NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Poly(ADP-ribose) polymerase (PARP-1) plays a crucial role in DNA repair and interacts with many DNA replication/repair factors, including the Proliferating Cell Nuclear Antigen (PCNA), a protein involved in many DNA transactions. The association between these proteins in vitro results in the inhibition of PARP-1 activity and of PCNA-dependent DNA synthesis carried out by DNA polymerase δ. In vivo, the interaction of the two proteins has been shown to increase after DNA damage. The activity of PCNA is regulated by p21waf1/cip1, which binds the interdomain connector loop of PCNA in the same region involved in the interaction of PCNA with DNA polymerase δ. This feature enables p21 to compete with DNA polymerase δ to regulate the differential inhibition of DNA replication vs. DNA repair. The finding that PARP-1 is associated with both PCNA and p21 suggests a possible cooperation of PARP-1 and p21 in regulating the functions of PCNA during DNA replication/repair.
Introduction
PARP-1 and PCNA share the property of working in association with a number of factors that are involved in the regulation of DNA metabolism. This feature legitimates the search for a physical interaction between PARP-1 and PCNA, which in fact has been demonstrated by in vitro assays and by coimmunoprecipitation experiments. Since both PARP-1 and PCNA play an active role in DNA repair and replication, a functional association between them has been postulated. Indeed, an inhibitory effect on the respective enzymatic activities, mediated by their association, was reported. Finally, due to the relevant role of p21waf1/cip1 in regulating the activity of PCNA, the existence of an interplay between PCNA-PARP-1, PCNA-p21 and p21-PARP-1 interactions was investigated.
PARP-1 Interacts with DNA Repair/Replication Proteins
PARP-1 catalyzes the conversion of β-NAD+ into ADP-ribose to produce polymers of ADP-ribose covalently bound to nuclear acceptors, including PARP-1 itself. The evidence for an active role of PARP-1 in DNA metabolism is supported by the observations that in vitro the activity of DNA polymerases is modulated by poly(ADP-ribosylation)1,2 and that PARP-1 is present in multiprotein DNA replication complexes (MRC),2-4 where it regulates the recruitment of some components of the DNA synthesome.2 When reconstituted systems were used, many potential acceptors of poly(ADP-ribose) (PAR) were identified; subsequently, several in vivo poly(ADP-ribosylated) proteins involved in DNA metabolism were found, including some factors that regulate chromatin structure, such as histones5 and DNA topoisomerases I6 and II.7 In this respect, it has been shown that poly(ADP-ribosylation) by PARP-1 contributes to regulate the catalytic activity of topoisomerase I8,9 and the expression of topoisomerase IIβ.10
The interaction between PARP-1 and other proteins is also mediated by the noncovalent binding of poly(ADP-ribose) through a canonical PAR-binding domain.11 The PAR-binding consensus sequence is present in a family of proteins involved in DNA damage recognition and repair, including XPA, XRCC1, DNA ligase III and DNA-PK.11 This motif has not been found in the amino acid sequence of PCNA but is present at the C-terminus of p21,11 where it coincides with the residues involved in its binding to the interdomain connector loop of PCNA.12,13 By a proteomic approach, heterogeneous nuclear ribonucleoproteins have been identified among the proteins that bind poly(ADP-ribose).14
After the pioneering work of Shall and coworkers15 reporting the negative effect of PARP inhibition on DNA repair, a large body of evidence supported the definition of PARP-1 as a guardian of the genome,16 thus including this protein in the category of DNA damage-sensor molecules.17,18 Recently, the generation of PARP-1-deficient mice allowed the definition of a more precise correlation between PARP-1 activity and DNA repair (reviewed in ref. 19). PARP-1 is mainly involved in the Base Excision Repair (BER) pathway,20-22 where it interacts with other components of the multiprotein repair complex, such as XRCC1, DNA ligase III and DNA polymerase β.23 Interestingly, the recruitment of DNA ligase III to DNA single strand breaks (SSB) is mediated by a zinc finger domain which is homologous to that of PARP-1. This feature can enable DNA ligase III to displace PARP-1 from the SSB, thus rendering DNA accessible to repair protein.24,25 An involvement of PARP-1 in the Nucleotide Excision Repair (NER) pathway has also been reported.26
