NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Asparaginylation of tRNA presents the unusual character that it can be performed by two distinct pathways. A direct, and probably modern one, consisting in direct attachment of Asn on tRNAAsn by asparaginyl-tRNA synthetase (AsnRS), and an indirect, likely ancestral one, in which a mischarged Asp-tRNAAsn, synthesized by a non-discriminating aspartyl-tRNA synthetase (AspRS), is converted into Asn-tRNAAsn by a tRNA-dependent amidotransferase (AdT). This chapter describes the structural and functional properties of enzymes and tRNAs involved in both pathways. AsnRS, the key player in the direct route, is grouped with AspRS and lysyl-tRNA synthetase (LysRS) in subclass IIb aminoacyl-tRNA synthetases displaying a similar modular organization. Despite a remarkable extent of structural and functional similarities, AsnRS and AspRS adapted their mode of recognition to ensure selective binding of the cognate amino acid. Conversely, the enzymes of the indirect pathway (AspRS and AdT) exhibit relaxed specificity. The non-discriminating AspRS is involved in tRNA aspartylation and asparaginylation, whereas AdT can ensure glutaminylation of tRNA in addition to asparaginylation. These features suggest the close functional interrelation between primitive pathways of tRNA aminoacylation and amino acid synthesis. The origin of AsnRS and the interrelation between the direct and indirect pathways of tRNA asparaginylation are investigated on the basis of the distribution of both pathways in the living kingdom and an extended phylogenic analysis.
Historical Overview
Asparaginylation of transfer RNA (tRNA) has first been evidenced in Lactobacillus arabinosus1 and in Escherichia coli,2,3 and later in eukaryotes (Neurospora crassa4 and rabbit liver5) and found associated to a 90-100 kDa protein. Asparaginyl-tRNA synthetase (AsnRS) purified from Bacillus stearothermophilus6,7 was characterized as an α2 homodimer (α = 51 KDa). The AsnRSs isolated from various organisms showed conservation of the oligomeric α2 structure.
The first studies on AsnRS were devoted to obtention of physicochemical and elementary kinetic information. Investigation of the structural properties of B. stearothermophilus AsnRS by centrifugation7 revealed sedimentation and diffusion coefficients of 6.6 S and 5.1 x 10-7 cm2.s. The Mr of 127 000 determined by centrifugation was in agreement with that determined by gel-filtration (120 000), suggesting a globular shape of the protein (Stokes radius 41.5 Å). Comparable values were reported for the enzyme from Chinese hamster ovary (CHO) cells. 8
The kinetic constants determined for AsnRSs from E. coli, Thermus thermophilus and mammalians are among the smallest values determined for this family of enzymes. AsnRS from E. coli displays KM values for asparagine (Asn) of 15 and 29 μM in ATP-PPi exchange and tRNA aminoacylation, and 500 and 76 μM for ATP.9 Comparable values were determined for the enzyme from CHO cells.8KM's for tRNAAsn were also determined for AsnRSs from T. thermophilus and CHO cells (0.048 and 0.060 μM).8,10 The sole known mechanistic information concerns the rate-limiting step of tRNA asparaginylation obtained by comparing the rate constants (kcat) of ATP-PPi exchange and tRNA aminoacylation. Since E. coli AsnRS catalyzes ATP-PPi exchange faster than tRNA charging (2.2 and 1.3 s-1 at 37 °C), either transfer of the activated amino acid (aa) on tRNA or dissociation of asparaginyl-tRNAAsn (Asn-tRNAAsn) determine the overall reaction rate.9 Preliminarykinetic investigations of eukaryotic AsnRS suggested involvement of tRNA in aa activation11 but AsnRSs, like most aminoacyl-tRNA synthetases (aaRS), activate aa in the absence of tRNA. Until now, no additional mechanistic data are available on this aminoacylation system. However, through obtention of vast number of AsnRS sequences and resolution of the 3D structure of one of them, more is known about structural properties of this aaRS.
