NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Concurrent with transcription, the ribosomal RNA precursor (pre-rRNA) is modified and associates with many of the ribosomal proteins. The modifications are guided by small nucleolar RNAs (snoRNAs), which base-pair with target sites in eukaryotic pre-rRNA and perhaps also play a role in pre-rRNA folding. A few snoRNAs are essential for rRNA processing; they appear to dock on the pre-rRNA substrate during its transcription but are not activated until the nascent transcript is completed and released from the DNA template. Pre-rRNA contains external transcribed spacers (5' ETS and 3' ETS) at both ends and internal transcribed spacers (ITS1 and ITS2) flanking either side of 5.8S RNA. The transcribed spacers may serve as docking sites for certain snoRNAs (such as U3) and their removal by processing might ensure that steps in rRNA folding cannot run in reverse. The first cleavages in metazoan pre-rRNA remove the 3'-ETS (site T1) and sometimes the 5'-end of the 5'-ETS (site A'). The next set of cleavages (sites A0, 1 and 2) require U3, U14 and U22 snoRNAs in vertebrates and give rise to 18S rRNA. In addition, E1 snoRNA (U17) is needed for cleavage at site 1 and E2 snoRNA for cleavage at site 2. A detailed mechanism of multiple base-pairings between U3 snoRNA and 18S rRNA is hypothesized to fold pre-rRNA appropriately for these cleavages and perhaps to position the cleavage factors. Site 3, which resides slightly upstream of the 5'-end of 5.8S rRNA, can be cleaved before or after sites A0, 1 and 2, depending on the system. Site 3 may link the 18S and 28S rRNA processing pathways, although each can proceed independently of the other. The late steps of rRNA processing involve cleavages of pre-rRNA at sites 3, 4', 4 and 5 to generate mature 5.8S and 28S rRNAs. These cleavages require U8 snoRNA in vertebrates, which is postulated to base-pair with the 5'-end of 28S rRNA within pre-rRNA, perhaps facilitating later base-pairing of 5.8S RNA with the 5'-end of 28S rRNA. snoRNAs and also the protein nucleolin may act as chaperones to fold rRNA. Evolutionary comparisons between kingdoms suggest that pre-rRNA base-pairing with snoRNAs in trans might have replaced intra-molecular pre-rRNA base-pairing in cis. The cleavage steps in rRNA processing could prevent rRNA folding reactions from running in reverse.
Introduction
The nucleolus has intrigued biologists for more than 200 years, since its presumed description by Fontana.1 Although there is debate over whether Fontana really did visualize the nucleolus,2 its documentation was clear by the 1830's (reviewed by refs. 2, 3). The subject of the nucleolus stimulated so much research that by 1898 Montgomery reviewed work in about 700 papers on this topic.4 In the past three decades, several books have been written about the nucleolus,5-7 and the compilation of information is now brought up to date in the present book.8
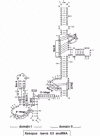
Pioneering research in the past half century has demonstrated that the nucleolus is the site of ribosome biogenesis. Initial clues came from the cytochemical studies of Caspersson9 and Brachet10 showing that the nucleolus contains RNA which they speculated might be related to cytoplasmic RNA. Other cytological studies in the same era noted that the granular component of the nucleolus11-13 looked similar to “Palade granules” (ribosomes) in the cytoplasm.14-15 Subsequent pulse-chase experiments16-18 and base composition comparisons19 proved that nucleolar RNA became stable, cytoplasmic RNA. Shortly thereafter, with the advent of the technique of RNA-DNA hybridization, it was shown in Drosophila and Xenopus that the nucleolus organizer region (the chromosomal locus associated with the nucleolus) contains the genes for ribosomal RNA (rRNA).20-23 Development of the method of in situ hybridization allowed the direct visualization by light microscopy of rRNA genes within the nucleoli of amphibia24-25 and flies.26 At the same time, Miller spreads of nucleolar DNA for electron microscopy revealed active transcription of rRNA genes.27 As shown in Figure 1a, these active transcription units resemble Christmas trees where each branch is a nascent rRNA precursor (pre-rRNA)28 bound to the trunk of the tree (DNA main axis) by a granule of RNA polymerase I.29-32 There is a tandem array of rRNA genes, each separated from the other by a non-transcribed spacer.
Internal Modifications: 2'-O-Methylation and Pseudouridylation
Either concurrently or immediately after synthesis of pre-rRNA, there are internal modifications at evolutionarily conserved regions in its 18S, 5.8S and 28S rRNA components.33-37 In Xenopus these comprise 10 base methylations, 105 2'-O-methylations (2'-O-Me) of ribose and about 100 pseudouridines (ψ); yeast pre-rRNA has approximately half as many internal modifications (see Chapters 12 and 13). The site for each modification is determined by a small nucleolar RNA (snoRNA)(see Chapter 13). The snoRNA base-pairs with the pre-rRNA, thereby appropriately positioning on the pre-rRNA substrate the enzyme which piggybacks along on the snoRNP complex. Members of the Box C/D snoRNA family guide formation of 2'-O-Me by the associated protein Nop1p/fibrillarin which appears to be the methylase.38-40 Similarly, members of the Box H/ACA snoRNA family guide ψ formation by the associated protein Cbf5p/dyskerin which is the pseudouridine synthase.41-43 It is conceivable that the base-pairing of the guide snoRNAs also plays a chaperone function in the folding of rRNA. A few snoRNAs to be discussed below are needed for cleavages in the pre-rRNA instead of, or in addition to, a role as a guide snoRNA for modification; this small group of snoRNAs is also implicated in acting in a chaperone capacity.
Addition of Ribosomal Proteins
The predominant initial pre-rRNA ranges in size from 37S (yeast) to 40S (Xenopus) to 45S (mammals). The pre-rRNA assembles with ~80 ribosomal proteins, and the sequence of events in mammals is as follow. Immuno-electron microscopy of Miller spreads revealed that some early binding ribosomal proteins associate with nascent pre-rRNA while it is being transcribed.44 Rapidly labeled 30S RNP isolated from nucleoli contains 45S pre-rRNA and seems to be a precursor to the 80S RNP particle.45-47 The 80S RNP contains 45S pre-rRNA, over two thirds of the 60S ribosomal proteins and about half of the 40S ribosomal proteins.48-53 Most of the remaining ribosomal proteins are not added until later when rRNA processing is completed. The 80S RNP is the precursor to the 55S RNP54-55 which constitutes 70 to 80% of the nucleolar population of pre-ribosomes, while 80S RNP constitutes only 10-20%.7 This may reflect the fact that cleavage of the 32S pre-rRNA intermediate, which is found in the 55S RNP, is a slow step in the kinetics of rRNA processing. Ultimately, the 55S RNP is converted into the mature 60S ribosomal subunit. Although the 80S RNP is also the precursor for the 40S ribosomal subunit, it has been harder to identify a direct precursor RNP of the 40S subunit in the nucleolus of vertebrate cells; probably it is rapidly transported to the cytoplasm.
Assembly of proteins with pre-rRNA seems to have some differences between mammals (see above) and yeast (see Chapter 12). Yeast pre-rRNA assembles with numerous ribosomal and non-ribosomal proteins (presumably needed for ribosome maturation) in a 90S particle, which is subsequently processed into 66S and 43S pre-ribosomes that form the 60S and 40S ribosomal subunits. Surprisingly, the yeast 90S precursor contains few proteins to form the 60S subunit, although it has many to form the 40S subunit.56-57 This suggests that ribosome assembly is biphasic in yeast, with formation of the 40S ribosomal subunit preceding that of the 60S subunit.
Ribosomal RNA Processing
After the addition of the 2'-O-Me and ψ internal modifications and assembly with some of the ribosomal proteins, cleavage of the rRNA precursor begins. Usually this occurs after the termination of transcription and release of the pre-rRNA. An exception to this is Dictyostelium rRNA which begins its processing during transcription, as seen by electron microscopy of Miller spreads.58
Non-Universal Processing Events in Eukaryotic Pre-rRNA
The major rRNA processing steps to be described below occur in all eukaryotes. However, a few additional cleavages are found in selected organisms. Most organisms do not have an intron in the 28S region of pre-rRNA, but when they do, splicing appears to be an early step before the standard cleavage events of rRNA processing. For example, splicing of the intron in Tetrahymena rRNA occurs during or shortly after transcription59-60 by autocatalytic self-splicing61 before further rRNA processing. In the case of Drosophila, there is polymorphism in the tandem array of rRNA genes, and only those lacking an intron are transcribed, while those containing an intron are transcriptionally silent.62-64
In some cases, certain “expansion segments” (sequences in mature rRNA that are highly variable in sequence and structure) are removed after the major steps of rRNA processing have been accomplished (reviewed by ref. 65). For example, in insects the 3'-end of 5.8S RNA is cleaved to generate a 2S fragment.66-67 Also, 28S rRNA of insects68-71 and some other lower eukaryotes such as Tetrahymena72 is cleaved near its center in “gap processing” of expansion segment 5, giving rise to 28Sa and 28Sβ. This expansion segment interrupts the binding site for a ribosomal protein (L25 in yeast). In yeast, mutation that disrupts L25 protein binding inhibits rRNA processing, and deletion of expansion segment 5 results in under-accumulation of the large subunit RNA,73 suggesting that removal of gap processing is coupled to events in canonical rRNA processing. Euglena presents an extreme example where removal of several expansion segments results in 16 discrete RNA species in the cytoplasmic ribosomes.74-75 Generally, the non-universal cleavages happen after the rest of rRNA processing, and it has been proposed that this occurs so as to not alter structures of the preceding pre-rRNA intermediates that may be required for the universal cleavages.76
Stepwise Order of the Universal Cleavage Steps
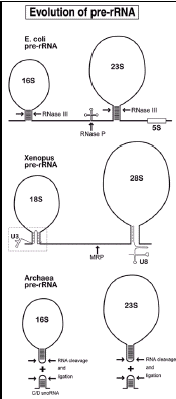
Early pulse-labelling experiments revealed that the 40-45S pre-rRNA is processed through various intermediates to form the mature rRNA species.77-79 Through a series of cleavages, the external and internal transcribed spacers (ETS, ITS) are removed from the precursor to liberate the mature 18S, 5.8S and 28S rRNAs. There is some flexibility to the order of the initial steps that ultimately result in mature 18S rRNA. As shown in Figure 1b, rRNA processing in Xenopus oocytes can follow either pathway A or pathway B. In pathway A, cleavage occurs first at site 3, separating the 5.8S and 28S rRNA coding regions in 32S pre-rRNA from the 18S rRNA coding region in 20S pre-rRNA. Pathway A is taken by HeLa cells for rRNA processing.84 Alternatively, in pathway B, cleavages at sites A0, 1 and 2 occur first to generate 18S rRNA before cleavage at site 3. Pathway B occurs in Xenopus somatic cells,85-86 mouse L cells,86 Drosophila87 and yeast (see Chapter 12). Sometimes, the pathway can even vary in the same cell type.86,88-90 In one example, a temperature-sensitive mutant of BHK cells followed pathway A at 33.5°C and pathway B at 38.5°C.88 Xenopus oocytes have either just pathway A or both pathways A and B.81 In the latter case, both pathways can co-exist in a single cell,81 indicating that the choice of pathway does not reflect some limiting component in the cell.