The surveillance network against DNA damage implies the interaction of PARP-1 with other factors. The interaction of PARP-1 with p53 has been widely described, thus supporting the existence of cooperation between these proteins in maintaining genome integrity (see Chapter 6). PARP-1 acts in concert with Werner syndrome protein (WRN), a protein involved in DNA replication and repair, which interacts directly with PARP-127-29 and with PCNA.30 It has been recently shown that PARP-1, WRN and Ku70/80, which is a component of DNA-PK holoenzyme, are stably associated.31 PARP-1 shares with Ku80 the ability to recognize and bind DNA double strand breaks (DSB), it stimulates the activity of DNA-PK and is associated with it;32,33 on the contrary, DNA-PK suppresses PARP activity, possibly by competing for the binding to DNA ends.34 The generation of double mutants for PARP-1/Ku80 revealed that the functions of both proteins are essential for DSB repair during mouse embryogenesis.35 PARP-1 was described to act synergistically also with another factor involved in the response to DNA strand breaks, i.e., ATM.36,37
PCNA: A Protein with Many Partners
PCNA is a ring-shaped homo-trimeric protein38 that functions on DNA as a clamping platform to recruit proteins involved in DNA metabolism.39 During DNA replication, PCNA is loaded onto DNA by the replication factor C (RFC), and then it clamps DNA polymerase δ (pol d) to the template (for a review see ref. 40). In the lagging strand, PCNA interacts also with other proteins, such as Flap endonuclease FEN-1,41 which removes RNA primers, and DNA ligase I,42 which seals newly synthesized DNA fragments.43 PCNA plays a role in chromatin assembly through the binding to CAF-144 and the interaction with DNA-5'-cytosine methyl transferase (DNAMT).45,46 An active role of PCNA during post-replication repair is supported by a number of reports indicating its interaction with mismatch repair proteins, i.e., MSH3 and MSH6,47 the uracil-DNA glycosylase 2,48 and the cyclin-like uracil-DNA glycosylase.49
The growing list of proteins interacting with PCNA includes DNA polymerases β,50 η,51 ι,52 κ53 and λ,54 terminal nucleotidyl transferase,55 the Werner syndrome helicase,30 AP endonucleases 1 and 2,56,57 DNA glycosylase MYH,58 the transcription factors Y-box binding protein59 and p300,60 the histone deacetylase HDAC,61 the apoptotic proteins MCL162 and ING1,63 and the cell cycle regulator Cdc25C.64 Recently, by proteomic- and genomic-based strategies, new PCNA-interacting proteins have been discovered, i.e., CHL12, involved in chromosome cohesion,65 and the 5'-3' DNA helicase RRM3.66
PCNA partners, including the CDK inhibitor p21, interact with the interdomain connector loop of PCNA through a motif of 8 aminoacids (QxxM/L/IxxFF/FY).67 PARP-1 shows a putative PCNA-binding consensus sequence (QDLIKMIF) at the position 669 within the catalytic domain that is essential for the conversion of NAD+ into ADP-ribose.
PCNA is present in the cell both as a free/detergent-soluble protein, and a chromatin-bound/ detergent-insoluble form which, encircling DNA, is directly involved in DNA synthesis. The native PCNA trimer has three potential binding sites,67 thus each PCNA ring may interact with more than one protein at the same time. In this respect, we have recently proposed a mechanism by which PCNA trimers bound to DNA during S phase are organized as distinct pools able to bind different partners in a mutually exclusive manner.68 During DNA repair, the multiple interactions established by PCNA could be regulated by p21 in a disassembly and/or recycling process of PCNA molecules at repair sites.69,70
P21 Regulates the Activity of PCNA
The cyclin-dependent kinase (CDK) inhibitor p21CDKN1A (also known as p21WAF1/Cip1) plays an important role in several cellular pathways in response to intracellular and extracellular stimuli. In particular, p53-dependent induction of p21 after DNA damage leads to inhibition of cell cycle progression and DNA replication. In addition, p21 is clearly involved in other processes like senescence, differentiation, and regulation of gene expression.71,72
Cell cycle arrest is performed by p21 not only through CDK inhibition, but also by direct binding to PCNA, thereby interfering with PCNA-dependent DNA synthesis.73,74 The dual effect on cell cycle regulatory proteins is mediated in the p21 sequence via distinct interaction sites for cyclin-Cdk complexes and for PCNA.73,74 Binding of p21 to PCNA occurs at the interdomain connector loop, i.e., the same region involved in the binding of several PCNA-interacting proteins.67 For some of them (e.g., DNA pol d, FEN-1, DNAMT), binding of p21 has been shown to result in their displacement from PCNA,75 thereby inhibiting DNA replication.76 In contrast, p21 does not inhibit PCNA-dependent DNA repair.77,78 Several lines of evidence suggest that p21 is actively involved in DNA repair; in fact, human p21-null cells have been shown to be more sensitive to DNA damage, and to be deficient in DNA repair.70,79,80
Recent work by our group has shown that in vitro p21 is also able to compete with PARP-1 and displace it from the interdomain connector loop of PCNA.81 In vivo, after MNNG-induced DNA damage, p21 interacts with PARP-1, but not with chromatin-bound PCNA, i.e., the form involved in DNA repair.81 These results suggest that PARP-1 may sequester p21, probably to avoid an untimely interaction with PCNA, which would inhibit DNA repair.
