NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Summary
The transition from the RNA to the DNA world was a major event in the history of life. The invention of DNA required the appearance of enzymatic activities for both synthesis of DNA precursors, retro-transcription of RNA templates and replication of singleand double-stranded DNA molecules. Recent data from comparative genomics, structural biology and traditional biochemistry have revealed that several of these enzymatic activities have been invented independently more than once, indicating that the transition from RNA to DNA genomes was more complex than previously thought. The distribution of the different protein families corresponding to these activities in the three domains of life (Archaea, Eukarya, and Bacteria) is puzzling. In many cases, Archaea and Eukarya contain the same version of these proteins, whereas Bacteria contain another version. However, in other cases, such as thymidylate synthases or type II DNA topoisomerases, the phylogenetic distributions of these proteins do not follow this simple pattern. Several hypotheses have been proposed to explain these observations, including independent invention of DNA and DNA replication proteins, ancient gene transfer and gene loss, and/or nonorthologous replacement. We review all of them here, with more emphasis on recent proposals suggesting that viruses have played a major role in the origin and evolution of the DNA replication proteins and possibly of DNA itself.
Introduction
All cellular organisms have double-stranded DNA genomes. The origin of DNA and DNA replication mechanisms is thus a critical question for our understanding of early life evolution. For some time, it was believed by some molecular biologist that life originated with the appearance of the first DNA molecule!1 Watson and Crick even suggested that DNA was possibly replicated without proteins, wondering “whether a special enzyme would be required to carry out the polymerization or whether the existing single helical chain could act effectively as an enzyme”.2 Such extreme conception was in line with the idea that DNA was the aperiodic crystal predicted by Schroedinger in his influential book “What's life”.3 Times have changed, and several decades of experimental work have convinced us that DNA synthesis and replication actually require a plethora of proteins.4 We are reasonably sure now that DNA and DNA replication mechanisms appeared late in early life history, and that DNA originated from RNA in an RNA/protein world. The origin and evolution of DNA replication mechanisms thus occurred at a critical period of life evolution that encompasses the late RNA world and the emergence of the Last Universal Cellular Ancestor (LUCA) to the present three domains of life (Eukarya, Bacteria and Archaea.).5-7 It is an exciting time to learn through comparative genomics and molecular biology about the details of modern mechanisms for precursor DNA synthesis and DNA replication, in order to trace their histories.
Origin of DNA
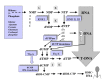
DNA can be considered as a modified form of RNA, since the “normal” ribose sugar in RNA is reduced into deoxyribose in DNA, whereas the “simple” base uracil is methylated into thymidine. In modern cells, the DNA precursors (the four deoxyribonucleoties, dNTPs) are produced by reduction of ribonucleotides di- or triphosphate by ribonucleotide reductases (fig. 1). The synthesis of DNA building blocks from RNA precursors is a major argument in favor of RNA preceding DNA in evolution. The direct prebiotic origin of is theoretically plausible (from acetaldehyde and glyceraldehyde-5-phosphate) but highly unlikely, considering that evolution, as stated by F. Jacob, works like a tinkerer, not an engineer.8,9
The first step in the emergence of DNA has been most likely the formation of U-DNA (DNA containing uracil), since ribonucleotide reductases produce dUTP (or dUDP) from UTP (or UDP) and not dTTP from TTP (the latter does not exist in the cell) (fig. 1). Some modern viruses indeed have a U-DNA genome,10 possibly reflecting this first transition step between the RNA and DNA worlds. The selection of the letter T occurred probably in a second step, dTTP being produced in modern cells by the modification of dUMP into dTMP by thymidylate synthases (followed by phosphorylation).11 Interestingly, the same kinase can phosphorylate both dUMP and dTMP.11 In modern cells, dUMP is produced from dUTP by dUTPases, or from dCMP by dCMP deaminases (fig. 1).11 This is another indication that T-DNA originated after U-DNA. In ancient U-DNA cells, dUMP might have been also produced by degradation of U-DNA (fig. 1).
The origin of DNA also required the appearance of enzymes able to incorporate dNTPs using first RNA templates (reverse transcriptases) and later on DNA templates (DNA polymerases). In all living organisms (cells and viruses), all these enzymes work in the 5' to 3' direction. This directionality is dictated by the cellular metabolism that produces only dNTP 5' triphosphates and no 3' triphosphates. Indeed, both purine and pyrimidine biosyntheses are built up on ribose 5 monophosphate as a common precursor. The sense of DNA synthesis itself is therefore a relic of the RNA world metabolism. Modern DNA polymerases of the A and B families, reverse transcriptases, cellular RNA polymerases and viral replicative RNA polymerases are structurally related and thus probably homologous (for references, see a recent review on viral RNA-dependent RNA polymerases.)12 This suggests that reverse transcriptase and DNA polymerases of the A and B families originated from an ancestral RNA polymerase that has also descendants among viral-like RNA replicases. However, there are several other DNA polymerase families (C, D, X, Y) whose origin is obscure (we will go back to this point below).
If DNA actually appeared in the RNA world, it was a priori possible to imagine that formation of the four dNTPs from the four rNTPs was initially performed by ribozymes. Most scientists, who consider that the reduction of ribose cannot be accomplished by an RNA enzyme, now reject this hypothesis.9,13-19 The removal of the 2' oxygen in the ribose involves indeed a complex chemistry for reduction that requires the formation of stable radicals in ribonucleotide reductases. Such radicals would have destroyed the RNA backbone of a ribozyme by attacking the labile phosphodiester bond of RNA. Accordingly, DNA could have only originated after the invention of modern complex proteins, in an already elaborated protein/DNA world. This suggests that RNA polymerases were indeed available at that time to evolve into DNA polymerases (as well as kinases to phosphorylate dUMP).
Three classes of ribonucleotide reductases (I, II and III) have been discovered so far (for a review, see refs. 9, 16-19) (fig. 1). Although they correspond to three distinct protein families, with different cofactors and mechanisms of action, these mechanisms are articulated around a common theme (radical based chemistry). In all cases, the critical step is the conversion of a cysteine residue into a catalytically essential thiol radical in the active center.18 Recent structural and mechanistic analyses of several RNR at atomic resolution have suggested that all ribonucleotide reductases originated from a common ancestral enzyme, favoring the idea that U-DNA was invented only once.17,18 It has been suggested that either class III (strictly anaerobic) or class II (anaerobic but oxygen tolerant) represent the ancestral form, and that new versions appeared in relation to different lifestyles by recruiting new mechanisms for radical activation (class III in strict anaerobes and class I in aerobes).9,18
The origin of U-DNA in a protein/RNA world logically implies that the second step in the synthesis of DNA precursors, the formation of the letter T, was catalyzed by ancestral thymidylate synthase. For a long time, it was believed that modern thymidylate synthases were all homologues of E. coli ThyA protein, indicating that the letter T was invented only once. However, comparative genomics has revealed recently that ThyA is absent in many archaeal and bacterial genomes, leading to the discovery of a new thymidylate synthase family (ThyX).19 ThyX and ThyA share neither sequence nor structural similarity between each other and have different mechanisms of action,19,20 indicating that thymidylate synthase activity was invented twice independently (fig. 1). T-DNA might have appeared either in two different U-DNA cells, or the invention of a second thymidylate synthase might have occurred in a cell already containing a T-DNA genome. The first possibility would indicate that T-DNA itself has been invented twice, thus suggesting a strong selection pressure to select for uracil modification. In the second case, one should imagine that the new enzyme (either ThyA or ThyX) brought a selective advantage over the previous one in the organism where it appeared first.
