NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Transcription and supercoiling of the template DNA are closely related each other. DNA supercoiling affects transcription and transcription affects supercoiling of the template DNA. Furthermore, packaging of genomic DNA into chromatin in eukaryotes raises another type of relation. DNA supercoiling can affect transcription through modulation of the chromatin structure.
Introduction
Typical double-stranded DNA consists of a helix with a pitch of one turn per 10.5 base pairs. Both underwinding and overwinding of the DNA double helix induce twisting and coiling of the helix unless the DNA strand can rotate freely. The coils thus formed are termed negative and positive supercoils, respectively (see Chapter 1 for details). Current views of the mechanisms underlying transcriptional regulation rely on a concept originally proposed by Jacob and Monod: regulation through cis-elements on DNA and trans-acting factors that bind to the elements.1 However, DNA supercoiling can modulate accessibility of trans-acting factors to cis-elements. Transcription is an asymmetric process: only one strand of the DNA double helix is copied into RNA. To achieve this, the double helix must be locally unwound. Therefore, negative supercoiling of DNA is thought to facilitate the step. Moreover, DNA supercoiling can affect transcription in chromatin context in eukaryotes. This chapter discusses roles of DNA supercoiling in transcriptional regulation.
Prokaryotic Transcription
In prokaryotes, there are two major topoisomerases that act toward opposite direction. 2 DNA gyrase can generate negative supercoils into relaxed DNA and relax positively supercoiled DNA. In contrast, topoisomerase I can relax negatively supercoiled DNA but not positively supercoiled DNA. The superhelical state of cellular DNA in prokaryotes appears to be under equilibrium between actions of these topoisomerases. Measurements of psoralen binding averaged globally across the Escherichia coli genome have detected unconstrained negative supercoils with a superhelical density of —0.05.3
Consistent with the finding of unconstrained negative supercoils in genomic DNA, it has been established that DNA supercoiling functions as a regulator of prokaryotic transcription.4 First, supercoiling affects transcription in vitro. Some promoters have an optimum level of supercoiling for their transcription and the level is different for different promoters. Second, supercoiling plays a regulatory role in vivo. Genetic studies have shown that mutations in topoisomerase I5 or DNA gyrase6 affect transcription in vivo.
Two rate limiting steps are known for prokaryotic transcription. One is formation of an RNA polymerase-DNA open complex and the other is promoter clearance. Because negative supercoiling favors the unwinding of the DNA double helix that is required for formation of the open complex, it is expected to increase the rate of transcription for promoters in which open complex formation is rate limiting. Indeed most genes are activated by increased negative supercoiling. However, transcription of gyr A and gyr B encoding the subunits of DNA gyrase is induced by relaxation of DNA. It has been proposed that promoter clearance but not open complex formation is the rate-limiting step for these promoters.7
The superhelical state of DNA is known to change depending on the growth conditions of cells.8 For example, nutrient downshift and stationary growth phase cause a decrease in the extent of negative supercoiling, while high osmolarity leads to an increase in the negative supercoiling of DNA. In agreement with these findings, transcription by RNA polymerase harboring the stationary phase-specific σs appears to be enhanced on templates with decreased superhelicity.9 DNA supercoiling also changes transiently during heat shock.10 The heat stress induces rapid relaxation of negative supercoils and then DNA topology returns back to the original state with negative supercoiling. In response to the relaxation, most genes are repressed but some specific or stress genes are induced.11
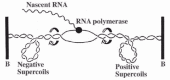
Transcription of a double helical DNA requires a rotation of an RNA polymerase and its nascent RNA chain relative to DNA. The velocity of the rotation is calculated to be a few hundred rounds per minute since a pitch of the helix is about 10 base pairs and the rate of RNA chain elongation is a few thousand nucleotides per minute. It seems difficult for the RNA polymerase and its nascent RNA to rotate around the template DNA at such a high velocity. Instead the DNA must turn around its axis during transcription. Then the tracking RNA polymerase generates positive supercoils in the template DNA ahead of it and negative supercoils behind it (fig. 1). These supercoils will be relaxed by DNA topoisomerases. Liu and Wang summarized the concept as the twin-supercoiled-domain model.12 The model predicts the followings. First, negative supercoils should accumulate in the absence of a negative supercoil-relaxing enzyme. Second, positive supercoils should accumulate in the absence of a positive supercoil-relaxing enzyme. These predictions are fulfilled by two earlier observations. First, pBR322 DNA harboring high degrees of negative supercoiling has been isolated from topA mutants of E. coli and Salmonella typhimurium, and the presence of the highly negatively supercoiled DNA was dependent on the transcription of the tetA gene.13 Second, highly positively supercoiled pBR322 DNA has been isolated from E. coli treated with gyrase inhibitors.14 According to the model, negative and positive supercoils would accumulate in the intergenic regions of two divergent and convergent transcription units, respectively. Such supercoils can in turn affect transcription.