Effect of the Interaction between PARP-1 and PCNA
We have recently shown that PARP-1 associates in vitro and in vivo with PCNA in HeLa cells and human fibroblasts.81 A typical pattern of coimmunoprecipitation is reported in Figure 1, which also shows the increase in the amount of coimmunoprecipitated PARP-1 and PCNA in MNNG-treated HeLa cells, thus suggesting that this association may play a role in the cell response to DNA damage by alkylating agents. This feature is not strictly mediated by poly(ADP-ribose) chains, since experiments carried out in the presence of 3-aminobenzamide, an inhibitor of poly(ADP-ribosylation), revealed that the association occurred also when PARP-1 activity was suppressed.81
We observed that PARP-1 affects PCNA-dependent pol δ activity in vitro and that this effect is more evident in the presence of an excess of PARP-1. From this finding, it is tempting to speculate that the association of PCNA with PARP-1 occurs at the same region involved in the interaction of PCNA with pol d. Since such interaction is essential for processive DNA synthesis, PARP-1 could act as a negative regulator of PCNA-dependent pol δ activity when replicative DNA synthesis has to be inhibited, e.g., under damage conditions. On the other hand, we have shown that PCNA inhibits PARP-1 activity in vitro, possibly through the physical interaction between the putative PCNA-binding sequence located within the catalytic domain of PARP-1. This feature suggests that PCNA could regulate negatively PARP-1 activity, thus allowing the inhibition of poly(ADP-ribosylation) reactions, to avoid an excessive consumption of NAD+ or the inappropriate modification of the structure and function of crucial proteins.
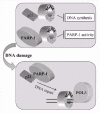
The region of the interdomain connector loop of PCNA that is involved in the association with pol δ is identical to the sequence interacting with the C-terminus of p21.12,13,75 Competition between pol δ and p21 for PCNA bindi_ has been proposed to regulate the differential inhibition of DNA replication vs. DNA repair.40,69 Interestingly, we have demonstrated that in vitro and in vivo the PCNA-binding region of p21 is also responsible for the direct association with PARP-1.81 As suggested by coimmunoprecipitation (fig. 1) and pull-down data,81 PARP-1 could compete with the C-terminal region of p21 for the binding to the interdomain connector loop of PCNA. Our findings showing that PARP-1 binds both PCNA and p21, and that both interactions increase upon DNA damage, suggest that in vivo PARP-1, PCNA and p21 undergo a dynamic exchange of partners to regulate PCNA functions during DNA replication/repair. A dynamic interaction of PCNA with its partners has also been demonstrated for the association with pol δ and DNA ligase I.68 In conclusion, these observations prompt us to propose a model in which, from one side, PARP-1 may inhibit PCNA-dependent DNA replication, while from the other side, sequestration of p21 by PARP-1 could enable PCNA to be recruited to DNA repair sites (fig. 2).
Acknowledgements
We acknowledge Drs. I Frouin, G Maga, M Denegri and F Riva (IGM-CNR) for their contribution to the experimental work presented here. Research was supported by the Italian CNR and MIUR (FIRB Project RBNE0132MY).