A major question is why was DNA selected to replace RNA? The traditional explanation is that DNA replaced RNA as genetic material because it is more stable and can be repaired more faithfully.4 Indeed, removal of the 2' oxygen of the ribose in DNA has clearly stabilized the molecule, since this reactive oxygen can attack the phophodiester bond (this explains why RNA is so prone to strand breakage). In addition, the replacement of uracil by thymine has made possible to correct the deleterious effect of spontaneous cytosine deamination, since a misplaced uracil cannot be recognized in RNA, whereas it can be pint-pointed as an alien base in DNA and efficiently removed by repair systems. Replacement of RNA by DNA as genetic material has thus opened the way to the formation of large genomes, a prerequisite for the evolution of modern cells.
The above scenario nicely explains why, through Darwinian competition, cell populations with DNA genomes finally eliminated cells with RNA genomes. However, this does not explain why the first organisms with a modified RNA (DNA-U), and later on with T-DNA, were successfully selected against the wild type organisms of that time? Indeed, the possibility to have a large genome or to repair cytosine deamination could not have been realized in that individual. In both cases, efficient DNA repair (to remove uracil from DNA) and replication proteins able to replicate large DNA genomes should have evolved first in order for the cell to take advantage of the presence of DNA.15 To explain the origin of DNA, it is thus necessary to consider an advantage that could have been directly selected in the organism in which the transition occurred.
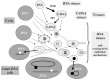
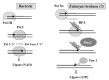
In order to solve this problem, it has recently been proposed that U-DNA first appeared in a virus, making this first U-DNA organism resistant to the RNAses of its host (fig. 2).6,7 Indeed, ribose reduction led to a drastic modification in the structure of the double helix (from the A to the B form) that explains why RNAses are usually inactive on DNA and DNAses inactive on RNA. Similarly, thymidylate synthase could have appeared later on in a virus with U-DNA, to makes its genome resistant to cellular U-DNAses (fig. 2). The same process would have lead to modifications observed in modern DNA viruses (further base methylation in many viral genomes or hydroxymethylation of cytosines in T-even bacteriophages). These modifications are clearly designed to protect viral DNA against host DNAses. Interestingly, thymidylate synthase of the ThyA family are homologous to the T-even bacteriophages DNA modification enzyme dCMP hydroxymethyl-transferases.21 Hydroxymethyl (HMC)-dCTP is directly incorporated into HMC-DNA by the viral polymerase (fig. 1).11 Restriction-modifications systems could be descendant of such viral mechanisms for genome protection; some of them being stolen later on by cells themselves.
If DNA replication and repair mechanisms also originated in viruses, it is easy to imagine that enzymes to correct cytosine deamination are of viral origin, and were later on transferred to cells, a prerequisite to understand the selective advantage of DNA cells over RNA cells in term of faithful replication (see a discussion of this problem in ref. 15). Several scenarios are possible for the transfer of a DNA genome from a virus to a cell: either a cell succeeded to capture several viral enzymes at once to change its genetic material from RNA to DNA, or a large DNA provirus, living in a carrier state inside an RNA cell, finally take over all functions of its host by retro-transcription, subsequently eliminating the labile RNA genomes.
The idea that viruses have played a critical role in the origin of DNA is in line with previous conception that retroviruses were relics of the RNA/DNA world transition.22 In particular, production of DNA from RNA genome in Hepadnavirus could reflect the ancient pathway leading from RNA to DNA.23 The invention of DNA by an RNA virus seems to be more likely than the invention of DNA by an RNA cell for protection against viral RNAses, because it has been probably easier for a virus, than for a cell, to change at once the chemical nature of its genome. This is exemplified by the fact that viruses have managed to multiply with very different types of genetic material (ssRNA, dsRNA, ssDNA, dsDNA, modified DNA) whereas, apart for localized methylation, all types of cells have the same kind of dsDNA genomes.
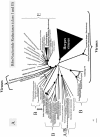
The hypothesis of a viral origin for DNA could explain why many DNA viruses encode their own ribonucleotide reductase and/or thymidylate synthase. This is usually interpreted as the recruitment of cellular enzymes by viruses, but, if DNA appeared in viruses, the opposite could be true as well. Many viral ribonucleotide reductases and thymidylate synthases branch far off from ribonucleotide reductases and thymidylate synthases of their hosts in phylogenetic trees, suggesting that the viral versions of these enzymes are indeed as ancient as their cellular versions (fig. 3A, 3B, 3C). Unfortunately, the direction of ancient transfer of these enzymes (either from cells to viruses or from viruses to cells) is difficult to determine, considering possible artifacts of long branch attraction that can be produced by differences in evolutionary rates between cellular and viral enzymes, i.e., viral sequences can be artificially separated from cellular ones because the latter have evolved more slowly and thus have conserved more common ancestral positions.
As previously mentioned, there is also a striking evolutionary connection at the structural level between most viral RNA dependent RNA replicases and some modern DNA polymerases.12,24 Interestingly, an ancient origin of viral DNA replication mechanisms (possibly predating cellular ones) (fig. 2) would explain why enzymes involved in viral DNA replication are often very different from their cellular counterparts (see ref. 25 for the case of DNA polymerases) (see below for further discussion of this point).
These speculations on the origin of DNA fit well with hypotheses on viral origin that consider no longer viruses as fragments of genetic materials recently escaped from their hosts, but as ancient players in life evolution, possibly predating the divergence between the three domains of life.26,27 The idea that viruses originated before LUCA has been recently supported by the discovery of structural and/or functional similarities between viruses infecting different cellular domains of life, such as those detected between some archaeal viruses (Lipothrixvirus and Rudivirus) and several large eukaryal DNA viruses (Poxviruses, ASFV, Chlorella viruses),28 between Adenoviruses (eukaryal virus) and bacterial Tectiviruses,29 or between eukaryal Flavivirus and bacterial Cystoviruses.30
Origin and Evolution of DNA Replication Mechanism
Viral DNA Replication Mechanisms
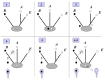
In contrast to cellular genomes, which are all made of double-stranded DNA, viral DNA genomes are very diverse; some viruses have circular or linear double-stranded DNA genomes, while others have circular single-stranded DNA genomes.11 Single-stranded DNA genomes are replicated via rolling circle replication with a double-stranded DNA intermediate, whereas double-stranded viral DNA genomes are replicated either via classical theta or Y-shaped replication (for circular and linear genomes, respectively), by rolling circle, or by linear strand displacement11 (for recent reviews on eukaryal viral DNA replication, see ref. 31). In addition, replication can be symmetric, with both strands replicated simultaneously, but also asymmetric (the two strand are replicated not simultaneously but one after the other) or semi-asymmetric (the initiation of DNA replication on one strand being delayed until the first one is already partly replicated) (fig. 1). Some viral replication mechanisms are also used by plasmids (rolling circle) and some plasmids encode DNA replication proteins homologous to viral ones (see below), suggesting that plasmids originated from ancient viruses that have lost their capsid genes.26
The initiation of viral DNA replication needs a specific viral encoded initiator protein that can be a site-specific endonuclease (rolling-circle replication) or a protein that trigger double-stranded unwinding. Interestingly, plasmid and viral endonucleases involved in rolling-circle replication are evolutionary related.32 The minimal recruitment for DNA chain elongation is a DNA polymerase. In contrast to RNA polymerases, all DNA polymerases (viral or cellular) need a 3'OH primer to initiate strand synthesis. This primer can be a tRNA (for reverse transcriptases), or a short RNA, either produced by a classical RNA polymerase (also involved in transcription) or a DNA primase. This use of RNA to initiate DNA synthesis is also often considered as a relic of the RNA world.