Eukaryotic Transcription
In eukaryotes, psoralen-binding assays on human HeLa and Drosophila Schneider cell lines have shown that the bulk of each genomic DNA is torsionally relaxed within nuclei.3 However, it did not necessarily exclude a possibility that there were negatively supercoiled micro domains within these genomes. Indeed unconstrained negative supercoils have been demonstrated in the hsp70 and 18S-ribosomal RNA genes of the Schneider cell line,15 and in the dihydrofolate reductase gene16 and the hygromycin resistance transgenes17 of cultured human cells.
To test whether the twin-supercoiled-domain model is applicable to eukaryotic transcription, Giaever and Wang constructed a yeast plasmid carrying the coding sequence of the E. coli topA gene placed downstream of an inducible yeast promoter.18 The plasmid DNA became positively supercoiled in yeast ΔtopI top2ts cells at the restrictive temperature when the E. coli DNA topoisomerase I was expressed. The generation of positive supercoils was observed only during transcription. Because neither one of the yeast DNA topoisomerases I and II can be functional under these conditions and because the E. coli enzyme can relax only negative supercoils, these results verify the model. Recently Matsumoto and Hirose19 have visualized transcription-coupled, unconstrained negative supercoils of DNA in approximately 150 loci on polytene chromosomes of Drosophila melanogaster. The results demonstrate that transcription-coupled negative supercoils of DNA exist within a cell even in the presence of active topoisomerases. These negative supercoils can affect transcription.
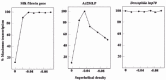
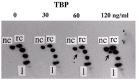
As described for prokaryotic transcription, supercoiling modulates in vitro transcription of eukaryotic genes.20 Hirose and Suzuki,21 and Mizutani et al22 have shown that transcription of the Bombyx mori fibroin gene increases and plateaus from templates of increasing negative supercoiling, and transcription from the adenovirus type 2 major late promoter (Ad2MLP) rises and then falls, while transcription of the Drosophila hsp70 gene remains unchanged (fig. 2). Dissection of transcription revealed that formation of a preinitiation complex on the fibroin gene or the Ad2MLP is slow on relaxed DNA and accelerated by negative supercoiling of DNA. On the contrary, the preinitiation complex assembled rapidly on the hsp70 gene irrespective of DNA topology. Tabuchi et al23 have demonstrated that binding of TATA element binding protein (TBP) to the TATA element induces underwinding of the duplex DNA (approximately 0.5 linking difference per bound TBP molecule as shown in fig. 3). The underwinding has been confirmed by crystal structure of a TBP-TATA element complex.24,25 The change was facilitated by negative supercoiling of DNA on the fibroin promoter and the Ad2MLP but not on the hsp70 promoter.23 These data reveal that although supercoiling can affect both prokaryotic and eukaryotic transcription in vitro, the critical steps are different: open complex formation in most genes of prokaryotes vs TBP binding to the TATA element in most genes of eukaryotes. In transcription from the Ad2MLP, promoter clearance is also facilitated by negative supercoiling of DNA.26 It is possible that clearance of the Ad2MLP goes up and then down with increasing negative supercoiling. Interestingly, the rate-limiting step in hsp70 transcription is not preinitiation complex formation but restart of a paused RNA polymerase to productive elongation.27 Probably hsp70 transcription becomes independent of DNA topology so that it can be induced immediately upon heat shock.