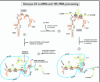
rRNA processing pathway choice seems to be influenced by U3 snoRNA.82 U3 snoRNA is the most abundant snoRNA and it is one of a few snoRNAs that are required for rRNA processing (also see Chapters 12 and 13). Chemical mutagenesis and phylogenetic comparisons of U3 snoRNA have resulted in a recently revised universal structural model that is conserved between yeast and higher organisms and consists of two domains (I and II) separated by two single-stranded “hinge” regions (fig. 2).91 A large complex containing U3 snoRNA and 28 proteins has recently been identified in yeast;95 most of the proteins are associated with domain II of U3. U3 snoRNA is a member of the Box C/D family of snoRNAs, but it functions solely in rRNA processing and not in 2'-O-methylation. The Box C/D signature motif is required for its nucleolar localization96-97 after traffic through the Cajal body.97-99 Domain II of U3 snoRNA plays a role in cleavage at site 3,81-82 but domain I and the hinge regions are also critically important for cleavages at sites A0, 1 and 2.82,91,100
The two domains of U3 appear to compete with one another for the first cleavage step in rRNA processing.82 When domain I of U3 snoRNA is removed in Xenopus oocytes, cleavage at sites A0, 1 and 2 is prevented and all U3 activity is channeled into its domain II function to cleave site 3 as the first processing step. Conversely, when domain II function is impaired, then cleavage at sites A0, 1 and 2 is preferred as the initial event. These results suggest that pathway choice is stochastic and depends upon the orientation of U3 snoRNP when it lands on the pre-rRNA substrate—if the conformation favors domain II action, then pathway A will result, and if the conformation favors domain I action, then pathway B will result. Why do some frogs have just pathway A, whereas most have pathways A + B? There are about 20 copies of the U3 snoRNA gene per genome of Xenopus, and sequence microheterogeneity exists between the gene copies.101 It could be that one variant of U3 is expressed more in frogs with just pathway A, and other U3 variants are expressed in frogs with pathways A + B.82
Formation of 18S rRNA
Although a few cleavages of pre-rRNA can be carried out in a test tube with nuclear extract, it has not yet been possible to recapitulate all the steps of rRNA processing in a sequential fashion in a cell-free system. Moreover, many ribosomal proteins are already assembled on pre-rRNA when processing occurs, yet in vitro reconstitution of rRNA with ribosomal proteins to form ribosomes has not yet been accomplished in eukaryotic systems. Therefore, much of what we know about rRNA processing mechanisms comes from in vivo studies in systems that can be experimentally manipulated—either yeast where genetic depletion and transformation with mutant forms of pre-rRNA and rRNA processing factors are possible (see Chapter 12) or Xenopus oocytes where endogenous snoRNA can be disrupted by RNase H after injection of antisense oligonucleotides, as first done for Xenopus U3 snoRNA,81 and subsequently mutated forms of the snoRNA can be injected.82
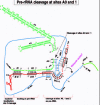
A detailed description follows of the role of U3 snoRNA in rRNA processing to form 18S rRNA, and a cartoon of the processing steps is depicted in Figure 3, with enhanced detail in the figures that follow.
snoRNA Docking on the External Transcribed Spacer
U3 snoRNP associates with pre-rRNA primarily through protein-protein interactions102 but also through base-pairing.103-104 In sedimentation ultracentifugation, U3 snoRNA is found over a broad range (ca 10S to 80S),105 suggesting its association with both the initial rRNA precursor and various pre-rRNA intermediates. Chemical modification of accessible sites suggests that most U3 snoRNP in Xenopus is free,92 in contrast to U3 snoRNP in yeast that is primarily associated with pre-rRNA.106 The association of U3 with pre-rRNA must be more stable or longer-lived than that of the other snoRNAs, as it is the only snoRNA that has been recovered by immunopreciptation of rRNA processing complexes.56-57
U3 snoRNA Hinge Base-Pairing with the ETS
In Xenopus, proper docking on pre-rRNA that will allow subsequent U3 function in processing requires base-pairing (proven by compensatory base changes) between the 3'-hinge region of U3 and complementary sequences in the 5'-ETS (hereafter called simply ETS), while base-pairing between the 5'-hinge of U3 and the ETS is auxiliary but not essential (fig. 4).107 Recall that the two hinge regions of U3 are single-stranded in the free U3 snoRNP (fig. 2) and therefore are available for base-pairing with the ETS. The 3'-hinge interaction with the ETS also has been validated in trypanosomes by cross-links111 and ETS deletions that obliterated 18S rRNA production.112 Interestingly, in trypanosomes there are two regions of the ETS (region 1b just downstream of cleavage site A' and region 3 upstream of cleavage site A0) that have been cross-linked with U3 snoRNA111 and are both complementary to the 3'-hinge of U3.112-113 Similar to the case in Xenopus, the ETS docking sites for the U3 3'-hinge are important for 18S rRNA production.113 In contrast, in budding yeast base-pairing between the 5'-hinge of U3 and the ETS is required.108 In both Xenopus and yeast, base-pairing with the ETS is possible for the 5'-hinge and the 3'-hinge, but which hinge in U3 is preferred as more important for association with the ETS varies.107 Compensatory base changes seem to have occurred during evolution, as base-pairing potential is maintained between the U3 hinges and the ETS although the sequence itself varies between distant species.91 Nonetheless, in closely related species, such as Xenopus laevis and Xenopus borealis, a few tracts of conserved sequences occur in the ETS, and these include the regions of the ETS that can base-pair with the U3 5'-hinge and 3'-hinge (tracts 5 and 3, respectively).114-115
Figure 4
U3 snoRNA docking on pre-rRNA ETS. See Figure 3 and text for details.
Nucleolin Helps U3 snoRNP to Dock on Pre-rRNA
The abundant phosphoprotein nucleolin (also called C23 or 100 kD protein) binds pre-rRNA as soon as it is transcribed116 and can be detected by electron microscopy of Miller spreads.117 Nucleolin is needed for traffic of U3 from Cajal bodies to the nucleolus98 and for the docking of U3 on the ETS.109 Interestingly, nucleolin promotes nucleic acid annealing,118-119 and therefore may facilitate the base-pairing between the U3 snoRNA hinges and the ETS. In fact, nucleolin binds to UCGA,120 located in an 11 nt (13 nt in mammals) evolutionarily conserved motif (ECM) GAUCGAUGUGG121-122 that is found in a single-stranded region of the ETS.123 This nucleolin binding site is near the ETS sequence that base-pairs with the 3'-hinge of U3 (fig. 5). The N-terminal end of nucleolin, containing multiple phosphorylation sites, is required for its interaction with U3 snoRNP, presumably through protein-protein contacts.109 The nucleolin-mediated base-pairing of the vertebrate U3 3'-hinge with the ETS is not the only contact that holds U3 snoRNP on pre-rRNA, as U3 snoRNP is still found associated with pre-rRNA in sucrose gradients of RNP particles after disruption of the U3-ETS base-pairing by mutagenesis (Borovjagin and Gerbi, unpublished observations), although U3 cannot function in rRNA processing after 3'-hinge mutation because it is not properly positioned on the pre-rRNA substrate.107 Moreover, U3 snoRNP cannot be recovered with a fragment of the ETS containing the consensus sequence and ~200 nucleotides downstream,124 suggesting that additional contacts between U3 snoRNP and pre-rRNA are needed.
Nucleolin plays still other roles in ribosomal biogenesis besides helping U3 snoRNP to dock on pre-rRNA. The central region of nucleolin has four RNA binding domains (RBD) which interact with pre-rRNA.116-117 The nucleolin recognition element (NRE) is a consensus sequence (U/G)CCCG(A/G) in a stem-loop structure with at least 4 bp in the stem and 7-14 nt in the loop.117,125 There are 33 putative NRE sites in human pre-rRNA, including 11 in the ETS.125 NMR spectroscopy suggested that two RBDs of nucleolin bind on opposite sides of the loop, forming a molecular clamp which brings the ends together to form the stem.126-127 This observation agrees with the earlier finding that nucleolin promotes annealing and the formation of helices.118-119 Thus, it may act as a chaperone to fold nascent pre-rRNA.126 Additionally, the N-terminal domain of nucleolin can interact with ribosomal proteins,128-129 and it is tempting to speculate that it may help to load them on pre-rRNA.
Besides help by nucleolin, it has been suggested from data in yeast that the protein complex of Imp3p, Imp4p and Mpp10p helps to guide or facilitate docking of U3 through the hinge-ETS base-pairing.130 Unlike most of the other proteins in the U3 snoRNP, the Imp3p, Imp4p and Mpp10p complex does not associate with domain II of U3. Instead, it has been proposed that it associates with the stem between the 5'-hinge and 3'-hinge (fig.2), since deletion (though, surprisingly, not sequence substitution) of this entire stem abolishes its binding to U3.131 The location between the hinge regions would position the complex favorably for its hypothesized role to facilitate the hinge-ETS base-pairing, but this proposal remains to be tested directly.
Cleavage at Site A' Is Not Universal
The association of U3 snoRNA with the ETS is sufficiently long-lived that it has been captured by psoralen cross-linking.111,122,132-134 In rat cells, the cross-links involved nucleotides in the area of Box A in U3 snoRNA (nt U13, C14 and U23).122 The region of the ETS cross-linked to U3 is nt 438-695 in human pre-rRNA132 and nt 767-1149 in rat pre-rRNA.122 Interestingly, this area of the ETS is cleaved in a primary processing event in mammalian pre-rRNA [cleavage at nt 415/ 422 in human,121 at nt 790/ 795 in rat122 and 651/657 in mouse121], indicated as site A' (formerly called site 0) in Figure 1b. Cleavage at site A' is dependent on U3, U14, E1 (=U17) and E3 snoRNAs and can be reproduced in a cell-free system.121,124,135 A nucleolar endoribonuclease has been described that cleaves the mouse A' site at nt 650.136-137 Although site A' cleavage is found in many organisms, it is less prevalent in Xenopus somatic cells, occurring in about 30% of the pre-rRNA, and occurs less than 1% if at all in Xenopus oocytes.80,138-139 Cleavage at site A' has not been seen in yeast pre-rRNA140 and should not be confused with site A0 cleavage in yeast pre-rRNA.141 Site A' cleavage is not an early processing event in trypanosomes112 or Xenopus.80,142 The A' cleavage site in the vertebrate ETS occurs 5-6 nt upstream of the 11 nt ECM121-122 that falls within the 28 bases found to be sufficient as a minimal substrate for A' cleavage.143 A degenerate match to the 11 nt ECM is also found in trypanosomes where it is also needed for site A' cleavage.113 As mentioned above, the 11 nt ECM binds nucleolin, and the ETS bases that pair with the U3 3'-hinge region are found just downstream of this consensus in mammalian pre-rRNA (fig. 5). In the case of Xenopus, the match to the 11 nt consensus lies further upstream,80 separated by about 200 nt from the region of the ETS that base-pairs with the U3 3'-hinge.91,107 As shown in Figure 5, the match to the ECM in Xenopus is adjacent to nt 105/106/107 where some cleavage has been observed in somatic cells.139 The significance of the cleavage at site A' is obscure, since correct maturation of the 5'-end of mammalian 18S rRNA occurs in vivo144 and in vitro145 in substrates lacking the processing sequence within the ETS.
Even though cleavage at site A' is not a prerequisite for further processing in pre-rRNA, the docking of U3 snoRNP in this area of the ETS is crucial for 18S rRNA production.91,107 A cross-link has been made between the 3' hinge of trypanosome U3 snoRNA and the ECM area of the ETS.111 Moreover, the association of U3 snoRNP with this region of the ETS correlates with the visualization by electron microscopy of terminal balls on the nascent pre-rRNA transcripts (fig.1a).95,138 The terminal balls contain fibrillarin146 and are likely to contain U3 snoRNA, although this has not been demonstrated directly. Nonetheless, depletion of U3 snoRNA or some of its associated proteins leads to loss of the terminal balls,95 consistent with the idea that terminal balls contain the U3 snoRNP complex. Since in vitro cleavage at site A' also requires U14, E1 (=U17) and E3 snoRNAs,121,124,135 it would be of interest to investigate if these other snoRNAs also are in the terminal balls. snoRNA disruption and rescue experiments in Xenopus oocytes revealed that U8 and U22 snoRNAs must be present during transcription of the nascent pre-rRNA if they are to function in subsequent processing.147 If generalized to other snoRNAs, this would suggest that snoRNAs dock on nascent pre-rRNA during its transcription and before it is released for processing, comparable to the scenario discussed above for U3 snoRNA. Since U8 snoRNA is required for 5.8S and 28S rRNA formation (see below) rather than 18S rRNA formation, it is unlikely that it will be part of the terminal balls seen by Miller spreads. Instead, U8, and perhaps the other snoRNAs, may each have their own docking sites on pre-rRNA.
Other Contacts between U3 snoRNP and the ETS
In addition to the psoralen cross-links between Box A of U3 and the ETS noted above, cross-links have also been found in mouse between nt C5, U6 and U8 of the GAC-Box A' element of U3 snoRNA and nt U1012 and U1013 of the ETS.134 Thus, sequences at the 5'-end of U3 snoRNA (GAC-Box A' and Box A) which are of functional importance for 18S rRNA production (see below) are sequestered on the nascent pre-rRNA through intimate association with the ETS. This may ensure that rRNA processing does not begin during transcription of the pre-rRNA.