Some primases have a strong DNA polymerase activity, suggesting that primases testify for the transition between RNA and DNA polymerases.33 The definition of a DNA polymerase is thus becoming less straightforward, as also demonstrated by the recent characterization of DNA polymerases of the Y family that are involved in DNA repair and synthesize very short patches of DNA (much like a primase)25,34 and by the discovery of structural similarities between eukaryal primase and DNA polymerases of the family X.35
As a consequence of the ancient metabolic pathway producing only 5' nucleotides, the strand moving in the 3' to 5' direction in symmetric or semi asymmetric replication has to be replicated backward in the form of short DNA pieces (Okazaki fragments) (fig. 3). These fragments are primed by DNA primase and later on assembled by a DNA ligase, after removal of the RNA primer by RnaseH or various exonuclease activities, sometimes associated to DNA polymerases. In some cases of asymmetric replication (Adenovirus, bacteriophage Φ29, mitochondrial linear plasmids), the DNA polymerases use a protein priming system to produce a free 3'OH for the DNA polymerase. All polymerases using this system belong to a subfamily of the DNA polymerase B family.25
Some DNA polymerases can perform strand displacement that is required for asymmetric DNA replication, while others, in order to improve the efficiency of this process associate with DNA helicases and/or single-stranded DNA binding proteins (ssb) to unwind the two DNA strands. The processivity of many viral DNA polymerases is further enhanced by specific processivity factors. In the case of T4, these include ring-shaped DNA clamps, and hand-shaped clamp-loader complexes that can open and close the ring-shaped DNA clamp around the DNA molecule.
In symmetric replication, the syntheses of the leading and lagging strands are coupled via an interaction between the primase and the helicase (fig. 4). In some bacteriophages (T7, P4) and eukaryal viruses (Herpes), this coupling is achieved by the fusion of the helicase and the primase activities into a single polypeptide.36,37 This is clearly a case of convergent evolution, since bacteriophages and Herpes primases belong to different protein families.38
The two DNA polymerases that replicate the lagging and the leading strands can be also physically linked. As a consequence, the lagging strand loops upon itself, and the two strands are replicated at once very rapidly, limiting the presence of single-stranded DNA to the fork vicinity. This is in striking contrast with asymmetric replication that requires complete denaturation of the two strands before replication of the lagging strand (fig. 4).
Some DNA viruses replicate their genome using only replication proteins encoded by their host (with the exception of initiator proteins). However, many large DNA viruses encode also several proteins involved in the elongation step of DNA replication. Some of them (e.g., T4-phages) have reached a high level of complexity in their DNA replication machinery, and consequently encode functional analogs for all proteins involved in cellular DNA replication (fig. 5).39
Considering that replication of double-strand RNA viruses is completely asymmetric, it is likely that DNA replication first occurred via the asymmetric mode and evolved toward fully symmetric theta mode via the semi-asymmetric mode (fig. 3). If viruses recruited their DNA replication mechanisms from the cells, as proposed in the “escaped theory” for viral origin, this means either that viruses originated from early DNA cells that have not yet reached the stage of the symmetric mode of replication, or that this mode has been modified in many viruses to produce simpler systems. The latter possibility cannot be excluded, since there is some plasticity in the evolution of DNA replication mechanisms, and this evolution is not necessarily unidirectional (fig. 4). For example, the replication of bacterial chromosome during conjugation can be changed from the symmetric theta mode to the asymmetric rolling-circle mode upon the integration of a conjugative plasmid.11
On the contrary, if DNA originated in viruses (7), one can even imagine that several DNA replication systems emerged and evolved independently from different lineages of RNA viruses. This hypothesis thus allows for a long period of DNA replication evolution purely in the viral world (fig. 2). This would nicely explain the existence of different version of functionally analogs but nonhomologous DNA replication proteins. The diversity of viral replication proteins can be exemplified by those of Pox virus, Herpes viruses or T4, that are completely different from each others, and are no more related to the archaeal/bacterial systems (in term of protein similarities) than these systems are related between each others.31,36,37,39 Recent sequencing of the 280 kbp bacteriophage phiKZ of Pseudomonas aeruginosa failed to identify virus-encoded DNA replication-associated proteins, suggesting that they may be strongly divergent from known homologous proteins.40 Finally, it is noteworthy that several families of proteins involved in DNA replication also appears restricted to the virus world, such as helicase of the superfamily III,41 the Herpes primases,38 or protein-primed DNA polymerases of the B family.25 Some linear mitochondrial plasmids also encode the latter enzyme, again suggesting a connection between viruses and plasmids. The recent discovery of a completely new family of DNA polymerase/primase encoded by the archaeal plasmid pRN2 once more emphasizes the potential of viruses and plasmids as source of novel DNA replication proteins.78 It is difficult to understand the existence of all these viral and/or plasmid specific DNA replication proteins in the framework of the “escaped theory” for the origin of viruses. On the contrary, in the viral origin hypothesis, these enzymes have simply originated in viruses and were never been transferred to the cells.
Cellular DNA Replication: Two Independent Inventions
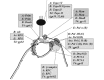
In all cells, DNA replication occurs by a symmetric (theta) mode of replication. The proteins involved and their mechanisms of action have been analyzed in much details during these last decades in several bacterial and eukaryal model systems.11,31,43 The basic principles of DNA replication are very similar in Bacteria and Eukarya, and probably in Archaea as well (fig. 5).44,45 For the initiation step, initiator proteins recognize specific DNA sequences at replication origin(s). A loading factor then brings the replication helicase to the initiation complex to start the assembly of the replisome. The movement of the replication forks involves the concerted action of primases, DNA helicases, ssb proteins, and at least two processive DNA polymerases (with clamp and clamp loading factors) to couple replication of the leading and lagging strands, allowing the efficient replication of large cellular genomes. In turn, type II DNA topoisomerases became essential to solve the topological problems due to the unwinding of the double-helix in such large molecules, counteracting the production of positive superturns ahead of the forks and allowing separation of daughter molecules. This mechanism of DNA replication strikingly resembles those of some large DNA bacteriophages, such as T4 (fig. 5).
Originally, the striking similarity between the enzymatic activities involved in bacterial and eukaryal DNA replication suggested that they originated from a common ancestral DNA replication mechanism already present in LUCA (in the nomenclature of the evolutionists, the bacterial and eukaryal DNA replication proteins were supposed to be orthologues, i.e., to have evolved in parallel to speciation from a common ancestor). In that case, bacterial, eukaryal and archaeal DNA replication proteins performing analogous function should be orthologous. However, comparative genomic analyses have shown that this is not the case (fig. 5).46-48 On the contrary, several critical DNA replication proteins identified in Bacteria by genetic and in vitro analyses have no homolog in Archaea or Eukarya, whereas others have only very distantly related homologues that are probably not orthologues. Similarly, most DNA replication proteins previously identified in Eukarya turned out to have readily detectable homologues only in Archaea.
The similarity between DNA replication proteins in Archaea and Eukarya is especially remarkable. It cannot be due to functional convergence since they have somewhat different modes of replication (unique origin and high-speed in Archaea, multiple origin and low speed in Eukarya),49 whereas Archaea and Bacteria have dissimilar replication proteins but identical replication mode (unique origin, high speed, hot spot of recombination at the replication terminus, and major genomic recombination events occurring between bi-directional replication forks.)49-50 The high level of similarities between the archaeal and eukaryal DNA replication proteins also cannot be explained by similar chromatin structure (as suggested by Cavalier-Smith),51 since most archaeal proteins involved in DNA replication are similar in the two archaeal phyla the Crenarchaeota and the Euryarchaeota, whereas the presence of eukaryal-like histones is restricted to the Euryarchaeota.