References
- 1.
- Eki T. Poly (ADP-ribose) polymerase inhibits DNA replication by human replicative DNA polymerase α, β and ε in vitro. FEBS Lett. 1994;356:261–266. [PubMed: 7805850]
- 2.
- Simbulan-Rosenthal CM, Rosenthal DS, Boulares H. et al. Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry. 1998;37:9363–9370. [PubMed: 9649317]
- 3.
- Dantzer F, Nasheuer HP, Vonesch JL. et al. Functional association of poly(ADP-ribose) polymerase with DNA polymerase α-primase complex: A link between DNA strand break detection and DNA replication. Nucl Acids Res. 1998;26:1891–1898. [PMC free article: PMC147507] [PubMed: 9518481]
- 4.
- Frouin I, Montecucco A, Biamonti G. et al. Cell cycle-dependent dynamic association of cyclin/ Cdk complexes with human DNA replication proteins. EMBO J. 2002;21:2485–2495. [PMC free article: PMC125998] [PubMed: 12006500]
- 5.
- Althaus FR, Richter C. ADP-ribosylation of proteins: Enzymology and biological significance. Springer Verlag, Berlin. 1987 [PubMed: 3118181]
- 6.
- Krupitza G, Cerutti P. ADP-ribosylation of ADPR-transferase and topoisomerase I in intact mouse epidermal cells JB6. Biochemistry. 1989;28:2034–2040. [PubMed: 2541771]
- 7.
- Scovassi AI, Mariani C, Negroni M. et al. ADP-ribosylation of nonhistone proteins in HeLa cells: Modification of DNA topoisomerase II. Exp Cell Res. 1993;206:177–181. [PubMed: 8387022]
- 8.
- Bauer PI, Chen H-J, Kenesi E. et al. Molecular interactions between poly(ADP-ribose) polymerase (PARP I) and topoisomerase I (Topo I): Identification of topology of binding. FEBS Lett. 2001;506:239–242. [PubMed: 11602253]
- 9.
- Malanga M, Althaus FR. Poly(ADP-ribose) reactivates stalled DNA topoisomerase I and induces DNA strand break resealing. J Biol Chem. 2004;279:5244–5248. [PubMed: 14699148]
- 10.
- Canitrot Y, de Murcia G, Salles B. Decreased expression of topoisomerase IIβ in poly(ADP-ribose) polymerase-deficient cells. Nucl Acids Res. 1998;26:5134–5138. [PMC free article: PMC147949] [PubMed: 9801310]
- 11.
- Pleschke JM, Kleczkowska HE, Strohm M. et al. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. [PubMed: 11016934]
- 12.
- Goubin F, Ducommun B. Identification of binding domains on the p21Cip1 cyclin-dependent kinase inhibitor. Oncogene. 1995;10:2281–2287. [PubMed: 7784076]
- 13.
- Gulbis JM, Kelman Z, Hurwitz J. et al. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. [PubMed: 8861913]
- 14.
- Gagné J-P, Hunter JM, Labrecque B. et al. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem J. 2003;371:331–340. [PMC free article: PMC1223283] [PubMed: 12517304]
- 15.
- Durkacz BW, Omidiji O, Gray DA. et al. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. [PubMed: 6243744]
- 16.
- Jeggo PA. DNA repair: PARP - another guardian angel? Curr Biol. 1998;8:R49–51. [PubMed: 9427640]
- 17.
- Bürkle A. Poly(ADP-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 2001;163:1–5. [PubMed: 11163101]
- 18.
- Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. [PubMed: 12829019]
- 19.
- Shall S, de MurciaG. Poly(ADP-ribose) polymerase-1: What have we learned from the deficient mouse model? Mutat Res. 2000;;460:1–15. [PubMed: 10856830]
- 20.
- Dantzer F, Schreiber V, Niedergang C. et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69–75. [PubMed: 10214912]
- 21.
- Lavrik OI, Prasad R, Sobol RW. et al. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J Biol Chem. 2001;276:25541–25548. [PubMed: 11340072]
- 22.
- Le PageF, Schreiber V, Dhérin C. et al. Poly(ADP-ribose) polymerase-1 (PARP-1) is required in murine cell lines for base excision repair of oxidative DNA damage in the absence of DNA polymerase β J Biol Chem. 2003;278:18471–18477. [PubMed: 12637553]
- 23.