The contacts of the 5'-end of U3 snoRNA with pre-rRNA change once transcription is completed and the newly synthesized pre-rRNA is released from the DNA. Concurrently, nucleolin appears to dissociate from pre-rRNA, as it is not found with 60-80S pre-ribosomal RNP particles.116,148 The exit of nucleolin may allow for a rearrangement of U3 snoRNP on its pre-rRNA substrate. It has been proposed that the U3 hinges act as an anchor on the ETS of pre-rRNA while U3 swivels into new positions.107 This anchor is reminiscent of the interaction between the tRNA anti-codon with the mRNA codon that serves to hold tRNA in place while it swivels from its entry/recognition site into the aminoacyl site on the ribosome.
Cleavage at Sites A0, 1 and 2 to Form 18S rRNA
Cleavage at sites A0, 1 (= A1 in yeast) and 2 (or the different site A2 in yeast) requires U3 snoRNA.82,100,141 In addition, cleavage at these sites require U14 snoRNA (in yeast149-150 and in Xenopus82,151), snR10152 and snR30153 in yeast and U22 snoRNA in Xenopus.154 Moreover, in higher eukaryotes, U17 snoRNA (=E1), is needed for site 1 cleavage and E2 for site 2 cleavage.155 Mechanistic details of how these snoRNA work remain unknown, and there is no evidence if they work together with U3 in a hypothetical processing particle containing several snoRNAs141,156 and dubbed the “processome”157 similar to the splicosome that contains several snRNAs for splicing, or if the snoRNAs work independently of one another (as in the newer use of the term “small subunit processome” meaning the large complex of U3 snoRNA and its associated proteins95). Normally, the cleavages at sites A0, 1 and 2 are coordinated, resulting in the production of mature 18S rRNA. However, recently it has been possible to uncouple these events in vivo by mutagenesis of U3 snoRNA in Xenopus.100 Previous studies using mini-substrates of pre-rRNA revealed that site 1 cleavage does not require the 3'-end of 18S rRNA144 and site 2 cleavage does not need the 5'-end of 18S rRNA,158 thus supporting the idea that cleavage at these sites can occur independently even though they usually occur close in time to each other in vivo.
Cleavage at Site A0
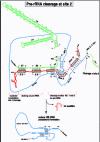
Site A0 has only recently been discovered in higher organisms.100 It reflects cleavage after A491 and A494 in the ETS and is 218 or 221 nt upstream of site 1 in Xenopus pre-rRNA,100 comparable to the position of site A0 in budding yeast (90 nt upstream of site A1)141 and trypanosomes (116 nt upstream of site A1).112 In all three organisms, site A0 is found near the base of a long stem and is opposite site 1 (or A1)(fig. 6). Due to this secondary structure, it was proposed that RNase III cleaved the stem at sites A0 and A1,160 but this idea was subsequently disproven in yeast.161
Depletion of U3 snoRNA in yeast revealed that cleavage at site A0 is U3-dependent.141 A cross-link has been made in budding yeast between U3 snoRNA and nt 655 in the yeast ETS,133 which is situated near the top of the stem that has sites A0 (nt 610) and site A1 (nt 699) at its base,162 further implicating U3 snoRNA for cleavage at sites A0 and A1. However, the portion of U3 snoRNA responsible for site A0 cleavage in yeast remains unknown, unlike the case for metazoa. Recently we showed that cleavage at site A0 in Xenopus pre-rRNA requires Box A of U3 snoRNA.100 When Box A of U3 is mutated and site A0 cleavage is inhibited, 20S pre-rRNA accumulates but is not processed into 18S rRNA.100 In this case, cleavage at sites 1 and 2 also is inhibited, suggesting that cleavage at site A0 precedes that at sites 1 and 2.100 Consistent with this idea, inhibition of cleavage at sites 1 and 2 by mutation of the GAC-box A' element in Xenopus allows cleavage at site A0; in this situation, a novel 19S pre-rRNA intermediate is found which extends from site A0 to site 3 (fig. 1b).100 Similarly, a novel 22S pre-rRNA intermediate is found in yeast when cleavage occurs at site A0 but not at sites A1 and A2.140 This circumstance can be created in yeast by mutation of Box A of U3 snoRNA,159 truncation of the U3-associated protein Mpp10p,163 or depletion of the U3-associated helicase Dhr1p.164
We suggest that Box A helps to position U3 snoRNP properly on pre-rRNA to allow U3-dependent cleavage at site A0 in Xenopus pre-rRNA. Intriguingly, bases at the 3'-end of Box A of U3 are complementary to bases 482AGAAA486 in the Xenopus ETS (fig. 6), and mutation of these bases in Xenopus U3 snoRNA inhibits site A0 cleavage.100 Site A0 has not yet been mapped in other metazoa besides Xenopus, but in trypanosomes where its position is known,112 it is also 8 nt downstream from 5 nt in the ETS that have the potential to base-pair with the same 3' portion of U3 Box A. Although this interaction cannot be drawn for yeast, it is noteworthy that mutation of U3 Box A does not inhibit site A0 cleavage in yeast,159 unlike the case in Xenopus.100 Regardless of whether this hypothesized base-pairing occurs between Box A and the ETS, Box A is positioned close to the A0 cleavage site by base-pairing of its 5'-end with a sequence near the 5'-end of 18S rRNA (fig. 6) to be discussed below.
Cleavage at Site 1
Most people believed that snoRNAs utilized for rRNA processing would base-pair with the transcribed spacers to assist in cleavage events. Therefore, it was a novel idea that Box A of U3 snoRNA might base-pair with sequences within the 18S rRNA portion of pre-rRNA,159 as depicted in Figure 6. This concept was stimulated by the observations that the snoRNAs that guide modifications base-pair with sequences within 18S and 28S of the pre-rRNA. It was proposed that the 5'-end of U3 Box A base-pairs with sequences in the loop of stem adjacent to the 5'-end of 18S rRNA while it is part of the rRNA precursor.159 In yeast, this interaction has been supported by relative sensitivities to chemical modification106 and has been proven by compensatory base changes.110 Furthermore, it was hypothesized that the 3'-end of U3 Box A base-pairs with internal sequences in the 18S coding region of pre-rRNA.159 In mature 18S rRNA, these internal sequences base-pair with the 5'-proximal loop to form a pseudoknot (fig. 7), and it was suggested that U3 Box A base-pairs with both of these regions to prevent premature pseudoknot formation.159 Thus, U3 snoRNA would act as a chaperone and the pseudoknot would not form until U3 snoRNA departs at the end of rRNA processing. Although the proposed interactions of U3 Box A with 18S rRNA in the rRNA precursor are evolutionarily conserved,159 compensatory mutations failed to prove the U3 base-pairing with the internal sequences in 18S.110 One possibility is that the 3'-end of U3 Box A might undergo a molecular switch in its pairing partners—pairing first with sequences in the ETS just upstream of the A0 cleavage site and subsequently pairing with the 18S internal sequences (fig. 5). In such a case, it would be necessary to create compensatory mutations at all three sites to restore 18S rRNA formation. Alternatively, base-pairing of the 5'-end of Box A with the experimentally proven 5'-proximal loop in the 18S region of the pre-rRNA would be sufficient to block premature pseudoknot formation, and the interaction of the 3'-end of U3 Box A with internal sequences in 18S might not exist. Additional experiments with compensatory mutations are needed to test these possibilities and to validate if they are applicable to higher eukaryotes.
The position of U3 Box A near site 1 is consistent with the observation in yeast that Box A is needed for site A1 cleavage.159 In Xenopus, mutation of U3 Box A inhibits cleavage at site A0 and the subsequent cleavages at sites 1 and 2,100 so it cannot be determined if the role of Box A on site 1 cleavage is direct or indirect for higher organisms. In addition, the GAC-Box A' element of Xenopus U3 snoRNA is required for cleavage at site 1.100 A novel 19S pre-rRNA is found, extending from site A0 to site 3, when cleavage at sites 1 and 2 are prevented by mutation in Xenopus GAC-Box A'.100 The sequences of Xenopus U3 Box A' are complementary to a region just downstream of the 5'-proximal stem of 18S rRNA (fig.5), and base-pairing would nicely position U3 Box A' close to site 1 where it is required for cleavage.100 Base-pairing between the GAC element of U3 snoRNA and 18S rRNA that has been hypothesized for yeast159 cannot be drawn for Xenopus, and the molecular interaction involving the GAC element, which is required for site 1 cleavage in metazoa, remains to be determined in higher eukaryotes.
The mechanism by which U3 snoRNA acts for cleavage at site 1 (and at other U3-dependent sites) is not yet clear. One possibility is that it is an RNA-based cleavage, with U3 snoRNA acting as a ribozyme. Another possibility is that a protein responsible for the cleavage is associated with U3, analogous to modification enzymes that piggyback with the guide snoRNA to be correctly positioned for action. A variant of this possibility is that the endoribonuclease may use U3 snoRNP as a landing pad. An appealing candidate for the latter is Rcl1p which associates transiently with U3 snoRNP in yeast,165 perhaps through association with the G protein Bms1p.166-167 Rcl1p is a member of the 3'-phosphate cyclase family that catalyze formation of a 2',3'-cyclic phosphodiester. This is extremely intriguing when considering the earlier report that in vitro cleavage at site 1 generates a 2',3'-cyclic phosphate as a first step and trimmed 3 nucleotides as a second step to generate the mature 5'-end of 18S rRNA.145,168
It is apparent from the model in Figure 6 that spacing between various functional elements in U3 snoRNA and in its pre-rRNA substrate should be critical to allow the hand and glove fit between U3 snoRNA and the pre-rRNA. Consistent with this, a specified distance of the 5'-proximal stem to site A1 is required for site A1 cleavage in yeast.169-170 Similarly, in Xenopus the distance between the 5'-hinge and 3'-hinge, between Box A and the 5'-hinge and the distance between domain I and domain II of U3 snoRNA is extremely important for 18S rRNA production.82 This suggests that the hinge-ETS base-pairing might be maintained while GAC-Box A' and Box A base-pair with sequences in the 18S region of pre-rRNA.82
Cleavage at Site 2
The 3'-end of Box A' and flanking nucleotides in Xenopus U3 snoRNA are required for cleavage at site 2,100 thereby liberating 18S rRNA from pre-rRNA. Mutation of individual or multiple bases within nt 11-14 of U3 snoRNA inhibits site 2 cleavage and results in a novel 18.5S rRNA intermediate that extends from site 1 to site 3.100 Thus, although cleavage at sites A0, 1 and 2 usually occur together, they can be temporally separated with cleavage occurring at site A0 before 1, and at site 1 before 2.
We have proposed that bases in the Box A' region of U3 snoRNA might switch base-pairing partners, replacing pairing near the 5'-proximal stem of 18S by pairing with complementary bases near the 3'-end of 18S rRNA within pre-rRNA (fig. 7).100 Interestingly, this area of U3 snoRNA contains ψ at nt 8 and at nt 12; ψ has been found in sequences involved in molecular switches such as in the spliceosome. A U3-associated helicase, such as Dhr1p identified in yeast,164 could facilitate such a switch in base-pairing partners. The proposed base-pairing would position U3 Box A' close to site 2 where it is required for cleavage (fig. 7). Moreover, insertions between U3 Box A' and Box A inhibits cleavage at both sites 1 and 2,100 suggesting that the interactions between these boxes and pre-rRNA may occur simultaneously. Thus, if U3 Box A pairs with the 5'-proximal loop of 18S while U3 Box A' pairs with sequences near the 3'-end of 18S, U3 could act as a molecular bridge to draw together the 5' and 3'-ends of 18S rRNA,100 suggesting yet another chaperone role for U3 snoRNA to fold 18S rRNA into its mature conformation.