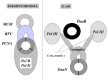
Five alternative hypotheses have been proposed to explain the evolutionary gap between the bacterial and the eukaryal/archaeal replication systems (fig. 6).
- the replication proteins of Bacteria and Archaea/Eukarya are actually orthologues, but they have diverged to such an extent that their homology cannot be detected anymore at the sequence level.46
- two different replication systems were present in the LUCA; one was retained in Bacteria, the other in Archaea/Eukarya.46
- The ancestral replication mechanism present in LUCA has been displaced either in Bacteria or in Archaea/Eukarya by a new one, corresponding to a nonorthologous displacement.46,52 More specifically, it has been suggested that the bacterial replication system, or part of the eukaryal one, are of viral origin.52 - 53
- Both bacterial, archaeal and eukaryal replication mechanisms are of viral origin and have been transferred to cells independently.7
The hypotheses 4 and 5 can be combined, if a first transfer from viruses to cells occurred before LUCA, and a second one displaced this ancestral cellular mechanism later on.
In addition several authors have proposed that the eukaryal nucleus itself originated from a large DNA virus (possibly an archaeal virus) that could be related to Poxviruses.54-55
The first hypothesis (the hidden orthology) can be clearly ruled out, since the bacterial and the archaeal/eukaryal versions of the two central players in the elongation step of DNA replication, the replicative polymerases and the primases, belong to different protein families.25,35,48 In the case of primases, structural analyses have shown that the bacterial and the eukaryal/archaeal versions are completely unrelated, the latter being member of the DNA polymerase X family.35 In the case of the replicase, the structure of the bacterial one (PolC/DnaE) has not yet been solved, but in-depth sequence analysis failed to detect any similarity with the superfamily of RNA polymerases, reverse transcriptase and DNA polymerases of the A and B families.48
In other cases (the replicative helicase, the single-stranded DNA binding proteins, the initiator proteins), comparative structural analyses and/or PSI-BLAST searches have shown that the bacterial and eukaryal/archaeal proteins belong to same superfamilies, since they share homologous domains. However, they are clearly not orthologues, since they belong to different families. For example, in the case the initiator protein (DnaA in Bacteria, Cdc6/Orc1 in Archaea and Eukarya) the bacterial and archaeal proteins share a common ATPase module of the same family (AAA+), but these modules are associated to different modules that are probably involved in DNA binding.57
The bacterial and archaeal/eukaryal versions of many DNA replication proteins have thus been certainly invented independently, probably by recruitment and modification of proteins previously involved in RNA replication and/or RNA gene regulation. However, a few DNA replication proteins (the clamp, the clamp loader, DNA ligase) could be orthologous in the three domains of life since they share sequence similarities that can be detected by elaborated PSI-BLAST analyses or structural similarity with unique fold and fold arrangement.48 Furthermore, they are more similar to each other's, from one domain to another, than to any other proteins. We should thus explain why different replication systems that have emerged independently use some homologous accessory proteins. It is possible that these proteins originated late in the history of DNA replication and were independently recruited by evolving DNA replication systems. Alternatively, they might have predated DNA replication itself and were independently used by different emerging systems.
In order to better understand the evolution of the DNA replication apparatus, it would be necessary to determine with some confidence when and where the independent inventions of the bacterial and the eukaryal/archaeal versions of nonorthologous DNA replication mechanisms occurred (either before or after LUCA, either in cells or in viruses?). We will discuss now several specific points of the above hypotheses (except hypothesis 1 that we have ruled out) in an attempt to answer some of these questions.
The Genome of LUCA (DNA or RNA)
In hypotheses 2 and 4, LUCA had a DNA genome, whereas in 3, LUCA had an RNA genome (hypothesis 5 can be accommodated with both possibilities) (fig. 6). The nature of the genome of LUCA is thus a major pending question. Obviously, LUCA had already a well-developed translation system (see other chapters), but the question of the status of transcription and replication in LUCA is by far more complex. The hypothesis of a primitive LUCA with an RNA genome was first formulated twenty years ago by Carl Woese.58 This hypothesis was mainly based on the prejudice of a very simple LUCA (a progenote). It is remarkable that Carl Woese correctly predicted in 1977, based on this idea, that DNA replication mechanisms should not be homologous in prokaryotes and eukaryotes (if prokaryotes are for this purpose assimilated to Bacteria).
The idea of a simple LUCA without DNA was strongly disputed by Forterre following the discovery in Archaea of DNA polymerases (family B) and type II DNA topoisomerases (gyrase) that were homologous to bacterial and eukaryal enzymes.59 However, it turned out that these were cases of lateral gene transfer.25,60 More recently, several authors also argued for the presence of DNA in LUCA, as some proteins using DNA as substrate are probably orthologues in the three domains of life.46,48 This is the case for the clamp, the clamp loader, and DNA ligase (as already mentioned), but also for DNA-dependent RNA polymerases, type I DNA topoisomerases of the A family, RecA-like recombinases, SMC proteins involved in chromosome condensation, and Mre11/Rad50 complex involved in homologous recombination. However, it is difficult to reject the RNA LUCA hypothesis simply based on this observation, because some of these proteins could have been already operational in the RNA world. For instance, cellular DNA-dependent RNA polymerases can also replicate the genome of RNA viruses,61-62 type I DNA topoisomerases of the A family can act as RNA topoisomerase,63 and the common ancestor of present-day DNA ligases could have been an RNA ligase involved in RNA processing.
The better argument in favor of a DNA-based LUCA could be actually the orthology of the clamp and clamp loader in the three domains, since RNA viruses apparently does not use clamp for their replication. However, the clamp and clamp loader of T-even bacteriophages being homologous to their cellular counterparts,64 another possibility is that Bacteria, Archaea/ Eukarya have recruited independently these homologous proteins from viruses related to T-even bacteriophages.
Perhaps a more direct strategy to decide whether LUCA had an RNA-based or a DNA-based genome could be to determine if ribonucleotide reductases and thymidylate synthases were already present in LUCA. Ribonucleotide reductases of class II and III are present in both Bacteria> and Archaea. Bacterial and archaeal class II ribonucleotide reductases form two monophyletic groups, suggesting that class II was present in LUCA (fig. 3A, fig. 3B). Similarly, ThyX is present in both Archaea and Bacteria,19 suggesting that ThyX could have been present in LUCA. The overall distribution of ribonucleotide reductases and thymidylate synthases thus seems to favor a LUCA with a T-DNA genome, in agreement with the presence of the clamp and clamp loader in LUCA. However, the presence of an orthologous protein in Bacteria and Archaea can be also explained by the monophyly of prokaryotes if the root of the universal tree is in the eukaryal branch (reference 65 and see discussion below). Furthermore, many viral sequences are interspersed with cellular sequences both in the ribonucleotide reductase and thymidylate synthase tree (fig. 3A, fig. 3B). Thus, one cannot exclude that these proteins have been transferred from viruses to the proto-archaeal and bacterial lineages shortly after their divergence from LUCA.
Finally, one should consider the possibility that LUCA had still an RNA genome but already contained retro-transcribed DNA. This hypothesis was proposed by Leipe and coworkers, in an attempt to reconcile the existence of two independent replication mechanisms with the possible presence of DNA in LUCA.48 We will thus discus now specifically the problem of the origin of the two DNA replication mechanisms.