- Caldecott KW, Aoufouchi S, Johnson P. et al. XRCC1 polypeptide interacts with DNA polymerase β and possibly poly(ADP-ribose) polymerase, and DNA ligase III is a novel molecular “nick sensor” in vitro. Nucl Acids Res. 1996;24:4387–4394. [PMC free article: PMC146288] [PubMed: 8948628]
- 24.
- Leppard JB, Dong Z, Mackey ZB. et al. Physical and functional interaction between DNA ligase IIIα and poly(ADP-ribose) polymerase 1 in DNA single-strand break repair. Mol Cell Biol. 2003;23:5919–5927. [PMC free article: PMC166336] [PubMed: 12897160]
- 25.
- Mackey ZB, Niedergang C, Ménissier-de Murcia J. et al. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:21679–21687. [PubMed: 10419478]
- 26.
- Flohr C, Bürkle A, Radicella JP. et al. Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein. Nucl Acids Res. 2003;31:5332–5337. [PMC free article: PMC203308] [PubMed: 12954769]
- 27.
- Adelfak C, Kontou M, Hirsch-Kauffmann M. et al. Physical and functional interaction of the Werner syndrome protein with poly-ADP ribosyl transferase. FEBS Lett. 2003;554:55–58. [PubMed: 14596914]
- 28.
- Lebel M, Lavoie J, Gaudreault I. et al. Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am J Pathol. 2003;162:1559–1569. [PMC free article: PMC1851180] [PubMed: 12707040]
- 29.
- von KobbeC, Harrigan JA, May A. et al. Central role for the Werner syndrome protein/ poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol. 2003;23:8601–8613. [PMC free article: PMC262662] [PubMed: 14612404]
- 30.
- Lebel M, Spillare EA, Harris CC. et al. The Werner syndrome gene product copurifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. [PubMed: 10608841]
- 31.
- Li B, Navarro S, Kasahara N. et al. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem. 2004;279:13659–13667. [PubMed: 14734561]
- 32.
- Morrison C, Smith GC, Stingl L. et al. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat Genet. 1997;17:479–482. [PubMed: 9398855]
- 33.
- Ruscetti T, Lehnert BE, Halbrook J. et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem. 1998;273:14461–14467. [PubMed: 9603959]
- 34.
- Ariumi Y, Masutani M, Copeland TD. et al. Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene. 1999;18:4616–4625. [PubMed: 10467406]
- 35.
- Henrie MS, Kurimasa A, Burma S. et al. Lethality in PARP/Ku80 double mutant mice reveals physiological synergy during early embryogenesis. DNA Repair. 2003;2:151–158. [PubMed: 12531386]
- 36.
- Ménissier-de Murcia J, Mark M, Wendling O. et al. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol Cell Biol. 2001;21:1828–1832. [PMC free article: PMC86747] [PubMed: 11238919]
- 37.
- Marecki JC, McCord JM. The inhibition of poly(ADP-ribose) polymerase enhances growth rates of ataxia telangiectasia cells. Arch Biochem Biophys. 2002;402:227–234. [PubMed: 12051667]
- 38.
- Krishna TSR, Kong X-P, Gary S. et al. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. [PubMed: 8001157]
- 39.
- Kelman Z, Hurwitz J. Protein-PCNA interactions: A DNA scanning mechanism? Trends Biochem Sci. 1998;23:236–238. [PubMed: 9697409]
- 40.
- Maga G, Hübscher U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J Cell Sci. 2003;116:3051–3060. [PubMed: 12829735]
- 41.
- Chapados BR, Hosfield DJ, Han S. et al. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. [PubMed: 14718165]
- 42.
- Levin DS, Bai W, Yao N. et al. An interaction between DNA ligase I and proliferating cell nuclear antigen: Implications for Okazaki fragment synthesis and rejoining. Proc Natl Acad Sci USA. 1997;94:12863–12868. [PMC free article: PMC24229] [PubMed: 9371766]
- 43.
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. [PubMed: 9759502]
- 44.
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. [PubMed: 10052459]
- 45.
- Chuang LS-H, Ian H-I, Koh T-W. et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. [PubMed: 9302295]
- 46.