Whether DNA supercoiling affects transcription in vivo is still elusive in eukaryotes. Although Dunaway and Ostrander have clearly shown that local domains of negative supercoiling activate the ribosomal RNA gene promoter in vivo,28 other promoters have not been tested through a similar approach. Supercoiling factor (SCF) is a protein capable of generating negative supercoils in relaxed DNA in conjunction with topoisomerase II.29 D. melanogaster SCF localizes to many interbands and puffs that are active sites of transcription30 (fig. 4), suggesting that SCF plays a role in formation of transcriptionally active chromatin. Recent study has shown that transcription of certain genes is compromised by targeting SCF with RNAi (Furuhashi and Hirose, unpublished). Because SWI/SNF-type chromatin remodeling factors require changes in DNA topology for their action,31 SCF and topoisomerase II may facilitate chromatin remodeling through generation of negative supercoils.
Formation of unusual DNA structure (see Chapter 1 for details) such as Z-form is significantly facilitated by negative supercoiling.32 Such unusual DNA structure can affect transcription. For example, Z-DNA in a promoter region has been suggested to participate in transcriptional activation in collaboration with a chromatin remodeling complex BAF.33 On the contrary, Z-form within a transcribable region would inhibit transcription elongation. Finally positive supercoiling of DNA has been reported to diminish transcription in vivo.34 These data indicate a possible role of DNA supercoiling in eukaryotic transcription in vivo.
Conclusion
DNA supercoiling can affect transcription in both prokaryotes and eukaryotes. While the critical step is open complex formation in most prokaryotic genes, TBP binding to the TATA element is affected in most eukaryotic genes. Furthermore, DNA supercoiling can affect transcription through chromatin remodeling in eukaryotes.
References
- 1.
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. [PubMed: 13718526]
- 2.
- Wang JC. DNA topoisomerases. Ann Rev Biochem. 1996;65:635–692. [PubMed: 8811192]
- 3.
- Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E.coli cells: analogous measurements in insect and human cells. Cell. 1980;21:773–783. [PubMed: 6254668]
- 4.
- Pruss GJ, Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989;56:521–523. [PubMed: 2645054]
- 5.
- Margolin P, Zumstein L, Sternglanz R. et al. The Escherichia coli supX locus is topA, the structural gene for DNA topoisomerase I. Proc Natl Acad Sci USA. 1985;82:5437–5441. [PMC free article: PMC390584] [PubMed: 2991925]
- 6.
- Rudd KE, Menzzel R. His operons of Escherichia coli and Salmonella typhimurium are regulated by DNA supercoiling. Proc Natl Acad Sci USA. 1987;84:517–521. [PMC free article: PMC304240] [PubMed: 3025879]
- 7.
- Menzel R, Gellert M. Modulation of transcription by DNA supercoiling: A deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc Natl Acad Sci USA. 1987;84:4185–4189. [PMC free article: PMC305049] [PubMed: 3035573]
- 8.
- Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol. 1992;6:425–433. [PubMed: 1313943]
- 9.
- Kusano S, Ding Q, Fujita N. et al. Promoter selectivity of Escherichia coli RNA polymerase Eσ70 and Eσ38 holoenzyme. J Biol Chem. 1996;271:1998–2004. [PubMed: 8567650]
- 10.
- Mizushima T, Natori S, Sekimizu K. Relaxation of supercoiled DNA associated with induction of heat shock proteins in Escherichia coli. Mol Gen Genet. 1993;238:1–5. [PubMed: 8097554]
- 11.
- Dorman CJ. Flexible response: DNA supercoiling, transcription and bacterial adaptation to environmental stress. Trends Microbiol. 1996;4:214–216. [PubMed: 8795154]
- 12.
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. [PMC free article: PMC299221] [PubMed: 2823250]
- 13.
- Pruss GJ, Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracyclin affects plasmid DNA supercoiling. Proc Natl Acad Sci USA. 1986;83:8952–8956. [PMC free article: PMC387052] [PubMed: 3024156]
- 14.
- Lockshon D, Morris DR. Positively supercoiled plasmid DNA is produced by treatment of Escherichia coli with DNA gyrase inhibitors. Nucl Acids Res. 1983;11:2999–3017. [PMC free article: PMC325943] [PubMed: 6304617]
- 15.
- Jupe ER, Sinden RR, Cartwright IL. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. EMBO J. 1993;12:1067–1075. [PMC free article: PMC413308] [PubMed: 8458324]
- 16.
- Ljungman M, Hanawalt PC. Localyzed torsional tension in the DNA of human cells. Proc Natl Acad Sci USA. 1992;89:6055–6059. [PMC free article: PMC49436] [PubMed: 1631091]
- 17.