When and Where DNA Replication Mechanisms Originated
If the bacterial and the archaeal/eukaryal versions of DNA replication proteins were already present in LUCA (hypotheses 2), (fig. 6), they might have appeared either successively in the same lineages ancestral to LUCA, or in different lineages (being later on mixed in LUCA or in one of its ancestor by cell fusion or gene transfer). In the first hypothesis, it is unclear how new DNA replication machinery could have been selected in any organism already containing a more evolved version? If the new version was more efficient by chance, why the old one survived? If the bacterial and the archaeal/eukaryal versions of DNA replication proteins appeared in different lineages, one can still imagine that they have evolved different properties explaining their coexistence in a single cell.
The hypothesis 3 implies that the two distinct sets of DNA replication proteins originated after LUCA, one in a proto-bacterium and one in a common lineage to Archaea and Eukarya (fig. 6). This is in nice agreement with the classical rooting of the universal tree of life in the bacterial branch. However, this rooting is highly disputed.66,67 It has been shown that phylogenetic data that support this rooting are not valid (which does not mean that this rooting is wrong) and other hypotheses have been proposed, such as an eukaryal rooting,65 or a fusion between a proto-bacterium and a proto-archaeon to give Eukarya.68
Even if the bacterial rooting is correct, the hypothesis 3 did not explain the distribution of some DNA replication proteins, such as type II DNA topoisomerases or DNA polymerases, that can be also divided into nonhomologous families. The case of type II DNA topoisomerases is illuminating. Although these elaborated enzymes catalyze a complicated reaction (the crossing of a double helix by another DNA duplex) two versions have been invented independently.59 This has been shown by the discovery in Archaea of an atypical topoisomerase (DNA topoisomerase VI, prototype of Topo IIB) whose modular organization and mechanism of action turned out to be distinct from classical type II DNA topoisomerases (Topo IIA.)69 The existence of two families of nonhomologous type II DNA topoisomerases is a priori in line with the independent invention of two sets of nonhomologous DNA replication proteins. However, the phylogenomic distribution of Topo IIA and Topo IIB did not fit with those of other DNA replication proteins. Topo IIB is present in Archaea and plants, whereas Topo IIA is present in Bacteria and Eukarya (with few recent transfers from Bacteria to Archaea). The situation is even more complex in the case of DNA polymerases, since seven nonorthologous families have already been recognized (A, B, C, D, X, Y, the pRN2 plasmid polymerase). Bacterial replicases are of the C family, whereas archaeal replicases are either from the B or D families, and eukaryal polymerases from the B family. In a tree of family B, archaeal and eukaryal DNA polymerases do not form a clade, but the three eukaryal replicases (α,δ and ε) are interspersed with archaeal DNA polymerases and many groups of DNA polymerases from bacteriophages and animal viruses (fig. 3C).25 These atypical distributions can be reconciled with the different hypotheses that have been proposed for the universal tree of life only by introducing ancient gene transfers and gene losses, as well as nonorthologous displacements, suggesting a scenario for the origin of DNA replication proteins more complex than hypothesis 3.
The hypothesis 4 (fig. 6) is based on the observation, from comparative genomics, that replacement of a protein belonging to a given family by a protein of similar function but belonging to another family (nonorthologous or even nonhomologous) has frequently occurred during genome evolution.47 In particular, phylogenomic analyses of replication proteins also revealed that nonorthologous displacement occurred during the evolution of Archaea. The archaeal Topo II (family Topo IIB) has thus been “recently” displaced in Archaea of the order Thermoplasmatales by bacterial DNA gyrase (Topo IIA).59 More ancient displacements have occurred between the two archaeal phyla. Thus, the eukaryal version of the single-strand binding (ssb) complex (RPA) that is present in Euryarchaea has been displaced in Crenarchaea by a novel form of ssb protein (or vice versa).70 Similarly, one should refer to nonorthologous displacement to explain why Crenarchaea and Euryarchaea have probably different DNA replicases (belonging to DNA polymerase B and D families, respectively).71 If nonorthologous displacement occurred during the diversification of Archaea, more drastic one (replacement of a nearly complete system by another, possibly carried by viral genomes) might well have occurred early on, during domain diversification, especially at a time when lateral gene transfer were more frequent and when primitive replication systems were probably even more plastic.
Interestingly, nonhomologous DNA polymerases of the B and D families interact at the replication forks with proteins that are homologous in Crenarchaea and Euryarchaea (fig. 5). Similarly, a small DNA polymerase subunit present in Archaea and Eukarya can interact with catalytic subunits of either phylogenetically unrelated DNA polymerases B or D.72 All these observations clearly indicate that nonorthologous displacement can affect interacting proteins of the replication complex at the forks. This could explain why the clamp and the clamp loader are still homologous in the bacterial and archaeal replication systems, if other elements have been displaced in the course of evolution.
Nonorthologous displacement can have also played an important role in modifying the relative rate of evolution of proteins that remained orthologues.65 For example, if several ancestral replication proteins have been displaced in Bacteria, proteins that remain orthologues in the three domains (such as the clamp and the clamp loader) will become more divergent in Bacteria since they have coevolved now with different partners, without regards to the real phylogenetic relationships between the three domains.
A Viral Origin for Cellular DNA Replication Proteins?
If DNA and DNA replication proteins originated in viruses,7 one can imagine that DNA replication mechanisms have been transferred from viruses to cells (fig. 2). This possibly occurred either at different stages of viral evolution (giving birth to various types of cellular lineages with different DNA replication modes), or only at the symmetric stage. In the latter case, the first DNA cells would have had an immediate advantage compared to RNA cells still using the asymmetric mode of RNA replication. In particular, the symmetric mode allows the replication of large cellular DNA genomes without accumulating excessive amount of unstable single-stranded DNA. Hypotheses 4 and 5 are in agreement with these ideas (also hypothesis 4 can also be accommodated with the idea that viral DNA replication proteins are of cellular origin but diverged extensively from their ancestral versions during viral evolution (before being transferred back to some cell lineages.)
Many of the problems that beset previous hypotheses can be solved in the framework of the viral origin of DNA and DNA replication theory. It is no more necessary to explain why two distinct sets of nonhomologous DNA replication proteins with similar function coexisted in the same cell (hypothesis 2), neither to refer to the universal tree of life based on rRNA (hypothesis 3). The existence of puzzling and contradictory phylogenies for many DNA replication proteins is now readily explained by suggesting that different elements of the replisome have been recruited independently from different viruses. Finally, the origin of the proteins involved in the nonorthologous displacement postulated in hypothesis 4 is clearly identified. The implication of viruses in a massive nonorthologous displacement is appealing, since DNA replication proteins encoded by DNA viruses often form gene clusters that facilitate their transfer in a single step from a virus to its host.
Figure 6 illustrates two scenarios for the viral origin of cellular DNA replication proteins. In the first case (hypotheses 5), all DNA replication proteins originated from viruses, after the separation of the archaeal and bacterial lineages, in agreement with an RNA based LUCA, whereas in the other (hypothesis 4-5) a first transfer occurred before LUCA, and a second one occurred in the bacterial branch (post-LUCA). The second step corresponds to the nonorthologous displacement of hypothesis 4.