- Zardo G, Reale A, Passananti C. et al. Inhibition of poly(ADP-ribosyl)ation induces DNA hypermethylation: A possible molecular mechanism. FASEB J. 2002;16:1319–1321. [PubMed: 12154007]
- 47.
- Kleczkowska HE, Marra G, Lettieri T. et al. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. [PMC free article: PMC312660] [PubMed: 11274057]
- 48.
- Otterlei E, Warbrick TA, Nagelhus T. et al. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. [PMC free article: PMC1171460] [PubMed: 10393198]
- 49.
- Muller-Weeks SJ, Caradonna S. Specific association of cyclin-like uracil-DNA glycosylase with the proliferating cell nuclear antigen. Exp Cell Res. 1996;226:346–355. [PubMed: 8806438]
- 50.
- Kedar PS, Kim SJ, Robertson A. et al. Direct interaction between mammalian DNA polymerase β and proliferating cell nuclear antigen. J Biol Chem. 2002;277:31115–31123. [PubMed: 12063248]
- 51.
- Haracska L, Johnson RE, Unk I. et al. Physical and functional interaction of human DNA polymerase η with PCNA. Mol Cell Biol. 2001;21:7199–7206. [PMC free article: PMC99895] [PubMed: 11585903]
- 52.
- Haracska L, Johnson RE, Unk I. et al. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc Natl Acad Sci USA. 2001;98:14256–14261. [PMC free article: PMC64669] [PubMed: 11724965]
- 53.
- Haracska L, Johnson RE, Unk I. et al. Stimulation of DNA synthesis activity of human DNA polymerase ? by PCNA. Mol Cell Biol. 2002;22:784–791. [PMC free article: PMC133560] [PubMed: 11784855]
- 54.
- Maga G, Villani G, Ramadan K. et al. Human DNA polymerase λ functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem. 2002;277:48434–48440. [PubMed: 12368291]
- 55.
- Ibe S, Fujita K, Toyomoto T. et al. Terminal deoxynucleotidyltransferase is negatively regulated by direct interaction with proliferating cell nuclear antigen. Genes Cells. 2001;6:815–824. [PubMed: 11554927]
- 56.
- Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. [PubMed: 11601988]
- 57.
- Tsuchimoto D, Sakai Y, Sakumi K. et al. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucl Acids Res. 2001;29:2349–2360. [PMC free article: PMC55700] [PubMed: 11376153]
- 58.
- Parker A, Gu Y, Mahoney W. et al. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J Biol Chem. 2001;276:5547–5555. [PubMed: 11092888]
- 59.
- Ise T, Nagatani G, Imamura T. et al. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 1999;59:342–346. [PubMed: 9927044]
- 60.
- Hasan S, Hassa PO, Imhof R. et al. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature. 2001;410:387–391. [PubMed: 11268218]
- 61.
- Milutinovic S, Zhuang Q, Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J Biol Chem. 2002;277:20974–20978. [PubMed: 11929879]
- 62.
- Fujise K, Zhang D, Liu J. et al. Regulation of apoptosis and cell cycle progression by MCL1. Differential role of proliferating cell nuclear antigen. J Biol Chem. 2000;275:39458–39465. [PubMed: 10978339]
- 63.
- Scott M, Bonnefin P, Vieyra D. et al. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci. 2001;114:3455–3462. [PubMed: 11682605]
- 64.
- Ando T, Kawabe T, Ohara H. et al. Involvement of the interaction between p21 and proliferating cell nuclear antigen for the maintenance of G2/M arrest after DNA damage. J Biol Chem. 2001;276:42971–42977. [PubMed: 11559705]
- 65.
- Ohta S, Shiomi Y, Sugimoto K. et al. A proteomic approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. J Biol Chem. 2002;277:40362–40367. [PubMed: 12171929]
- 66.
- Schmidt KH, Derry KL, Kolodner RD. Saccharomyces cerevisiae RRM3, a 5' to 3' DNA helicase, physically interacts with proliferating cell nuclear antigen. J Biol Chem. 2002;277:45331–45337. [PubMed: 12239216]
- 67.
- Warbrick E. PCNA binding through a conserved motif. BioEssays. 1998;20:195–199. [PubMed: 9631646]
- 68.
