the nonconserved core sequence in binding site affinity. Virology. 1990;174:557–575. [PubMed: 2154890]
- 22.
- Inokuchi K, Nakayama A, Hishinuma F. Sequence-directed bends of DNA helix axis at the upstream activation sites of α-cell-specific genes in yeast. Nucleic Acids Res. 1988;16:6693–6711. [PMC free article: PMC338326] [PubMed: 3043374]
- 23.
- Ohyama T, Hashimoto S. Upstream half of adenovirus type 2 enhancer adopts a curved DNA conformation. Nucleic Acids Res. 1989;17:3845–3853. [PMC free article: PMC317863] [PubMed: 2734105]
- 24.
- Falvo JV, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFNβ gene enhancer by transcription factors and the architectural protein HMG I(Y). Cell. 1995;83:1101–1111. [PubMed: 8548798]
- 25.
- Chang YN, Jeang KT, Chiou CJ. et al. Identification of a large bent DNA domain and binding sites for serum response factor adjacent to the NFI repeat cluster and enhancer region in the major IE94 promoter from simian cytomegalovirus. J Virol. 1993;67:516–529. [PMC free article: PMC237389] [PubMed: 8380090]
- 26.
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002162593-2620 [PubMed: 12381659]
- 27.
- Kahn JD, Crothers DM. Protein-induced bending and DNA cyclization. Proc Natl Acad Sci USA. 1992;89:6343–6347. [PMC free article: PMC49497] [PubMed: 1321436]
- 28.
- Parvin JD, McCormick RJ, Sharp PA. et al. Pre-bending of a promoter sequence enhances affinity for the TATA-binding factor. Nature. 1995;373:724–727. [PubMed: 7854460]
- 29.
- Ohyama T. Bent DNA in the human adenovirus type 2 E1A enhancer is an architectural element for transcription stimulation. J Biol Chem. 1996;271:27823–27828. [PubMed: 8910380]
- 30.
- Delic J, Onclercq R, Moisan-Coppey M. Inhibition and enhancement of eukaryotic gene expression by potential non-B DNA sequences. Biochem Biophys Res Commun. 1991;181:818–826. [PubMed: 1755861]
- 31.
- Kim J, Klooster S, Shapiro DJ. Intrinsically bent DNA in a eukaryotic transcription factor recognition sequence potentiates transcription activation. J Biol Chem. 1995;270:1282–1288. [PubMed: 7836392]
- 32.
- Nishikawa J, Amano M, Fukue Y. et al. Left-handedly curved DNA regulates accessibility to cis-DNA elements in chromatin. Nucleic Acids Res. 2003;31:6651–6662. [PMC free article: PMC275550] [PubMed: 14602926]
- 33.
- Miyano M, Kawashima T, Ohyama T. A common feature shared by bent DNA structures locating in the eukaryotic promoter region. Mol Biol Rep. 2001;28:53–61. [PubMed: 11710566]
- 34.
- Cress WD, Nevins JR. A role for a bent DNA structure in E2F-mediated transcription activation. Mol Cell Biol. 1996;16:2119–2127. [PMC free article: PMC231199] [PubMed: 8628278]
- 35.
- Schild C, Claret FX, Wahli W. et al. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 1993;12:423–433. [PMC free article: PMC413225] [PubMed: 8440235]
- 36.
- Kim J, de HaanG, Shapiro D. DNA bending between activator sequences increases transcriptional synergy. Biochem Biophys Res Commun. 1996;226:638–644. [PubMed: 8831669]
- 37.
- Porter ME. The DNA polymerase δ promoter from Plasmodium falciparum contains an unusually long 5' untranslated region and intrinsic DNA curvature. Mol Biochem Parasitol. 2001;114:249–255. [PubMed: 11378205]
- 38.
- Wada-Kiyama Y, Kiyama R. An intrachromosomal repeating unit based on DNA bending. Mol Cell Biol. 1996;16:5664–5673. [PMC free article: PMC231566] [PubMed: 8816479]
- 39.
- Fiorini A, Basso LRJr, Paçó-Larson ML. et al. Mapping of intrinsic bent DNA sites in the upstream region of DNA puff BhC4-1 amplified gene. J Cell Biochem. 2001;83:1–13. [PubMed: 11500949]
- 40.
- Angermayr M, Oechsner U, Gregor K. et al. Transcription initiation in vivo without classical transactivators: DNA kinks flanking the core promoter of the housekeeping yeast adenylate kinase gene, AKY2, position nucleosomes and constitutively activate transcription. Nucleic Acids Res. 2002;30:4199–4207. [PMC free article: PMC140550] [PubMed: 12364598]
- 41.
- Eckdahl TT, Anderson JN. Bent DNA is a conserved structure in an adenovirus control region. Nucleic Acids Res. 1988;16:2346. [PMC free article: PMC338233] [PubMed: 3357789]
- 42.
- Wada-Kiyama Y, Kiyama R. Periodicity of DNA bend sites in human ε-globin gene region. J Biol Chem. 1994;269:22238–22244. [PubMed: 8071350]
- 43.
- Dlakic M, Harrington RE. DIAMOD: display and modeling of DNA bending. Bioinformatics. 1998;14:326–331. [PubMed: 9632827]
- 44.
- Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–376. [PubMed: 7482707]
- 45.
- Kabsch W, Sander C, Trifonov EN. The ten helical twist angles of B-DNA. Nucleic Acids Res. 1982;10:1097–1104. [PMC free article: PMC326224] [PubMed: 7063417]
Publication Details
Author Information and Affiliations
Authors
Takashi Ohyama.Copyright
Publisher
Landes Bioscience, Austin (TX)
NLM Citation
Ohyama T. Curved DNA and Transcription in Eukaryotes. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.