Many protein phylogenies support the idea of ancient transfers of replicative proteins between cells and viruses. As previously mentioned, it is striking that the various subtypes of eukaryal DNA polymerases in the B family (α, δ, ε) are not grouped together in phylogenetic trees but interspersed with archaeal DNA polymerases, bacteriophage T4 DNA polymerases, and various groups of viral DNA polymerases.25 Furthermore, DNA polymerase δ branches off a group of viruses including Iridovirus in phylogenetic trees of DNA polymerases B (fig. 3C).25,53 The phylogenetic tree of type II DNA topoisomerases of the A family can also be explain by a viral origin. Indeed, both bacterial and eukaryal Topo IIA emerged independently from a group of viral sequences that include both bacteriophages and eukaryal viruses (fig. 3D).59
These phylogenies clearly indicate that ancient transfers actually occurred between viruses and cells. Unfortunately, as in the case of thymidylate synthases and ribonucleotide reductases, the direction of these transfers (from cells to viruses or from viruses to cells) cannot be determined with complete confidence, since viral lineages have usually long branches that can be attracted by outgroup sequences, artificially separating viruses from cellular domains.
However, it is noteworthy that such a global replacement of cellular proteins by viral-encoded functional analogs actually occurred in the course of mitochondial evolution. Indeed, both the mitochondrial RNA polymerase that initiate replication of the H-strand, the mitochondrial DNA polymerase γ, and the mitochondrial primase that initiates on the L strand, are phylogenetically related to viral homologues of the T3/T7 superfamily (refs. 25, 73, and unpublished result from this laboratory), clearly indicating that nonorthologous displacement of cellular DNA replication proteins by viral ones is possible. It is remarkable that the present-day mitochondrial genome in yeast and mammals replicate via a semi-asymmetric mode, instead to replicate via the symmetric mode of the proteobacterial genomes, suggesting that a virus (or a plasmid) take over DNA replication in mitochondria both in term of proteins and replication mode.
Evolution of Specific Mechanisms Associated to Cellular DNA Replication: Two Case Studies
Further progress in our understanding of the origin and evolution of DNA and DNA replication apparatus will certainly benefit from the sequencing of new genomes, especially from protists and viruses. However, it will be also critical to deepen our understanding of the “historical logic” hidden in various facets of the replication process itself. This will require more experimental data on a great variety of systems to get new insights from comparative molecular biology. We will finish this chapter by discussing two examples that illustrate this point.
The first one refers to the different sets of proteins performing the synthesis and processing of Okazaki fragments in Bacteria, and Eukarya (fig. 7).74 In Bacteria>, DNA polymerase III directly used the RNA primers synthesized by the primase DnaG (a monomer) to produce at once full-length Okazaki fragment (about 1000 base pairs). A single protein, DNA Polymerase I, can both eliminate the RNA primer via its 5' to 3' exonuclease module, and fill the gap with its polymerase activity. The mechanism of Okazaki fragment synthesis and processing in Eukarya appears to be more complex and less “rational”, even bizarre. The RNA primer synthesized by the eukaryal primase is first elongated in vivo by DNA polymerase α to produce a short RNA-DNA primer (about 30 base pairs) that is extended into a full length Okazaki fragment (about 100-150 base pairs) by DNA polymerases δ. The role of DNA polymerase α is puzzling, since DNA polymerase δ can extend an RNA primer to a full length Okazaki fragment in vitro.54 Furthermore, DNA polymerase α lacks the editing 3' to 5' exonuclease activity required for faithful DNA synthesis. As a consequence, the processing of Okazaki fragments in Eukarya requires the removal of the RNA primer and, also, of the stretch of DNA containing possible errors that has been synthesized by DNA Polymerase α (fig. 8). This involves the successive action of three proteins: RPA, Dna2 and FEN-1. DNA polymerase δ first displaces the RNA primer and the DNA portion synthesized by DNA Polymerase α. The displaced single-stranded DNA is then covered with RPA that both inhibits further progression by DNA polymerase δ and recruits the Dna2 protein. The displaced strand can then be cleaved by the endonuclease activity of Dna2, leaving a short single-stranded tail, which is finally degraded by the flap-endonuclease FEN-1.
Interestingly, RPA, Dna2 and FEN-1 are conserved in Archaea, despite the apparent absence of DNA Polymerase α ortholog in archaeal genomes! In the traditional view of evolution (from simple prokaryotes to complex eukaryotes), the eukaryal replication system is an improved version of the archaeal one. What is the significance of this? What improvement is gained from the introduction of an unfaithful DNA polymerase in the system? Could it be that the archaeal system is in fact derived from the eukaryotic one, and that the mechanism of Okazaki fragment processing in Archaea is a relic of the time when Pol α was still present? To answer this question, we should know more about the role of the Pol α and other actors in both the eukaryal and the archaeal systems! In general, the archaeal DNA replication system is not only a simpler version of the eukaryal one (with a smaller number of polypeptides to perform the same function) but also a more efficient one. For example, the rate of elongation is as fast in Archaea as in Bacteria,49 although the sizes of Okazaki fragments are similar in Archaea and Eukarya (much shorter than in Bacteria, see fig. 7).75
The second story refers to the structure of the bacterial clamp loader.76 We have seen that, although the clamps and clamp loaders are homologues in the three domains, they interact with nonhomologous replicative proteins in Bacteria on one side and Archaea/Eukarya on the other (in particular with DNA polymerases of different families). In E. Coli, the clamp loader contains subunits called τ, γ, δ, δ' that are homologous to archaeal/eukaryal RFC proteins. The same gene (dnaX) encodes for the γ and τ subunits. The protein τ (71 kDa) is the full-length protein, whereas γ is a truncated version (47 kDa) due to a translational frameshift followed by a stop codon. The C-terminal amino-acid extension of 24 kDa in τ has been added to the clamp loader during bacterial evolution since it has no homolog in archaeal or eukaryal RFC proteins. This extension allows the dimeric clamp loader to connect the clamp loader to the helicase (DnaB) and the two replicases (Pol III) (fig. 9). What is the reason for this? One can argue that it helps to structure the bacterial replisome, or that it compensates for the absence of one of the two types of bacterial replicase (PolC) that are found in other Bacteria (e.g., Bacillus subtilis).77 On the other hand, in the hypothesis of nonorthologous displacement of ancestral replication proteins by DnaB and Pol III, one could imagine that this C-terminal extension is the trick found by these proteins to force the cellular clamp loader to interact with them, instead of interacting with the ancestral cellular machinery (much like the P protein of bacteriophage γ force the bacterial initiator protein DnaA to interact with it instead to DnaC). However, this scenario is challenged by the restricted distribution of this C-terminal extension in the bacterial domain. Clearly, we would like to know more about the different types of replisomes that are present in Bacteria and to understand how they are evolutionary related to figure out the signification of such oddities as the dnaX gene!
Conclusion and Future Prospects
Up to now, most scientists interested in the studying of DNA replication have not been apparently concerned by the problem of the origin and evolution of this central cellular mechanism. The problem of the origin of DNA is also largely ignored, with few exceptions. It is striking that recent hypotheses on the evolution of DNA replication have been proposed by evolutionists involved in comparative genomics, and not by people actively involved at the bench in the molecular study of DNA replication. The same is also true for transcription. In contrast, scientists working on translation have a long lasting interest in the origin and evolution of the genetic code and the translation apparatus. The central role of RNA in both the origin of life theory and the mechanism of protein synthesis can explain this trends, reinforced by the role that 16S/18S rRNA have played in evolutionary constructions. However, thanks to comparative genomics, scientists working on DNA replication on various model systems should now be encouraged to grasp a new cultural attitude and realize that their work is not only important to understand the functioning of modern cells, identify new drugs targets, or design new products for biotechnology, but that it is also critical for understanding the history of life. They should not only try to adapt their findings to current evolutionary theories, but also try to detect possibilities to check the validity of these theories in these findings. As we have seen in this chapter, there is no lack of alternative, and sometimes contradictory, hypotheses. We have emphasized the importance that viruses could have played in the story since their role is usually ignored or underestimated. In any case, viral replication systems should not be only considered as simple model system, giving possible clue to more complex cellular ones, but as mechanisms interesting to study on their own, as witnesses of critical aspects of early life evolution. The availability of many more replication protein sequences from viruses of the three domains of life and new methods to analyze viral protein phylogenies will possibly help to critically test some of the hypotheses we propose. Their comparison with other replication systems will certainly be productive at the end, if done with an evolutionary oriented mind.