Base-pairing between U3 Box A' and the 3'-end of 18S rRNA cannot be found in yeast, but in yeast cleavage at site D to form the 3'-end of 18S rRNA occurs in the cytoplasm,171 presumably in a U3-independent manner. The cis-acting sequences needed for site D cleavage in yeast are in close proximity to the site.172 A somewhat greater number of nucleotides (60 nt of 18S and 553 nt of ITS1) are needed at the 18S rRNA/ITS1 boundary for site 2 cleavage as studied in minigenes transfected into mouse cells.173 Furthermore, the endonucleolytic cleavage at site 2 has been carried out in a cell free system, and resulted in cleavage at the mature 3'-end of 18S rRNA and at a site ~55 nt downstream.174 These cleavages correspond to those detected by S1 nuclease after transfection of mouse rDNA in hamster cells.158 The role played by U3 and other snoRNAs (U14, U22, E2) for site 2 cleavage remains to be elucidated. It is intriguing that vertebrate U13 snoRNA105 is also complementary to sequences at the 3'-end of 18S rRNA,175 but whether U13 plays a role in site 2 cleavage remains to be investigated.
Formation of 5.8S and 28S rRNA
Cleavage at Site 3
Although site 3 is generally shown abutting the 5'-end of 5.8S RNA in maps of pre-rRNA from higher organisms, in fact cleavage seems to occur somewhat upstream (fig. 1b). S1 nuclease mapping demonstrated that site 3 is 161-163 nt upstream of the 5'-end of rat 5.8S RNA.176 Similarly, site 3 cleavage in Xenopus pre-rRNA occurs about 100 nt before the 3'-end of ITS1,100,154 refining earlier results.177 Xenopus 20S, 19S and 18.5S pre-rRNAs all share the same 3' terminus located ~100 nt before the 3'-end of ITS1.100 The data are consistent with a model where endonucleolytic cleavage occurs at this position and subsequent trimming by a 5'-exonuclease results in the true 5'-end of 5.8S RNA indicated as site 3' in Figure 1b, comparable to Rat1p and/or Xrn1p exonuclease trimming in yeast (see Chapter 16). Site 3' is found as the 5'-end of 32S pre-rRNA in mouse,178 rat176 and Xenopus,154 but the presumed exonucleolytic degradation of the ITS1 tail between sites 3 and 3' seems by be slowed down when U22 snoRNA is disrupted.154
Cleavage at site 3 is dependent on several snoRNAs. The first one to be discovered was U3 snoRNA, where its disruption decreased (but did not eliminate) cleavage at site 3 and consequently there were reduced levels of 20S and 32S pre-rRNAs.81-82 The effect is dependent on U3 snoRNA, since efficient site 3 cleavage can be rescued by injection of U3 snoRNA.82 Domain II of U3 snoRNA is sufficient for its role in site 3 cleavage.82 As discussed above, U3 snoRNA is required for 18S rRNA formation and does not play a direct role in 28S rRNA production. This raises the possibility that U3-dependent site 3 cleavage in Xenopus may be analogous to U3-dependent site A2 cleavage in yeast which occurs within ITS1 and is part of the processing pathway to form 18S rRNA.141 In addition, E3 snoRNA is required for site 3 cleavage.155 Moreover, U8 snoRNA is required for site 3 cleavage in Xenopus pre-rRNA.177,179 In contrast to U3 snoRNA, U8 snoRNA is required for 5.8S and 28S rRNA formation but not for 18S rRNA.177,179
Because of its apparent link between the 18S and 28S rRNA processing pathways in Xenopus, it is plausible that site 3 in higher organisms is a composite of sites A2 and A3 in yeast (fig. 1b). Site A3 is downstream of site A2 in the yeast ITS1, and cleavage at site A3 requires the MRP snoRNA in vivo180-182 and in vitro.183 Endonucleolytic cleavage at site A3 is followed by 5'-exonucleolytic digestion of 76 nt of ITS1 until the 5'-end of a short form of 5.8S RNA which predominates in wild type yeast. However, when MRP is depleted in yeast, the end point of exonucleolytic digestion differs and a longer form of 5.8S RNA predominates, which has 7 nt more at its 5'-end.180-182 Long and short forms of 5.8S RNA also exist in vertebrates, differing by 6-7 nt at the 5'-end with the short form predominating178 just as in yeast. Although MRP snoRNA exists in higher organisms, efforts by several groups to disrupt it by injection of antisense oligonucleotides into Xenopus oocytes have been unsuccessful, and thus its putative function for site 3 cleavage has not yet been established for metazoan systems.
Consistent with the hypothesis that site 3 might link the 18S and 28S rRNA processing pathways in higher organisms, these two pathways are linked in yeast by the protein Rrp5p184-188 whose dual function is reflected in its bipartite structure (see Chapter 12). In yeast, Rrp5p genetically interacts with snR10,189 but the homologous snoRNA has not yet been identified in metazoa.
U8 snoRNA Is Required for 5.8S and 28S rRNA Formation
Formation of 5.8S and 28S rRNA is impaired by disruption of U8 snoRNA in Xenopus oocytes.177 This is the only snoRNA found so far that is needed for the 28S rRNA processing pathway, and yeast seems to lack a homologous snoRNA. Like U3 snoRNA, U8 snoRNA also is required for cleavage at several sites in pre-rRNA—specifically, sites 3, 4, 4', 5 and T1.177 The 5' domain of U8 snoRNA is important for its function in rRNA processing.179
The first of the U8-dependent cleavages is at site T1, and it occurs shortly after cleavage at site A' in mammals.192 rRNA transcription terminates at site T2 (or even further downstream at site T3 in Xenopus193) and is rapidly processed to site T1 at the 3'-end of the 28S rRNA coding region in pre-rRNA. The sequence between sites T1 and T2 is called the 3'-ETS and ranges in length from 210 nt in yeast194-195 to 235 nt in Xenopus193 to 565 nt in mouse.196-197 The initial step in 3'-end formation of 28S rRNA in mouse involves removal of 10 nt just upstream of site T2197 in pre-rRNA prior to subsequent events that result in T1 as the 3'-terminus. 3'-end processing has been carried out in vitro with yeast cell extracts and synthetic rRNA substrate, and micrococcal nuclease sensitive RNA is not required.198 However, recall that yeast lacks U8 snoRNA.
The next U8-dependent cleavage occurs at site 3 near the end of ITS1 to form 32S pre-rRNA which is a long-lived intermediate. 32S pre-rRNA then undergoes cleavages to form 5.8S and 28S rRNA. U8 snoRNA is needed to form not only the 5'-end but also the 3'-end of 5.8S RNA. It is well documented in many organisms that cleavage within ITS2 (site 4' in fig. 1b) produces a precursor of 5.8S RNA that is longer at its 3'-end (yeast,199 Drosophila,200 mouse,90,178 rat201-203). This 5.8S precursor is 12S in size in Xenopus oocytes and contains ca. 200 nt of ITS2.177 Cleavage at site 4' fails to happen when U8 snoRNA is disrupted,177 and it is possible that failure to observe cleavage at sites 4 and 5 reflects inhibition of cleavage at site 4' when U8 snoRNA is not present. Sites 4 and 5 may be formed by exonucleolytic trimming away from site 4' as in yeast rRNA processing (see Chapter 12), or by subsequent endonucleolytic cleavages at sites 4 and 5. Many of these exonucleases have been identified in yeast and include the exosome and additional proteins to form the 3'-end of 5.8S RNA (see Chapter 12) and Rat1p and/or Xrn1p to form the 5'-end of 28S rRNA.204 Up to 502 nt 28S rRNA sequences at its 5'-end are needed for site 5 cleavage, as studied in mammalian rDNA minigenes.173 This span of 28S sequence includes areas complementary to both the 5' and 3'-ends of 5.8S RNA, but truncating the 28S further to only 217 nt abolishes cleavage at site 5.173 Moreover, when a stable 2'-O-methyl oligoribonucleotide complementary to the 3'-end of 5.8S RNA is injected into Xenopus oocytes, production of 28S rRNA is inhibited.147 Both of these results suggest that pairing between 5.8S and 28S rRNA may be required for cleavage at site 5 for rRNA processing. Surprisingly, and in contrast to this idea, site 5 cleavage occurs in mammalian rDNA minigenes in the absence of 5.8S sequence.173 However, up to 326 nt of mammalian ITS2 is required,173 and since this probably includes site 4', it suggests that site 5 cleavage may be dependent upon site 4'. The 5'-end of mammalian nuclear 28S rRNA is heterogeneous, extending a few nucleotides beyond the mature 5'-terminus.178,205
E. coli pre-rRNA lacks an ITS2, and the 5'-end of 23S rRNA is homologous to 5.8S RNA.76,206-207 Thus, the ITS2 appears to be an insertion that breaks the large subunit RNA into two pieces (5.8S and 28S rRNA). Ultimately, both ends of 5.8S RNA base-pair with the 5' area of 28S rRNA,208-210 thereby holding these two fragments together in eukaryotes after the ITS2 has been removed by processing. It has been proposed that U8 snoRNA acts as a chaperone to prevent premature base-pairing between 5.8S and 28S rRNA,190 as diagrammed in Figure 8. In this model,1900 0the 5'-end of U8 base-pairs with the 5'-end of 28S rRNA, preventing this region from base-pairing with 5.8S RNA. The potential base-pairing between U8 snoRNA and 28S rRNA is necessary but not sufficient for rRNA processing, and human-Xenopus chimeric U8 constructs indicate that sequences beyond the 5'-end of U8 snoRNA also appear to be necessary for processing.190 An evolutionarily conserved bulge in 28S rRNA that is not paired with U8 might act as a nucleation site for initiation of base-pairing with the 3'-end of 5.8S RNA.190 This model remains to be proven by compensatory base changes, but, if correct, it would position U8 very close to cleavage sites 4 and 5 which are dependent on U8.
Evolution of rRNA Processing
Pre-rRNA is found in all three kingdoms of life. Analogy exists between these pre-rRNAs. For example, RNase P is used to remove the intergenic tRNA from bacterial pre-rRNA, and its close relative, MRP snoRNA, is required for cleavage at site A3 in the yeast ITS1.211 There are other structural similarities between pre-rRNAs from the various kingdoms (fig. 9). In eubacteria, exemplified by E. coli, 16S and 23S rRNA each reside at the top of long base-paired stems where processing begins by RNase III cleavage.212-213 The two stems are also found in pre-rRNA from archaebacteria,216 but are considerably shorter in yeast217 and do not exist in Xenopus pre-rRNA. It has been hypothesized that base-pairing between sequences in cis in eubacterial pre-rRNA has been replaced in eukaryotes by base-pairing in trans between snoRNAs and the termini of the rRNA coding regions in pre-rRNA.83,211,214 As discussed above, U3 snoRNA might act as a molecular bridge to bring together the two ends of the 18S coding region in pre-rRNA.100 Similarly, U8 snoRNA can base-pair with the 5'-end of the 28S coding region in pre-rRNA.190 Recently it was reported that cleavage and ligation of the stem below 16S rRNA in archaebacteria results in the formation of a Box C/D-like snoRNA from the base of the stem and a covalently closed circular 16S rRNA.215 Similarly, a small RNA, though lacking snoRNA motifs, is produced from the base of the stem leading to 23S rRNA in archaebacteria. Thus, in two kingdoms Box C/D snoRNAs are implicated in bringing together the 5' and 3'-ends of rRNA in the precursor.
It is not yet clear why all three kingdoms bother to have pre-rRNA, rather than just transcribing the mature forms of rRNA. The transcribed spacers seem to have some role. In fact, deletion of ITS2 is lethal in yeast.218 We propose that the transcribed spacers are required for the proper folding of rRNA. In eukaryotes they are the docking site for snoRNAs like U3 that fulfill chaperone functions. In Schizosaccharomyces pombe they are also the binding site for various polypeptides that are part of the “Ribosome Assembly Complex” (“RAC”)219-220 that might be involved in assembly and folding of the pre-ribosome before cleavages begin, reminiscent of the binding and putative function of nucleolin. RAC protein also directs the removal of the 3' ETS in S. pombe.221 In archaebacteria, transcribed spacers in pre-rRNA are used instead of U3 snoRNA to base-pair with 16S rRNA and prevent premature pseudoknot formation.222
Several questions remain. Do the snoRNAs that guide modifications in pre-rRNA also act as chaperones for rRNA folding? Do the snoRNAs required for rRNA processing simply fold the rRNA precursor appropriately so that exposed regions are cleaved by exogenous nucleases? Or, do the nucleases piggyback along with the snoRNAs (or use the snoRNAs as a landing pad), analogous to the modification enzymes that piggyback along with the guide snoRNAs? Is there a required order for folding rRNA in the precursor, suggesting that snoRNAs should act as an ordered series? What are the roles in ribosome biogenesis of the numerous nucleolar proteins identified by proteomics, some of which are associated with the pre-ribosome?56-57,223-227 Why do some proteins that affect ribosome biogenesis also affect the cell cycle228-234 including DNA replication?235,236
The ribosome appears to function through a series of conformational changes. Thus, it is crucial that its RNA component be properly folded—a task that appears to be carried out by snoRNAs, ribosomal proteins and other proteins. Successful completion of the various folding steps may be demarcated by cleavage events to prevent the folding reaction from running in reverse. In this view, rRNA processing exists to ensure correct folding of an amazing macomolecular machine, the ribosome.