- Riva F, Savio M, Cazzalini O. et al. Distinct pools of proliferating cell nuclear antigen associated to DNA replication sites interact with p125 subunit of DNA polymerase δ or DNA ligase I. Exp Cell Res. 2004;293:357–367. [PubMed: 14729473]
- 69.
- Prosperi E. Multiple roles of the proliferating cell nuclear antigen: DNA replication, repair, and cell cycle controlIn: Meijer L, Guidet S, Philippe M, eds.Progress in Cell Cycle ResearchNew York: Plenum Press,19973193–210. [PubMed: 9552415]
- 70.
- Stivala LA, Riva F, Cazzalini O. et al. p21waf1/cip1-null human fibroblasts are deficient in nucleotide excision repair downstream the recruitment of PCNA to DNA repair sites. Oncogene. 2001;20:563–570. [PubMed: 11313988]
- 71.
- Dotto GP. p21WAF1/Cip1: More than a break to the cell cycle? Biochim Biophys Acta. 2000;1471:M43–M46. [PubMed: 10967424]
- 72.
- Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: A function for each cell compartment? Trends Cell Biol. 2003;13:65–70. [PubMed: 12559756]
- 73.
- Chen J, Jackson PK, Kirschner MW. et al. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature. 1995;374:386–388. [PubMed: 7885482]
- 74.
- Luo Y, Hurwitz J, Massague J. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature. 1995;375:159–161. [PubMed: 7753174]
- 75.
- Warbrick E, Lane DP, Glover DM. et al. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: A potential mechanism to coordinate DNA replication and repair. Oncogene. 1997;14:2313–2321. [PubMed: 9178907]
- 76.
- Cazzalini O, Perucca P, Riva F. et al. p21CDKN1A does not interfere with loading of PCNA at DNA replication sites, but inhibits subsequent binding of DNA polymerase δ at the G1/Sphase transition. Cell Cycle. 2003;2:596–603. [PubMed: 14504476]
- 77.
- Shivji MK, Ferrari E, Ball K. et al. Resistance of human nucleotide excision repair synthesis in vitro to p21Cdn1. Oncogene. 1998;17:2827–2838. [PubMed: 9879989]
- 78.
- Savio M, Stivala LA, Scovassi AI. et al. p21waf1/cip1 protein associates with the detergent-insoluble form of PCNA concomitantly with disassembly of PCNA at nucleotide excision repair sites. Oncogene. 1996;13:1591–1598. [PubMed: 8895503]
- 79.
- McDonald IIIER, Wu GS, Waldman T. et al. Repair defect in p21WAF1/CIP1 -/- human cancer cells. Cancer Res. 1996;56:2250–2255. [PubMed: 8625293]
- 80.
- Sheikh MS, Chen YQ, Smith ML. et al. Role of p21waf1/cip1/sdi1 in cell death and DNA repair as studied using a tetracycline-inducible system in p53-deficient cells. Oncogene. 1997;14:1875–1882. [PubMed: 9150394]
- 81.
- Frouin I, Maga G, Denegri M. et al. Human proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1 and p21waf1/cip1: A dynamic exchange of partners. J Biol Chem. 2003;278:39265–39268. [PubMed: 12930846]
- Dynamic Interaction between PARP-1, PCNA and p21waf1/cip1 - Madame Curie Bioscie...Dynamic Interaction between PARP-1, PCNA and p21waf1/cip1 - Madame Curie Bioscience Database
- DNA: Alternative Conformations and Biology - Madame Curie Bioscience DatabaseDNA: Alternative Conformations and Biology - Madame Curie Bioscience Database
- NMD in Drosophila: A Snapshot into the Evolution of a Conserved mRNA Surveillanc...NMD in Drosophila: A Snapshot into the Evolution of a Conserved mRNA Surveillance Pathway - Madame Curie Bioscience Database
- A Prelude to Translational (Re)Initiation - Madame Curie Bioscience DatabaseA Prelude to Translational (Re)Initiation - Madame Curie Bioscience Database
- Adding Complexity to Phagocytic Signaling: Phagocytosis-Associated Cell Response...Adding Complexity to Phagocytic Signaling: Phagocytosis-Associated Cell Responses and Phagocytic Efficiency - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...