Acknowledgments
Work on DNA replication in our laboratory was supported by grants from l'Association de Recherche contre le Cancer (ARC) and the Programme de Recherche Fondamentale en Microbiologie et Maladies infectieuses et parasitaires (PRFMMIP) of the Ministère de Education et de la Recherche.
References
- 1.
- Monod J. Chance and Necessity: An Essay on the Natural Philosophy of Modern BiologyNew York: Knopf,1971 .
- 2.
- Watson JD, Crick FHC. The structure of DNA. Cold Spring Harbor Symp Quant Biol. 1953;18:123–113. [PubMed: 13168976]
- 3.
- Shroedinger E. What is life, the physical aspect of the living cell. Cambridge. 1944
- 4.
- Lazcano A, Guerrero R, Margulis L. et al. The evolutionary transition from RNA to DNA in early cells. J Mol Evol. 1988;27:283–290. [PubMed: 2464698]
- 5.
- Olsen GJ, Woese CR. Archaeal genomics: an overview. Cell. 1997;89:991–994. [PubMed: 9215619]
- 6.
- Forterre P. 2001, Genomic and early cellular evolution. The origin of the DNA world. CR Acad Sci Paris Life Sciences. 2001;324:1067–1076. [PubMed: 11803805]
- 7.
- Forterre P. Origin of DNA and DNA genomes. Curr Opin in Microbiol. 2002;5:525–532. [PubMed: 12354562]
- 8.
- Jacob F. Evolution and thinkering. Science. 1997;196:1161–1166. [PubMed: 860134]
- 9.
- Poole AM, Logan DT, Sjöberg B-M. The evolution of ribonucleotide reductase: much ado about oxygen. J Mol Evol. 2002;55: 180–196. [PubMed: 12107594]
- 10.
- Takahashi I, Marmur J. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature. 1963;197:794–5. [PubMed: 13980287]
- 11.
- Kornberg A, Baker T. DNA replicationNew York: Freeman and Company,1992 .
- 12.
- Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. [PubMed: 12016304]
- 13.
- Freeland SJ, Knight R, Landweber LF. Do proteins predate DNA? Science. 1999;286:690–692. [PubMed: 10577226]
- 14.
- Poole A, Penny D, Sjöberg B-M. Methyl-RNA: Evolutionary bridge between RNA and DNA? Chemistry and Biology. 2000;7:207–216. [PMC free article: PMC7172353] [PubMed: 11137821]
- 15.
- Poole A, Penny D, Sjöberg B-M. Confounded cytosine! Tinkering and the evolution of DNA. Nature Rev Mol Cell Biol. 2001;2:147–151. [PubMed: 11252956]
- 16.
- Stubbe JA. Ribonucleotide reductases: The link between an RNA and a DNA world? Current Opin Structural Biol. 2000;10:731–773. [PubMed: 11114511]
- 17.
- Eklund H, Uhin U, Farnegardh M. et al. Structutre and function of the radical enzyme ribonucleotide reductase. Prog Biophys Mol Biol. 2001;77:177–268. [PubMed: 11796141]
- 18.
- Fontecave M, Mulliez E, Logan DT. Deoxyribonucleotide synthesis in anaerobic microorganisms: the class III ribonucleotide reductase. Prog Nucleic Acid Res and Mol Biol. 2002;72:95–128. [PubMed: 12206460]
- 19.
- Myllykallio H, Lipowski G, Leduc D. et al. An Alternative Flavin-Dependent Mechanism for Thymidylate Synthesis. Science. 2002;297:105–107. [PubMed: 12029065]
- 20.
- Murzin AG. DNA building blocks reinvented. Science. 2002;297:61–62. [PubMed: 12029066]
- 21.
- Song HK, Sohn SH, Suh SW. Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex. EMBO J. 1999;18:1104–1113. [PMC free article: PMC1171202] [PubMed: 10064578]
- 22.
- Lazcano A, Valverde V, Hernandez G. et al. On the early emergence of reverse transcription : theeoretical basis and experimental evidence. J Mol Evol. 1992;35:524–536. [PubMed: 1282161]
- 23.
- Wintersberger U, Wintersberger E. RNA makes DNA : A speculative view of the evolution of DNA replication mechanisms. Trends in Genet. 1987;3:198–202.
- 24.
- Ng KK, Cherney MM, Vazquez AL. et al. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J Biol Chem. 2002;277:1381–1387. [PubMed: 11677245]
- 25.
- Filee J, Forterre P, Sen-Lin T. et al. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol. 2002;54:763–773. [PubMed: 12029358]
- 26.
- Forterre P. New hypotheses about the origins of viruses, prokaryotes and eukaryotesIn: Trân Thanh Vân JK, Mounolou JC, Shneider J and Mc Kay C, eds.Gif-sur-Yvette, France: Editions Frontières 1992221–234.
- 27.
- Bamford DH, Burnett RM, Stuart DI. Evolution of viral structure. Theor Popul Biol. 2002;61:461–470. [PubMed: 12167365]
- 28.
- Peng X, Blum H, Qhe Q. et al. Sequence and replication of genomes of the archaeal Rudivirus SIRV1 and SIRV2: relationships to the archaeal Lipothrixvirus SIFV and some eukaryal viruses. Virology. 2001;291:226–234. [PubMed: 11878892]
- 29.
- Benson SD, Bamford JKH, Bamford D. et al. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. [PubMed: 10499799]
- 30.
- Butcher SJ, Grimes JM, Makeyev EV. et al. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. [PubMed: 11242087]
- 31.
- De PamphilisML. DNA replication in eukaryotic cells. Cold spring Harbor Laboratory Press. 1996
- 32.
- Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. [PMC free article: PMC312478] [PubMed: 1630899]
- 33.
- Bocquier AA, Liu L, Cann IK. et al. Archaeal primase: bridging the gap between RNA and DNA polymerases. Curr Biol. 2001;11:452–456. [PubMed: 11301257]
- 34.
- Ohmori H, Friedberg EC, Fuchs RP. et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. [PubMed: 11515498]
- 35.
- Kirk BW, Kuchta RD. Arg304 of human DNA primase is a key contributor to catalysis and NTP binding: primase and the family X polymerases share significant sequence homology. Biochemistry. 1999;38:7727–7736. [PubMed: 10387012]
- 36.
- Kato M, Frick DN, Lee J. et al. A complex of the bacteriophage T7 primase-helicase and DNA polymerase directs primer utilization. J Biol Chem. 2001;276:21809–1820. [PubMed: 11279245]
- 37.
- Lehman IR, Boehmer PE. Replication of herpes simplex virus DNA. J Biol Chem. 1999;274:28059–18062. [PubMed: 10497152]
- 38.
- Dracheva S, Koonin EV, Crute JJ. Identification of the primase active site of the herpes simplex virus type 1 helicase-primase. J Biol Chem. 1995;270:14148–14153. [PubMed: 7775476]
- 39.