Acknowledgements
This chapter is dedicated to James T. McIlwain upon his retirement, with respect, affection and gratitude for his continuous support and forbearance. We thank Elaine Butler for her outstanding help with the references, and NIH for grants GM20261 (previously) and GM61945 (currently) supporting our studies on rRNA.
References
- 1.
- Fontana F. Traité sur le vénim de la vipère. Florence.
- 2.
- Franke WW. Matthias Jacob Schleiden and the definition of the cell nucleus. Eur J Cell Biol. 1988;47:145–156. [PubMed: 3072200]
- 3.
- Gerbi SA, Lange TS. About the cover. Mol Biol Cell. 1998;9cover
- 4.
- Montgomery TH. Comparative cytological studies with special regard to the morphology of the nucleolus. J Morphol. 1898;15:265–560.
- 5.
- Busch H, Smetana K. The Nucleolus. New York: Academic Press. 1970
- 6.
- Jordan EG, Cullis CA. The Nucleolus. Cambridge: Cambridge University Press. 1982
- 7.
- Hadjiolov AA. The Nucleolus and Ribosome Biogenesis. Vienna: Springer-Verlag KG. 1985 .
- 8.
- Olson MOJ.ed.The NucleolusAustin: Landes Bioscience/Eurekah.com,2004 .
- 9.
- Caspersson TO. Cell Growth and Function. New York: WW Norton and Co. 1950. p. 185.
- 10.
- Brachet J. 1957. Biochemical Cytology. New York: Academic Press; p. 535.
- 11.
- Porter KR. Electron microscopy of basophilic components of cytoplasm. J Histochem Cytochem. 1954;2:346–371. [PubMed: 13192325]
- 12.
- Gall JG. Small granules in the amphibian oocyte nucleus and their relationship to RNA. J Biophys Biochem Cytol. 1956;2(suppl):393–395. [PMC free article: PMC2229703] [PubMed: 13357575]
- 13.
- Swift H. Studies on nuclear fine structure. Brookhaven Symp Biol. 1959;12:134–152. [PubMed: 13836127]
- 14.
- Palade GE. A small particulate component of the cytoplasm. J Biophys Biochem Cytol. 1955;1:59–68. [PMC free article: PMC2223592] [PubMed: 14381428]
- 15.
- Palade GE, Siekevitz P. Liver microsomes. J Biophys Biochem Cytol. 1956;2:171–200. [PMC free article: PMC2223971] [PubMed: 13319380]
- 16.
- Woods PS, Taylor JH. Studies of ribonucleic acid metabolism with tritium-labeled cytidine. Lab Invest. 1959;8:309–318. [PubMed: 13621629]
- 17.
- Perry RP, Hell A, Errera M. The role of the nucleolus in ribonucleic acid and protein synthesis. I. Incorporation of cytidine into normal and nucleolar inactivated HeLa cells. Biochim Biophys Acta. 1961;49:47–57. [PubMed: 13734590]
- 18.
- Perry RP. The cellular sites of ribosomal and 4S RNA. Proc Nat Acad Sci. 1962;48:2179–2186. [PMC free article: PMC221141] [PubMed: 16591032]
- 19.
- Edström JE, Grampp W, Schor N. The intracellular distribution and heterogeneity of ribonucleic acid in starfish oocytes. J Biophys Biochem Cytol. 1961;11:549–557. [PMC free article: PMC2225141] [PubMed: 13889253]
- 20.
- Ritossa F, Spiegelman S. Localization of DNA complementary to rRNA in the nucleolus organizer region of Drosophila melanogaster. Proc Nat Acad Sci. 1965;53:737–745. [PMC free article: PMC221060] [PubMed: 14324529]
- 21.
- Birnstiel ML, Wallace H, Sirlin JL. et al. Localization of the ribosomal DNA complements in the nucleolar organizer region of Xenopus laevis. Natl Cancer Inst Monogr. 1966;23:431–448. [PubMed: 5963987]
- 22.
- Ritossa F, Atwood K, Lindsley D. et al. On the chromosomal distribution of DNA complementary to ribosomal and soluble RNA. Natl Cancer Inst Monogr. 1966;23:449–472. [PubMed: 5963988]
- 23.
- Wallace HR, Birnstiel ML. Ribosomal cistrons and the nucleolar organizer. Biochim Biophys Acta. 1966;114:296–310. [PubMed: 5943882]
- 24.
- Gall JG, Pardue ML. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Nat Acad Sci. 1969;63:378–383. [PMC free article: PMC223575] [PubMed: 4895535]
- 25.
- John HA, Birnstiel ML, Jones KW. RNA-DNA hybrids at the cytological level. Nature. 1969;223:582–587. [PubMed: 5799530]
- 26.
- Pardue ML, Gerbi SA, Eckhardt RA. et al. Cytological localization of DNA complementary to rRNA in polytene chromosomes of Diptera. Chromosoma. 1970;29:269–290. [PubMed: 4908207]
- 27.
- Miller Jr. OL, Beatty BR. Visualization of nucleolar genes. Science. 1969;164:955–957. [PubMed: 5813982]
- 28.
- Angelier N, Hemon D, Bouteille M. Mechanisms of transcription in nucleoli of amphibian oocytes as visualized by high resolution autoradiography. J Cell Biol. 1979;80:277–290. [PMC free article: PMC2110331] [PubMed: 457746]
- 29.
- Miller Jr. OL, Hamkalo BA. Electron microscopy of active genes. FEBS Symp. 1972a;23:367–378.
- 30.
- Miller Jr. OL, Hamkalo BA. Visualization of RNA synthesis on chromosomes. Int Rev Cytol. 1972b;33:1–25. [PubMed: 4562602]
- 31.
- Franke WW, Scheer U, Spring H. et al. Organization of nucleolar chromatinIn: Busch H, ed.The Cell NucleusNew York: Academic Press,197949–95.
- 32.
- Franke WW, Scheer U, Spring H. et al. Morphology of transcriptional units of rDNA. Exp Cell Res. 1976;100:233–244. [PubMed: 945768]
- 33.
- Khan MSN, Salim M, Maden BEH. Extensive homologies between the methylated nucleotide sequences in several vertebrate rRNAs. Biochem J. 1978;169:531–542. [PMC free article: PMC1183826] [PubMed: 417718]
- 34.
- Brand RC, Gerbi SA. Fine structure of ribosomal RNA. II. Distribution of methylated sequences within Xenopus laevis rRNA. Nucleic Acids Res. 1979;7:1497–1511. [PMC free article: PMC342323] [PubMed: 388355]
- 35.
- Atmadja J, Brimacombe R, Maden BEH. Xenopus laevis 18S ribosomal RNA: Experimental determination of secondary structure elements and locations of methyl groups in the secondary structure model. Nucleic Acids Res. 1984;12:2649–2667. [PMC free article: PMC318697] [PubMed: 6424099]
- 36.
- Maden BEH. Identification of the locations of the methyl groups in 18S ribosomal RNA from Xenopus laevis and man. J Mol Biol. 1986;189:681–699. [PubMed: 3783688]
- 37.
- Maden BEH. Location of methyl groups in 28S rRNA of Xenopus laevis and man: Clustering in the conserved core of the molecule. J Mol Biol. 1988;201:289–314. [PubMed: 3418702]
- 38.
- Niewmierzycka A, Clarke S. S-adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J Biol Chem. 1999;274:814–824. [PubMed: 9873020]
- 39.
- Wang H, Boisvert D, Kim KK. et al. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 _solution. EMBO J. 2000;19:317–323. [PMC free article: PMC305568] [PubMed: 10654930]
- 40.
- Galardi S, Fatica A, Bachi A. et al. Purified box C/D snoRNPs are able to reproduce site-specific 2'-O-methylation of target RNA in vitro. Mol Cell Biol. 2002;22:6663–6668. [PMC free article: PMC134041] [PubMed: 12215523]
- 41.
- Lafontaine DLJ, Bousquet-Antonelli C, Henry Y. et al. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. [PMC free article: PMC316522] [PubMed: 9472021]
- 42.
- Zebarjadian Y, King T, Fournier MJ. et al. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–7472. [PMC free article: PMC84741] [PubMed: 10523634]
- 43.
- Hoang C, Ferre-D'Amaré AR. Co-crystal structure of a tRNA ψ55 pseudouridine synthase: A nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. [PubMed: 11779468]
- 44.
- Chooi WY, Leiby KR. An electron microscopic method for localization of ribosomal proteins during transcription of ribosomal DNA: A method for studying protein assembly. Proc Nat Acad Sci. 1981;78:4823–4827. [PMC free article: PMC320262] [PubMed: 6795622]
- 45.
- Bachellerie JP, Martin-Prevel C, Zalta J. Cinétique de l'incorporation d'uridine (3H) dans les fractions subnucleolaires de cellules d'hepatome ascitique du rat. Biochimie. 1971;53:383–389. [PubMed: 4327552]
- 46.
- Bachellerie JP, Nicoloso M, Zalta JP. Early nucleolar preribosomal RNA-protein in mammalian cells. Eur J Biochem. 1975;55:119–129. [PubMed: 170095]
- 47.
- Rodrigues-Pousada C, Cyrne ML, Hayes D. Characterization of preribosomal ribonucleoprotein particles from Tetrahymena pyriformis. Eur J Biochem. 1979;102:389–397. [PubMed: 118876]
- 48.
- Kuter DJ, Rodgers A. The protein composition of HeLa ribosomal subunits and nucleolar precursor particles. Exp Cell Res. 1976;102:205–212. [PubMed: 976341]
- 49.
- Auger-Buendia M-A, Longuet M. Characterization of proteins from nucleolar preribosomes of mouse leukaemia cells by two-dimensional polyacrylamide gel electrophoresis. Eur J Biochem. 1978;85:105–114. [PubMed: 639809]
- 50.
- Auger-Buendia M-A, Longuet M, Tavitian A. Kinetic studies on ribosomal protein assembly in preribosomal particles and ribosomal subunits of mammalian cells. Biochim Biophys Acta. 1979;563:113–128. [PubMed: 497202]
- 51.
- Fujisawa T, Imai K, Tanaka V. et al. Studies on the protein components of 110S and total ribonucleoprotein particles of rat liver. J Biochem (Tokyo). 1979;85:277–286. [PubMed: 762048]
- 52.
- Lastick SM. The assembly of ribosomes in HeLa cell nucleoli. Eur J Biochem. 1980;113:175–182. [PubMed: 7460944]
- 53.
- Todorov I, Noll F, Hadjiolov AA. The sequential addition of ribosomal proteins during the formation of the small ribosomal subunit in Friend erythroleukemia cells. Eur J Biochem. 1983;131:271–275. [PubMed: 6572588]
- 54.
- Craig NC. Ribosomal RNA synthesis in eucaryotes and its regulation. MTP Int Rev Sci Ser 1 Biochem. 1974;6:255–288.
- 55.
- Warner JR. The assembly of ribosomes in eucaryotesIn: Nomura M, Tissières A, Lengyel P, eds.RibosomesCold Spring Harbor: Cold Spring Harbor Laboratory Press,1974461–488.
- 56.
- Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. [PubMed: 12067653]
- 57.
- Grandi P, Rybin V, Baβler J. et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell. 2002;10:105–115. [PubMed: 12150911]
- 58.