- Kramer PR, Sinden RR. Measurement of unrestrained negative supercoiling and topological domain size in living cells. Biochemistry. 1997;36:3151–3158. [PubMed: 9115991]
- 18.
- Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. [PubMed: 2847873]
- 19.
- Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila J Cell Sci, in press . [PubMed: 15252118]
- 20.
- Hirose S, Ohta T. DNA supercoiling and eukaryotic transcription-Cause and effect. Cell Struct Funct. 1990;15:133–135. [PubMed: 2204487]
- 21.
- Hirose S, Suzuki Y. In vitro transcription of eukaryotic genes is affected differently by the degree of DNA supercoiling. Proc Natl Acad Sci USA. 1988;85:718–722. [PMC free article: PMC279626] [PubMed: 2829200]
- 22.
- Mizutani M, Ura K, Hirose S. DNA superhelicity affects the formation of transcription preinitiation complex on eukaryotic genes differently. Nucl Acids Res. 1991;19:2907–2911. [PMC free article: PMC328250] [PubMed: 1647522]
- 23.
- Tabuchi H, Handa H, Hirose S. Underwinding of DNA on binding of yeast TFIID to the TATA element. Biochem Biophys Res Commun. 1993;192:1432–1438. [PubMed: 8507207]
- 24.
- Kim Y, Geiger JH, Hahn S. et al. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. [PubMed: 8413604]
- 25.
- Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recongizing the minor groove of a TATA element. Nature. 1993;365:520–527. [PubMed: 8413605]
- 26.
- Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. [PubMed: 8490964]
- 27.
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D.melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. [PubMed: 3136931]
- 28.
- Dunaway M, Ostrander EA. Local domains of supercoiling activate a eukaryotic promoter in vivo. Nature. 1993;361:746–748. [PubMed: 8441472]
- 29.
- Ohta T, Hirose S. Purification of a DNA supercoiling factor from the posterior silk gland of Bombyx mori. Proc Natl Acad Sci USA. 1990;87:5307–5311. [PMC free article: PMC54312] [PubMed: 2164676]
- 30.
- Kobayashi M, Aita N, Hayashi S. et al. DNA supercoiling factor localizes to puffs on polytene chromosomes in Drosophila melanogaster. Mol Cell Biol. 1998;18:6737–6744. [PMC free article: PMC109257] [PubMed: 9774687]
- 31.
- Gavin I, Horn PJ, Peterson CL. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol Cell. 2001;7:97–104. [PubMed: 11172715]
- 32.
- Nordheim A, Lafer EM, Peck LJ. et al. Negatively supercoiled plamids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982;31:309–318. [PubMed: 7159926]
- 33.
- Liu R, Liu H, Chen X. et al. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;105:309–318. [PubMed: 11509180]
- 34.
- Gartenberg MR, Wang JC. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci USA. 1992;89:11461–11465. [PMC free article: PMC50571] [PubMed: 1333610]
- 35.
- Leng F, McMacken R. Potent stimulation of transcription-coupled DNA supercoiling by sequence-specific DNA-binding proteins. Proc Natl Acad Sci USA. 2002;99:9139–9144. [PMC free article: PMC123107] [PubMed: 12093906]
- Possible Roles of DNA Supercoiling in Transcription - Madame Curie Bioscience Da...Possible Roles of DNA Supercoiling in Transcription - Madame Curie Bioscience Database
- Proteases and Their Cognate Inhibitors of the Serine and Metalloprotease Subclas...Proteases and Their Cognate Inhibitors of the Serine and Metalloprotease Subclasses, in Testicular Physiology - Madame Curie Bioscience Database
- Roles of Poly(ADP-Ribose) Metabolism in the Regulation of Centrosome Duplication...Roles of Poly(ADP-Ribose) Metabolism in the Regulation of Centrosome Duplication and in the Maintenance of Neuronal Integrity - Madame Curie Bioscience Database
- Enzyme-RNA Substrate Recognition in RNA-Modifying Enzymes - Madame Curie Bioscie...Enzyme-RNA Substrate Recognition in RNA-Modifying Enzymes - Madame Curie Bioscience Database
- DNA Demethylation - Madame Curie Bioscience DatabaseDNA Demethylation - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...