- Trakselis MA, Mayer MU, Ishmael FT. et al. Dynamic protein interactions in the bacteriophage T4 replisome. Trends Biochem Sci. 2001;26:566–572 Review. [PubMed: 11551794]
- 40.
- Mesyanzhinov VV, Robben J, Grymonprez B. et al. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J Mol Biol. 2002;317:1–19. [PubMed: 11916376]
- 41.
- Lakshminarayan MI, Aravind L, Koonin E. Common origin of four large families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. [PMC free article: PMC114758] [PubMed: 11689653]
- 42.
- Waga S, Stillman B. The 46, 48DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. [PubMed: 9759502]
- 43.
- Keck JL, Berger JM. DNA replication at high resolution. Chem Biol. 2000;7(3):R63–71. [PubMed: 10712935]
- 44.
- Bohlke K, Pisani FM, Rossi M. et al. Archaeal DNA replication: spotlight on a rapidly moving field. Extremophiles. 2002;6:1–14. [PubMed: 11878556]
- 45.
- Matsunaga F, Forterre P, Ishino Y. et al. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc Natl Acad Sci USA. 2001;98:11152–11157. [PMC free article: PMC58699] [PubMed: 11562464]
- 46.
- Edgell DF, Doolittle WF. Archaea and the origin[s] of DNA replication proteins. Cell. 1997;89:995–998. [PubMed: 9215620]
- 47.
- Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA. 1996;93:10268–10273. [PMC free article: PMC38373] [PubMed: 8816789]
- 48.
- Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res 199927|3389-3401. [PMC free article: PMC148579] [PubMed: 10446225]
- 49.
- Myllykallio H, Lopez P, Lopez-Garcia P. et al. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. [PubMed: 10864870]
- 50.
- Zivanovic Y, Lopez P, Philippe H. et al. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 2002;30:1902–1910. [PMC free article: PMC113857] [PubMed: 11972326]
- 51.
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol. 2002;52:7–76. [PubMed: 11837318]
- 52.
- Forterre P. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol Microbiol. 1999;33:457–465. [PubMed: 10417637]
- 53.
- Villarreal LP, DeFilippis A. Hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J Virol. 2000;74:7079–7084. [PMC free article: PMC112226] [PubMed: 10888648]
- 54.
- Takemura M. Poxviruses and the origin of the eukaryotic nucleus. J Mol Evol. 2001;52:419–425. [PubMed: 11443345]
- 55.
- Bell PJL. Viral eukaryogenesis: Was the ancestor of the nucleus a complex DNA virus? J Mol Evol. 200;5:251–256. [PubMed: 11523012]
- 56.
- Keck JL, Roche DD, Lynch AS. et al. Structure of the RNA polymerase domain of E. coli primase. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. [PubMed: 10741967]
- 57.
- Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21:4763–4773. [PMC free article: PMC126292] [PubMed: 12234917]
- 58.
- Woese CR, Fox GE. The concept of cellular evolution. J Mol Evol. 1977;1:1–6. [PubMed: 903983]
- 59.
- Forterre P, Benhachenou N, Confalonieri F. et al. The nature of the last universal ancestor and the universal tree of life, still open questions. Biosystem. 1993;28:15–32-LUCA DNA. [PubMed: 1337989]
- 60.
- Gadelle D, Filée J, Bulher C. et al. Phylogenomics of type II DNA topoisomerases. Bioassay. 2003;25:232–242. [PubMed: 12596227]
- 61.
- MacNaughton TB, Shi ST, Modahl LE. et al. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA Polymerases. J Virol. 2002;76:3920–3927. [PMC free article: PMC136092] [PubMed: 11907231]
- 62.
- Moraleda G, Taylor J. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J Virol. 2001;75:10161–10169. [PMC free article: PMC114590] [PubMed: 11581384]
- 63.
- Wang H, Di GateRJ, Seeman NC. An RNA topoisomerase. Proc Natl Acad Sci USA. 1996;93:9477–9482. [PMC free article: PMC38453] [PubMed: 8790355]
- 64.
- Davey MJ, Jeruzalmi D, Kuriyan J. et al. Motors and switches: AAA+ machines within the replisome. Nat Rev Mol Cell Biol. 2002;3:826–835. [PubMed: 12415300]
- 65.
- Brinckman H, Philippe H. Archaea sister group of Bacteria ? Indications from tree reconstruction artifacts in ancient phylogenies. Mol Biol Evol. 1999;16:817–825. [PubMed: 10368959]
- 66.
- Forterre P, Philippe H. Where is the root of the universal tree of life? Bioessays. 1999;21:871–879. [PubMed: 10497338]
- 67.
- Gribaldo S, Philippe H. Ancient phylogenetic relationships. Theor Pop Biol. 2002;61:391–408. [PubMed: 12167360]
- 68.
- Lopez-Garcia P, Moreira D. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci. 1999;24:88–93. [PubMed: 10203753]
- 69.
- Bergerat A, de MassyB, Gadelle D. et al. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. [PubMed: 9121560]
- 70.
- Wadsworth RI, White MF. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 2001;29:914–920. [PMC free article: PMC29618] [PubMed: 11160923]
- 71.
- Myllykallio H, Forterre P. Mapping of a chromosome replication origin in an archaeon: Response. Trends Microbiol. 2000;8:537–539. [PubMed: 11201257]
- 72.
- Makiniemi M, Pospiech H, Kilpelainen S. et al. A novel family of DNA-polymerase- associated B subunits. Trends Biochem Sci. 1999;24:14–16. [PubMed: 10087916]
- 73.
- Moreira D. Multiple independent horizontal transfers of informational genes from bacteria to plasmids and phages implications for the origin of bacterial replication machinery. Mol Microbiol. 2000;35:1–5. [PubMed: 10632872]
- 74.
- MacNeill SA. DNA replication: partners in the Okazaki two-step. Current Biol. 2001;11:R842–R844. [PubMed: 11676941]
- 75.
- Matsunaga F, Norais C, Forterre P. et al. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Report. 2003;4:154–158. [PMC free article: PMC1315830] [PubMed: 12612604]
- 76.
- O'Donnell M, Jeruzalmi D, Kuriyan J. Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr Biol. 2000;11:R935–R946. [PubMed: 11719243]
- 77.
- Dervyn E, Suski C, Daniel R. et al. Two essential DNA polymerases at the bacterial replication fork. Science. 2001;294:1716–1719. [PubMed: 11721055]
- 78.
- Lipps G, Röther S, Hart C. et al. A novel type of replicative enzyme barbouring ATPase, primase and DNA polymerase activity. EMBO J. 2003;22:2516–2525. [PMC free article: PMC156004] [PubMed: 12743045]
- Origin and Evolution of DNA and DNA Replication Machineries - Madame Curie Biosc...Origin and Evolution of DNA and DNA Replication Machineries - Madame Curie Bioscience Database
- Polyomavirus Allograft Nephropathy: Clinico-Pathological Correlations - Madame C...Polyomavirus Allograft Nephropathy: Clinico-Pathological Correlations - Madame Curie Bioscience Database
- STATs and Infection - Madame Curie Bioscience DatabaseSTATs and Infection - Madame Curie Bioscience Database
- Regulation of Medical Devices - Madame Curie Bioscience DatabaseRegulation of Medical Devices - Madame Curie Bioscience Database
- Calreticulin in Cytotoxic Lymphocyte-Mediated Cytotoxicity - Madame Curie Biosci...Calreticulin in Cytotoxic Lymphocyte-Mediated Cytotoxicity - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...