- Grainger RM, Maizels N. Dictysostelium ribosomal RNA is processed during transcription. Cell. 1980;20:619–623. [PubMed: 7418001]
- 59.
- Cech TR, Rio DC. Localization of transcribed regions on extrachromosomal rRNA genes of Tetrahymena thermophila by R-loop mapping. Proc Nat Acad Sci. 1979;76:5051–5055. [PMC free article: PMC413077] [PubMed: 291921]
- 60.
- Din N, Keffenberger W, Eckert W. The intervening sequence in the 26S rRNA coding region of T. thermophila is transcribed within the largest stable precursor for rRNA. Cell. 1979;18:525–532. [PubMed: 498282]
- 61.
- Kruger K, Grabowski PJ, Zaug AJ. et al. Self-splicing RNA: Autoexcision and autocyclization of the rRNA intervening sequences of Tetrahymena. Cell. 1982;31:147–157. [PubMed: 6297745]
- 62.
- Long EO, Dawid IB. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell. 1979;18:1185–1196. [PubMed: 117903]
- 63.
- Jolly DJ, Thomas CA. Nuclear RNA transcripts from Drosophila melanogaster ribosomal RNA genes containing introns. Nucleic Acids Res. 1980;8:67–84. [PMC free article: PMC327243] [PubMed: 6243780]
- 64.
- Kidd SJ, Glover DM. Drosophila melanogaster ribosomal DNA containing type II insertions is variably transcribed in different strains and tissues. J Mol Biol. 1981;151:645–662. [PubMed: 6276565]
- 65.
- Gerbi SA. Expansion segments: Regions of variable size that interrupt the universal core secondary structure of ribosomal RNAIn: Zimmermann RA, Dahlberg AE, eds.Ribosomal RNA—Structure, Evolution, Processing and Function in Protein BiosynthesisBoca Raton: CRC Press,199671–87.
- 66.
- Pavlakis GN, Jordan BR, Wurst RM. et al. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and the processing site between them. Nucleic Acids Res. 1979;7:2213–2238. [PMC free article: PMC342381] [PubMed: 118436]
- 67.
- Jordan BR, Latil-Damotte M, Jourdan R. Coding and spacer sequences in the 5.8S-2S region of Sciara coprophila ribosomal DNA. Nucleic Acids Res. 1980;8:67–84. [PMC free article: PMC324175] [PubMed: 7433100]
- 68.
- Delanversin G, Jacq B. Séquence de la région de la coupure centrale du précurseur de l'ARN ribosomique 26S de Drosophile. CR Acad Sci Ser III. 1983;296:1041–1044. [PubMed: 6313147]
- 69.
- Ware VC, Renkawitz R, Gerbi SA. rRNA processing: removal of only nineteen bases at the gap between 28Sa and 28Sβ rRNAs in Sciara coprophila. Nucleic Acids Res. 1985;13:3581–3597. [PMC free article: PMC341260] [PubMed: 2989775]
- 70.
- Fujiwara H, Ishikawa H. Molecular mechanisms of introduction of the hidden break into the 28S rRNA of insects: Implications based on structural studies. Nucleic Acids Res. 1986;14:6393–6401. [PMC free article: PMC311653] [PubMed: 3018670]
- 71.
- Delanversin G, Jacq B. Sequence and secondary structure of the central domain of Drosophila 26S rRNA: A universal model for the central domain of the large rRNA containing the region in which the central break may happen. J Mol Evol. 1989;28:403–417. [PubMed: 2501502]
- 72.
- Engberg J, Nielsen H, Lenaers G. et al. Comparison of primary and secondary 26S rRNA structures in two Tetrahymena species: Evidence for a strong evolutionary and structural constraint in expansion segments. J Mol Evol. 1990;30:514–521. [PubMed: 2115930]
- 73.
- van BeekveltCA, Kooi EA, de Graaff-Vincent M. et al. Domain III of Saccharomyces cerevisiae 25S ribosomal RNA: Its role in binding of ribosomal protein L25 and 60S subunit formation. J Mol Biol. 2000;296:7–17. [PubMed: 10656814]
- 74.
- Schnare MN, Gray MW. Sixteen discrete RNA components in the cytoplasmic ribosomes of Euglena gracilis. J Mol Biol. 1990;215:73–83. [PubMed: 2118960]
- 75.
- Schnare MN, Cook JR, Gray MW. Fourteen internal transcribed spacers in the circular ribosomal DNA of Euglena gracilis. J Mol Biol. 1990;215:85–91. [PubMed: 2118961]
- 76.
- Clark CG, Gerbi SA. Ribosomal RNA evolution by fragmentation of the 23S progenitor: Maturation pathway parallels evolutionary emergence. J Mol Evol. 1982;18:329–336. [PubMed: 7120427]
- 77.
- Scherrer K, Latham H, Darnell JE. Demonstration of an unstable RNA and of precursor to ribosomal RNA in HeLa cells. Biochemistry. 1963;49:240–248. [PMC free article: PMC299789] [PubMed: 13991616]
- 78.
- Warner JR, Girard M, Latham H. et al. Ribosome formation in HeLa cells in the absence of protein synthesis. J Mol Biol. 1966;19:373–382. [PubMed: 5969072]
- 79.
- Udem SA, Warner JR. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972;65:227–242. [PubMed: 4557192]
- 80.
- Savino R, Gerbi SA. Preribosomal RNA processing in Xenopus oocytes does not include cleavage within the external transcribed spacer as an early step Biochimie 199173805–812.[Erratum in Biochimie 1996 78295] [PubMed: 1764525]
- 81.
- Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. [PMC free article: PMC551956] [PubMed: 2357971]
- 82.
- Borovjagin AV, Gerbi SA. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J Mol Biol. 1999;286:1347–1363. [PubMed: 10064702]
- 83.
- Gerbi SA, Borovjagin AV, Ezrokhi M. et al. Ribosome biogenesis: Role of small nucleolar RNA in maturation of eukaryotic rRNA. Cold Spring Harbor Symp Quant Biol. 2001;LXVI:575–590. [PubMed: 12762059]
- 84.
- Wellauer PK, Dawid IB. Secondary structure maps of RNA. Processing of HeLa rRNA. Proc Nat Acad Sci. 1973;70:2827–2831. [PMC free article: PMC427118] [PubMed: 4517935]
- 85.
- Loening UE, Jones KW, Birnstiel ML. Properties of the rRNA precursor in Xenopus laevis: Comparison to the precursor in mammals and in plants. J Mol Biol. 1969;45:353–366. [PubMed: 5367033]
- 86.
- Wellauer PK, Dawid IB. Secondary structure maps of rRNA and rDNA. I. Processing of Xenopus laevis rRNA and structure of single-stranded rDNA. J Mol Biol. 1974;89:379–395. [PubMed: 4444053]
- 87.
- Levis R, Penman S. Processing steps and methylation in the formation of the rRNA in cultured Drosophila cells. J Mol Biol. 1978;121:219–238. [PubMed: 660652]
- 88.
- Winicov I. Alternate temporal order in rRNA maturation. J Mol Biol. 1976;100:141–155. [PubMed: 942591]
- 89.
- Dudov KP, Dabeva MD, Hadjiolov AA. et al. Processing and migration of ribosomal RNA in the nucleolus and nucleoplasm of rat liver nuclei. Biochem. J. 1978;171:375–383. [PMC free article: PMC1183966] [PubMed: 656051]
- 90.
- Bowman LH, Rabin B, Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981;9:4951–4966. [PMC free article: PMC327491] [PubMed: 7312622]
- 91.
- Borovjagin AV, Gerbi SA. The spacing between functional cis-elements of U3 snoRNA is critical for rRNA processing. J Mol Biol. 2000;300:57–74. [PubMed: 10864498]
- 92.
- Jeppesen C, Stebbins-Boaz B, Gerbi SA. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988;16:2127–2148. [PMC free article: PMC338204] [PubMed: 3357768]
- 93.
- Watkins NJ, Ségault V, Charpentier B. et al. A common core RNP structure shared between the small nucleolar Box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. [PubMed: 11081632]
- 94.
- Reddy R, Busch H. Small nuclear RNA and RNA processing. Prog Nucleic Acid Res Mol Biol. 1983;30:127–162. [PubMed: 6198692]
- 95.
- Dragon F, Gallagher JE, Compagnone-Post PA. et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. [PubMed: 12068309]
- 96.
- Lange TS, Ezrokhi M, Borovjagin AV. et al. Nucleolar Localization Elements of Xenopus laevis U3 snoRNA. Mol Biol Cell. 1998b;9:2973–2985. [PMC free article: PMC25574] [PubMed: 9763456]
- 97.
- Narayanan A, Speckmann W, Terns R. et al. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–2147. [PMC free article: PMC25425] [PubMed: 10397754]
- 98.
- Verheggen C, Mouaikel J, Thiry M. et al. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 2001;20:5480–5490. [PMC free article: PMC125276] [PubMed: 11574480]
- 99.
- Verheggen C, Lafontaine DL, Samarsky D. et al. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. [PMC free article: PMC126019] [PubMed: 12032086]
- 100.
- Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA GAC-box A' and box A sequences play distinct functional roles in rRNA processing. Mol Cell Biol. 2001;21:6210–6221. [PMC free article: PMC87338] [PubMed: 11509664]
- 101.
- Savino R, Hitti Y, Gerbi SA. Genes for Xenopus laevis U3 small nuclear RNA. Nucleic Acids Res. 1992;20:5435–5442. [PMC free article: PMC334353] [PubMed: 1437561]
- 102.
- Epstein P, Reddy R, Busch H. Multiple states of U3 RNA in Novikoff hepatoma nucleoli. Biochemistry. 1984;23:5421–5425. [PubMed: 6210104]
- 103.
- Prestayko AW, Tonato M, Busch H. Low molecular weight RNA associated with 28S nucleolar RNA. J Mol Biol. 1970;47:505–515. [PubMed: 5418169]
- 104.
- Zieve G, Penman S. Small RNA species of the HeLa cell: Metabolism and subcellular localization. Cell. 1976;8:19–31. [PubMed: 954090]
- 105.
- Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. [PMC free article: PMC401391] [PubMed: 2531075]
- 106.
- Méreau A, Fournier A, Grégoire A. et al. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNA: Protein-RNA contacts and base-pair interactions with pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. [PubMed: 9356246]
- 107.
- Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. submitted. 2003 [PMC free article: PMC1370586] [PubMed: 15146078]
- 108.
- Beltrame M, Tollervey D. Base-pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. [PMC free article: PMC394519] [PubMed: 7556076]
- 109.
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO. J1998;17:1476–1486. [PMC free article: PMC1170495] [PubMed: 9482744]
- 110.
- Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5' end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19:6012–6019. [PMC free article: PMC84488] [PubMed: 10454548]
- 111.
- Hartshorne T. Distinct regions of U3 snoRNA interact at two sites within the 5' external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res. 1998;26:2541–2554. [PMC free article: PMC147592] [PubMed: 9592135]
- 112.
- Hartshorne T, Toyofuku W. Two 5'-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999;27:3300–3309. [PMC free article: PMC148563] [PubMed: 10454637]
- 113.
- Hartshorne T, Toyofuku W, Hollenbaugh J. Trypanosoma brucei 5'ETS A'-cleavage is directed by 3'-adjacent sequences, but not two U3 snoRNA-binding elements, which are all required for subsequent pre-small subunit rRNA processing events. J Mol Biol. 2001;313:733–749. [PubMed: 11697900]
- 114.
- Furlong JC, Forbes J, Robertson M, Maden BEH. The external transcribed spacer and preceding region of Xenopus borealis rDNA: Comparison with the corresponding region of Xenopus laevis rDNA. Nucleic Acids Res. 1983;11:8183–8196. [PMC free article: PMC326574] [PubMed: 6672764]
- 115.
- Ajuh PM, Heeney PA, Maden BEH. Xenopus borealis and Xenopus laevis 28S ribosomal DNA and the complete 40S ribosomal precursor RNA coding units of both species. Proc Royal Soc ser B. 1991;245:65–71. [PubMed: 1682930]
- 116.
- Herrera A, Olson MOJ. Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry. 1986;25:6258–6264. [PubMed: 3790520]
- 117.
- Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F. et al. Nucleolin is a sequence specific RNA-binding protein—characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. [PubMed: 8676391]
- 118.
- Sipos K, Olson MOJ. Nucleolin promotes secondary structure in ribosomal RNA. Biochem Biophys Res Comm. 1991;177:673–678. [PubMed: 2049089]
- 119.
- Hanakahi LA, Bu Z, Maizels N. The C-terminal domain of nucleolin accelerates nucleic acid annealing. Biochemistry. 2000;39:15493–15499. [PubMed: 11112535]
- 120.
- Ginisty H, Serin G, Ghisolfi-Nieto L. et al. Interaction of nucleolin with an evolutionarily conserved pre-ribosomal RNA sequence is required for the assembly of the primary processing complex. J Biol Chem. 2000;275:18845–18850. [PubMed: 10858445]
- 121.
- Kass S, Craig N, Sollner-Webb B. The primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987;7:2891–2898. [PMC free article: PMC367908] [PubMed: 3670298]
- 122.
- Stroke IL, Weiner AM. The 5' end of U3 snRNA can be cross-linked in vivo to the external transcribed spacer of rat ribosomal RNA precursors. J Mol Biol. 1989;210:497–512. [PubMed: 2614831]
- 123.
- Michot B, Bachellerie J-P. Secondary structure of the 5' external transcribed spacer of vertebrate pre-rRNA. Presence of phylogenetically conserved features. Eur J Biochem. 1991;195:601–609. [PubMed: 1999184]
- 124.
- Kass S, Tyc K, Steitz JA. et al. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. [PubMed: 2156625]
- 125.
- Serin G, Joseph G, Faucher C. et al. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie. 1996;78:530–538. [PubMed: 8915542]
- 126.
- Allain FH-T, Bouvet P, Dieckmann TJ. et al. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. [PMC free article: PMC305906] [PubMed: 11118222]
- 127.
- Bouvet P, Allain FH, Finger LD. et al. Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin. J Mol Biol. 2001;309:763–775. [PubMed: 11397095]
- 128.
- Bouvet P, Diaz JJ, Kindbeiter K. et al. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–19029. [PubMed: 9668083]
- 129.
- Sicard H, Faubladier M, Noillac-Depeyre J. et al. The role of the Schizosaccharomyces pombe gar2 protein in nucleolar structure and function depends on the concerted action of its highly charged N terminus and its RNA-binding domains. Mol Biol Cell. 1998;9:2011–2023. [PMC free article: PMC25453] [PubMed: 9693363]
- 130.
- Wehner KA, Gallagher JEG, Baserga SJ. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol Cell Biol. 2002;22:7258–7267. [PMC free article: PMC139808] [PubMed: 12242301]
- 131.
- Wormsley S, Samarsky DA, Fournier MJ. et al. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA. 2001;7:904–919. [PMC free article: PMC1370138] [PubMed: 11421365]
- 132.
- Maser RL, Calvet JP. U3 snRNA can be psoralen cross-linked in vivo to the 5' external transcribed spacer of pre-ribosomal RNA. Proc Nat Acad Sci. 1989;86:6523–6527. [PMC free article: PMC297876] [PubMed: 2771939]
- 133.
- Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. [PMC free article: PMC556602] [PubMed: 1563354]
- 134.
- Tyc K, Steitz JA. A new interaction between the mouse 5' external transcribed spacer of pre-rRNA and U3 snRNA detected by psoralen crosslinking. Nucleic Acids Res. 1992;20:5375–5382. [PMC free article: PMC334344] [PubMed: 1437554]
- 135.
- Enright CA, Maxwell ES, Eliceiri GL. et al. B. 5' ETS rRNA processing facilitated by four small RNAs: U14, E3, U17 and U3 RNA 199621094–1099.[Erratum in RNA 1996 21318] [PMC free article: PMC1369439] [PubMed: 8903340]
- 136.
- Eichler DC, Eales SJ. Isolation and characterization of a single-stranded specific endoribonuclease from Ehrlich cell nucleoli. J Biol Chem. 1982;257:14384–14389. [PubMed: 7142216]
- 137.
- Shumard CM, Eichler DC. Ribosomal RNA processing: limited cleavages of mouse preribosomal RNA by a nucleolar endoribonuclease include the early +650 processing site. J Biol Chem. 1988;263:19346–19352. [PubMed: 3198630]
- 138.
- Mougey EB, O'Reilly M, Osheim Y. et al. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993a;7:1609–1619. [PubMed: 8339936]
- 139.
- Mougey EB, Pape LK, Sollner-Webb B. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5' external transcribed spacer of Xenopus precursor rRNA. Mol Cell Biol. 1993b;13:5990–5998. [PMC free article: PMC364653] [PubMed: 8413202]
- 140.
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Ann Rev Genet. 1999;33:261–311. [PubMed: 10690410]
- 141.
- Hughes JMX, Ares Jr. M. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. [PMC free article: PMC453175] [PubMed: 1756730]
- 142.
- Labhart P, Reeder RH. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986;45:431–443. [PubMed: 3453104]
- 143.
- Kass S, Sollner-Webb B. The first pre-rRNA processing event occurs in a large complex: Analysis by gel retardation, sedimentation, and UV cross linking. Mol Cell Biol. 1990;10:4920–4931. [PMC free article: PMC361110] [PubMed: 2388630]
- 144.
- Vance VB, Thompson AE, Bowman LH. Transfection of mouse ribosomal DNA into rat cells: Faithful transcription and processing. Nucleic Acids Res. 1985;13:7499–7513. [PMC free article: PMC322058] [PubMed: 2997749]
- 145.
- Hannon GJ, Maroney PA, Branch A. et al. Accurate processing of human pre-rRNA in vitro. Mol Cell Biol. 1989;9:4422–4431. [PMC free article: PMC362525] [PubMed: 2586517]
- 146.
- Scheer U, Benavente R. Functional and dynamic aspects of the mammalian nucleolus. BioEssays. 1990;12:14–20. [PubMed: 2181998]
- 147.
- Peculis BA. snoRNA nuclear import and potential for cotranscriptional function in pre-rRNA processing. RNA. 2001;7:207–219. [PMC free article: PMC1370079] [PubMed: 11233978]
- 148.
- Bourbon H, Bugler B, Caizergues-Ferrer M. et al. Role of phosphorylation on the maturation pathways of a 100 kDa nucleolar protein. FEBS Lett. 1983;155:218–222. [PubMed: 6852233]
- 149.
- Li HV, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. [PMC free article: PMC360983] [PubMed: 2406561]
- 150.
- Liang WQ, Fournier MJ. U14 base-pairs with 18S rRNA: A novel snoRNA interaction required for rRNA processing. Genes Dev. 1995;9:2433–2443. [PubMed: 7557394]
- 151.
- Lange TS, Borovjagin A, Maxwell ES. et al. Conserved Boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 1998a;17:3176–3187. [PMC free article: PMC1170656] [PubMed: 9606199]
- 152.
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. [PMC free article: PMC553900] [PubMed: 3327689]
- 153.
- Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. [PMC free article: PMC359567] [PubMed: 8455623]
- 154.
- Tycowski KT, Shu M-D, Steitz JA. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. [PubMed: 7985025]
- 155.
- Mishra RK, Eliceiri GL. Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc Nat Acad Sci. 1997;94:4972–4977. [PMC free article: PMC24615] [PubMed: 9144174]
- 156.
- Girard J-P, Lehtonen H, Caizergues-Ferrer M. et al. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. [PMC free article: PMC556499] [PubMed: 1531632]
- 157.
- Fournier MJ, Maxwell ES. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993;18:131–135. [PubMed: 8493724]
- 158.
- Raziuddin, Little RD, Labella T. et al. Transcription and processing of RNA from mouse ribosomal DNA transfected into hamster cells. Mol Cell Biol. 1989;9:1667–1671. [PMC free article: PMC362585] [PubMed: 2725522]
- 159.
- Hughes JMX. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small subunit RNA. J Mol Biol. 1996;259:645–654. [PubMed: 8683571]
- 160.
- Abou ElelaS, Igel H, Ares Jr. M. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. [PubMed: 8620530]
- 161.
- Kufel J, Dichtl B, Tollervey D. yeast Rnt1p is required for cleavage of the pre-ribosomal RNA 3' ETS but not the 5' ETS. RNA. 1999;5:909–917. [PMC free article: PMC1369815] [PubMed: 10411134]
- 162.
- Intine RVA, Good L, Nazar RN. Essential structural features in the Schizosaccharomyces pombe pre-rRNA 5' external transcribed spacer. J Mol Biol. 1999;286:695–708. [PubMed: 10024444]
- 163.
- Lee SJ, Baserga SJ. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component. Mpp10p. Proc Nat Acad Sci. 1997;94:13536–13541. [PMC free article: PMC28341] [PubMed: 9391061]
- 164.
- Colley A, Beggs JD, Tollervey D. et al. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol Cell Biol. 2000;20:7238–7246. [PMC free article: PMC86278] [PubMed: 10982841]
- 165.
- Billy E, Wegierski T, Nasr F. et al. Rcl1p, the yeast protein similar to the RNA 3'-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 2000;19:2115–2126. [PMC free article: PMC305690] [PubMed: 10790377]
- 166.
- Gelperin D, Horton L, Beckman J. et al. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA. 2001;7:1268–1283. [PMC free article: PMC1370171] [PubMed: 11565749]
- 167.
- Wegierski T, Billy E, Nasr F. et al. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA. 2001;7:1254–1267. [PMC free article: PMC1370170] [PubMed: 11565748]
- 168.
- Yu Y-T, Nilsen TW. Sequence requirements for maturation of the 5' terminus of human 18S rRNA in vitro. J Biol Chem. 1992;267:9264–9268. [PubMed: 1577760]
- 169.
- Venema J, Henry Y, Tollervey D. Two distinct recognition signals define the site of endonucleolytic cleavage at the 5' end of yeast 18S rRNA. EMBO J. 1995;14:4883–4892. [PMC free article: PMC394586] [PubMed: 7588617]
- 170.
- Sharma K, Venema J, Tollervey D. The 5' end of the 18S rRNA can be positioned from within the mature rRNA. RNA. 1999;5:678–686. [PMC free article: PMC1369795] [PubMed: 10334338]
- 171.
- Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem. 1973;248:1412–1416. [PubMed: 4568815]
- 172.
- van BeekveltCA, Jeeninga RE, van't Riet J. et al. Identification of cis-acting elements involved in 3'-end formation of Saccharomyces cerevisiae 18S rRNA. RNA. 2001;7:896–903. [PMC free article: PMC1370137] [PubMed: 11421364]
- 173.
- Hadjiolova KV, Normann A, Cavaillé J. et al. Processing of truncated mouse or human rRNA transcribed from ribosomal minigenes transfected into mouse cells. Mol Cell Biol. 1994;14:4044–4056. [PMC free article: PMC358770] [PubMed: 8196643]
- 174.
- Shumard CM, Torres C, Eichler DC. In vitro processing at the 3'-terminal region of pre-18S rRNA by a nucleolar endonuclease. Mol Cell Biol. 1990;10:3868–3872. [PMC free article: PMC360897] [PubMed: 2370856]
- 175.
- Cavaillé J, Hadjiolov AA, Bachellerie J-P. Processing of mammalian rRNA precursors at the 3' end of 18S rRNA. identification of cis-acting signals suggests the involvement of U13 small nucleolar RNA. Eur J Biochem. 1996;242:206–213. [PubMed: 8973634]
- 176.
- Hadjiolova KV, Georgiev OI, Nosikov VV. et al. Mapping of the major early endonuclease cleavage site of the rat precursor to rRNA within the internal transcribed spacer sequence of rDNA. Biochim Biophys Acta. 1984c;782:195–201. [PubMed: 6326837]
- 177.
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. [PubMed: 8513505]
- 178.
- Bowman LH, Goldman WE, Goldberg GI. et al. Location of the initial cleavage sites in mouse pre-rRNA. Mol Cell Biol. 1983;3:1501–1510. [PMC free article: PMC369996] [PubMed: 6621536]
- 179.
- Peculis BA, Steitz JA. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 1994;8:2241–2255. [PubMed: 7958892]
- 180.
- Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. [PMC free article: PMC364865] [PubMed: 8247008]
- 181.
- Chu S, Archer RH, Zengel JM. et al. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Nat Acad Sci. 1994;91:659–663. [PMC free article: PMC43008] [PubMed: 8290578]
- 182.
- Henry Y, Wood H, Morrissey JP. et al. The 5' end of yeast 5.8S rRNA is generated by exonuclease from an upstream cleavage site. EMBO J. 1994;13:2452–2463. [PMC free article: PMC395111] [PubMed: 7515008]
- 183.
- Lygerou Z, Allmang C, Tollervey D. et al. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. [PubMed: 8602511]
- 184.
- Allmang C, Henry Y, Morrissey JP. et al. Processing of yeast pre-rRNA at sites A2 and A3 is linked. RNA. 1996;2:63–73. [PMC free article: PMC1369351] [PubMed: 8846297]
- 185.
- Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article: PMC452314] [PubMed: 8896463]
- 186.
- Torchet C, Jacq C, Hermann-Le DenmatS. Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA. 1998;4:1636–1652. [PMC free article: PMC1369731] [PubMed: 9848659]
- 187.
- Eppens NA, Rensen S, Granneman S. et al. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA. 1999;5:779–793. [PMC free article: PMC1369804] [PubMed: 10376877]
- 188.
- Eppens EA, Faber AW, Rondaij M. et al. Deletions in the S1 domain of Rrp5p cause processing at a novel site in ITS1 of yeast pre-rRNA that depends on Rex4p. Nucleic Acids Res. 2002;30:4222–4231. [PMC free article: PMC140538] [PubMed: 12364601]
- 189.
- Torchet C, Hermann-Le DenmatS. High dosage of the small nucleolar RNA snR10 specifically suppresses defects of a yeast rrp5 mutant. Mol Genet Genomics. 2002;268:70–80. [PubMed: 12242501]
- 190.
- Peculis BA. The sequence of the 5' end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol Cell Biol. 1997;17:3702–3713. [PMC free article: PMC232222] [PubMed: 9199304]
- 191.
- Reddy R, Henning D, Busch H. Primary and secondary structure of U8 small nuclear RNA. J Biol Chem. 1985;260:10930–10935. [PubMed: 2411727]
- 192.
- Gurney T. Characterization of mouse 45S ribosomal RNA subspecies suggests that the first processing cleavage occurs 600 + 100 nucleotides from the 5' end and the second 500 + 100 nucleotides from the 3' end of a 13.9 kb precursor. Nucleic Acids Res. 1985;13:4905–4919. [PMC free article: PMC321834] [PubMed: 4022778]
- 193.
- Labhart P, Reeder RH. A point mutation uncouples RNA 3'-end formation and termination during ribosomal gene transcription in Xenopus laevis. Genes Dev. 1990;4:269–276. [PubMed: 1692557]
- 194.
- van der Sande CAFM, Kulkens T, Kramer AB. et al. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 1989;17:9127–9146. [PMC free article: PMC335119] [PubMed: 2685755]
- 195.
- Johnson SP, Warner JR. Termination of transcription of ribosomal RNA in Saccharomyces cerevisiae. Mol Cell Biochem. 1991;104:163–168. [PubMed: 1921996]
- 196.
- Grummt I, Maier U, Öhrlein A. et al. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985;43:801–810. [PubMed: 4075406]
- 197.
- Kuhn A, Grummt I. 3'-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989;3:224–231. [PubMed: 2714650]
- 198.
- Yip MT, Holland MJ. In vitro RNA processing generates mature 3' termini of yeast 35 and 25S ribosomal RNAs. J Biol Chem. 1989;264:4045–4051. [PubMed: 2645284]
- 199.
- Veldman GM, Klootwijk J, van Heerikhuisen H. et al. The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA genes of the yeast ribosomal RNA operon Possible implications for the interaction between 5.8S and 26S rRNA and the processing of the primary transcript|Nucleic Acids Res 1981b94847–4862. [PMC free article: PMC327484] [PubMed: 7312619]
- 200.
- Long EO, Dawid IB. Alternate pathways in the processing of ribosomal RNA precursor in Drosophila melanogaster. J Mol Biol. 1980;138:873–878. [PubMed: 6774101]
- 201.
- Dudov KP, Hadjiolova KV, Kermekchiev MB. et al. A 12S precursor to 5.8S rRNA associated with rat liver 28S rRNA. Biochim Biophys Acta. 1983;739:79–84. [PubMed: 6550497]
- 202.
- Reddy R, Rothblum LI, Subrahmanyam CS. et al. The nucleotide sequence of 8S RNA bound to preribosomal RNA of Novikoff hepatoma. The 5' end of 8S RNA is 5.8S RNA. J Biol Chem. 1983;258:584–589. [PubMed: 6401296]
- 203.
- Hadjiolova KV, Georgiev OI, Nosikov VV. et al. Localization and structure of endonuclease cleavage sites involved in the processing of the rat 32S precursor to ribosomal RNA. Biochem J. 1984b;220:105–116. [PMC free article: PMC1153599] [PubMed: 6331404]
- 204.
- Geerlings T, Vos JC, Raué HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5'-3' exonucleases. RNA. 2000;6:1698–1703. [PMC free article: PMC1370040] [PubMed: 11142370]
- 205.
- Hadjiolova KV, Georgiev OI, Hadjiolov AA. Excess 5'-terminal sequences in the rat nucleolar 28S ribosomal RNA. Exp Cell Res. 1984a;153:266–269. [PubMed: 6329794]
- 206.
- Nazar RN. A 5.8S rRNA-like sequence in prokaryotic 23S rRNA. FEBS Lett. 1980;119:212–214. [PubMed: 6159235]
- 207.
- Jacq B. Sequence homologies between eukaryotic 5.8S rRNA and the 5' end of prokaryotic 23S rRNA: Evidences for a common evolutionary origin. Nucleic Acids Res. 1981;9:2913–2932. [PMC free article: PMC326902] [PubMed: 7024907]
- 208.
- Pace NR, Walker TA, Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977;16:5321–5328. [PubMed: 921935]
- 209.
- Nazar RN, Sitz TO. Role of the 5'-terminal sequence in the RNA binding site of yeast 5.8S rRNA. FEBS Lett. 1980;115:71–76. [PubMed: 6771162]
- 210.
- Michot B, Bachellerie J-P, Raynal F. Sequence and secondary structure of mouse 28S rRNA 5'-terminal domain. Organization of the 5.8S:28S rRNA complex. Nucleic Acids Res. 1982;10:5273–5283. [PMC free article: PMC320870] [PubMed: 6292836]
- 211.
- Morrissey J, Tollervey D. Birth of the snoRNPs: the evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem Sci. 1995;20:78–82. [PubMed: 7701567]
- 212.
- Young RA, Steitz JA. Complementary sequences 1700 nucleotides apart form a RNAse III cleavage site in E. coli ribosomal precursor RNA. Proc Nat Acad Sci. 1978;75:3593–3597. [PMC free article: PMC392831] [PubMed: 358189]
- 213.
- Bram RJ, Young RA, Steitz JA. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980;19:393–401. [PubMed: 6153577]
- 214.
- Gerbi SA. Small nucleolar RNA. Biochem Cell Biol. 1995;73:845–858. [PubMed: 8722000]
- 215.
- Tang TH, Rozhdestvensky TS, Clouet d'Orval B. et al. RNomics in Archaea reveals a further link between splicing of arachaeal introns and rRNA processing. Nucleic Acids Res. 2002;30:921–930. [PMC free article: PMC100335] [PubMed: 11842103]
- 216.
- Potter S, Durovic P, Dennis PP. Ribosomal RNA precursor processing by a eukaryotic U3 small nucleolar RNA-like molecule in an archeon Science 19952681056–1060.[retraction in Science 1997 2771189] [PubMed: 7538698]
- 217.
- Veldman GM, Klootwijk J, deRegt VC. et al. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981a;9:6935–6952. [PMC free article: PMC327652] [PubMed: 7335496]
- 218.
- Musters W, Planta RJ, Van Heerikhuizen H. et al. Functional analysis of the transcribed spacers of Saccharomyces cerevisiae ribosomal DNA: It takes a precursor to form a ribosomeIn: Hill WE, Dahlberg A, Garrett RA, Moore PB, Schlessinger D, Garrett JR, eds.The Ribosome—Structure, Function and EvolutionWashington DC: American Society for Microbiology Press,1990435–441.
- 219.
- Lalev AI, Nazar RN. A chaperone for ribosome maturation. J Biol Chem. 2001;276:16655–16659. [PubMed: 11279180]
- 220.
- Lalev AI, Abeyrathne PD, Nazar RN. Ribosomal RNA maturation in Schizosaccharomyces pombe is dependent on a large ribonucleoprotein complex of the internal transcribed spacer 1. J Mol Biol. 2000;302:65–77. [PubMed: 10964561]
- 221.
- Spasov K, Perdomo LI, Evakine E. et al. RAC protein directs the complete removal of the 3' external transcribed spacer by the Pac1 nuclease. Mol Cell. 2002;9:433–437. [PubMed: 11864615]
- 222.
- Dennis PP, Russell AG, Moniz de Sá M. Formation of the 5' end pseudoknot in small subunit ribosomal RNA: Involvement of U3-like sequences. RNA. 1997;3:337–343. [PMC free article: PMC1369486] [PubMed: 9085841]
- 223.
- Harnpicharnchai P, Jakovljevic J, Horsey E. et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. [PubMed: 11583614]
- 224.
- Saveanu C, Bienvenu D, Namane A. et al. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. [PMC free article: PMC125736] [PubMed: 11707418]
- 225.
- Anderson JS, Lyon CE, Fox AH. et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. [PubMed: 11790298]
- 226.
- Fatica A, Cronshaw AD, Dlakic M. et al. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol Cell. 2002;9:341–351. [PubMed: 11864607]
- 227.
- Nissen TA, Baßler J, Petfalski E. et al. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. [PMC free article: PMC129079] [PubMed: 12374754]
- 228.
- Fonagy A, Swiderski C, Dunn M. et al. Antisense-mediated specific inhibition of P120 protein expression prevents G1- to S-phase transition. Cancer Res. 1992;52:5250–5256. [PubMed: 1394129]
- 229.
- Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol Cell Biol. 2000;20:5516–5528. [PMC free article: PMC86002] [PubMed: 10891491]
- 230.
- Visintin R, Amon A. The nucleolus: The magician's hat for cell cycle tricks. Curr Opin Cell Biol. 2000;12:752. [PubMed: 11063944]
- 231.
- Volarevic S, Stewart MJ, Ledermann B. et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. [PubMed: 10856218]
- 232.
- Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G1/S transition. Mol Cell Biol. 2001a;21:4246–4255. [PMC free article: PMC87085] [PubMed: 11390653]
- 233.
- Pestov DG, Stockelman MG, Strezoska Z. et al. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 2001b;17:3621–3630. [PMC free article: PMC55883] [PubMed: 11522832]
- 234.
- Strezoska Z, Pestov DG, Lau LF. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J Biol Chem. 2002;277:29617–29625. [PubMed: 12048210]
- 235.
- Zhang Y, Yu Z, Fu X. et al. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell. 2002;109:849–860. [PubMed: 12110182]
- 236.
- Du YC, Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. [PubMed: 12110181]
- Pre-Ribosomal RNA Processing in Multicellular Organisms - Madame Curie Bioscienc...Pre-Ribosomal RNA Processing in Multicellular Organisms - Madame Curie Bioscience Database
- Regulation of T Cell Immunity by OX40 and OX40L - Madame Curie Bioscience Databa...Regulation of T Cell Immunity by OX40 and OX40L - Madame Curie Bioscience Database
- Overview Αβ Metabolism: From Alzheimer Research to Brain Aging Control - Madame ...Overview Αβ Metabolism: From Alzheimer Research to Brain Aging Control - Madame Curie Bioscience Database
- Metastatic Genitourinary Malignancies - Madame Curie Bioscience DatabaseMetastatic Genitourinary Malignancies - Madame Curie Bioscience Database
- Lipocalins in Arthropoda: Diversification and Functional Explorations - Madame C...Lipocalins in Arthropoda: Diversification and Functional Explorations - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...