The first AsnRS sequenced was that from E. coli12 but was one of the last aaRSs sequenced from this organism. Sequence analysis revealed extensive similarities with AspRSs,12,13 and to a lower extent, also with LysRSs,14 indicating a strong structural and probably also phylogenetic interrelation between these aaRSs. Crystals suitable for analysis were obtained from T. thermophilus AsnRS,15 making this enzyme the twelfth representative of the 20 aaRSs whose 3D structure was solved.16
Knowledge of an increasing number of genomic sequences allowed comparison of the structural properties of AsnRS from various origins. Their polypeptide chains comprise 447 residues (for AsnRS of Lactobacillus lacti the smallest one, of Mr 50 808) to 638 residues (for AsnRS of Arabidopsis thaliana the largest one, of Mr 71 319), with pI values ranging from 4.8 to 9.3. Like in other aaRSs, the eukaryotic chains contain an appended N-terminal (Nt) extension and are longer than the prokaryotic ones. The sequences display the 3 consensus motifs characterizing class II aaRSs.17 Comparison of the 3D structure of T. thermophilus AsnRS with those of other class II aaRSs shows a modular organization highly similar to that of AspRSs and LysRSs, but different from that of other class II aaRSs. In AsnRSs, AspRSs and LysRSs, grouped in subclass IIb, part of the Nt-appendix (approximatively 140 residues) is organized in a β-barrel involved in recognition of the tRNA anticodon.18 Like other class II aaRSs, AsnRS attaches the amino acid on the 3'OH of the terminal adenosine of tRNA.19,20
Recent biochemical investigations have shown that, in prokaryotes, Asn-tRNAAsn can be formed without AsnRS, by a mechanism similar to the indirect pathway of glutaminyl-tRNAGln (Gln-tRNAGln) formation, originally described in Bacillus megaterium.21-23 Analysis of whole genome sequences shows absence of an AsnRS gene in a number of prokaryotes, suggesting a wider use of this atypical pathway in Asn-tRNAAsn formation than initially expected. The existence of alternate pathways of tRNA asparaginylation, one direct and probably modern, and another one, indirect and likely primitive, shed a new light on the evolution of this aminoacylation system.
Structure and Properties of Asparaginyl-tRNA Synthetases
The 3D Structure of Thermus thermophilus Asparaginyl-tRNA Synthetase
Among AsnRSs of various origins assayed for crystallization, only the enzyme from T. thermophilus gave crystals suitable for analysis.15 This agrees with the remarkable ability of thermostable proteins to crystallize.24 The enzyme expressed in E. coli crystallized in polyethylene glycol 6000 solutions and its structure was solved from isomorphous unanium derivatives of crystals diffracting at 2.6 Å of resolution.15,16 Soaking of these crystals in ATP•Mg2+ solution provoked disordering of the P6422 crystal and resulted in a new crystal form with a P6522 hexagonal space group. In this crystal, the dimer becomes the asymmetric subunit, as a probable consequence from asymmetric binding of ATP on each subunit. This crystal reverted to the initial form diffracting at 2.95 Å when the ATP sites from both subunits were fully occupied. In contrast to other class II aaRSs, the three Mg2+ ions were clearly localized, probably because of the low salt concentration of the crystallization medium. Cocrystallization of the enzyme with ATP, Mg2+ and Asn resulted in obtention of crystals containing asparaginyl-adenylate (Asn∼AMP) diffracting only at 3.2 Å.16 Finally the structure of AsnRS complexed to the activated aa was solved from crystals containing the non hydrolyzable 5'-O-(N-asparaginyl-sulfamoyl)adenosine analog diffracting at 2.65 Å.16
The modular organization of each subunit resembles that of AspRS and LysRS and consists in a Nt β-barrel connected by a small hinge region to the larger C-terminal (Ct) domain built upon an αβ fold (fig. 1).16
The N-terminal domain (residues 1 to 97) is formed by a five-stranded β-barrel (strands S1 to S5), in which strands S3 and S4 are separated by a α helix (H0). The 3D structures of yeast AspRS25 and T. thermophilus AspRS26 and LysRS27 complexed with the tRNA indicate, by analogy, that this domain binds the tRNA anticodon. This β-barrel which characterizes subclass IIb aaRSs is functionally equivalent to the mixed αβ fold located in the C-terminus of subclass IIa aaRSs, except for seryl-tRNA synthetase deprived of this domain. The Nt segment, preceding strand S1, and of variable length in subclass IIb aaRSs, is significantly longer in eukaryotes than in prokaryotes. It includes a 310 helix (G1) of 7 residues in the thermophilic enzyme.
The hinge region (residues 98 to 123), formed by a turn followed by two a helices (H1 and H2), connects the Nt to the Ct domain. This region resembles structurally that of LysRSs whereas in AspRSs, the longer and more globular domain contains additional helices.
The C-terminal domain (residues 124 to 438) is built upon a αβ fold including the three class-defining motifs. It displays the catalytic site formed by a six-stranded antiparallel β-sheet (A2, A3 and A5 to A8) and by the dimer interface which includes motif 1. This β-sheet is interrupted between motifs 2 and 3 by an insertion of 62 residues (256 to 318) in AsnRS from T. thermophilus. This module, inserted between helices H7 and H8, is formed by four helices (H6 to H9) and a β-strand (A4). The topology of this extra-domain resembles that of eukaryotic AspRSs which is significantly smaller than in prokaryotic AspRSs. The interfaces of helices H3, H5 and H6 contain interacting Asp, Glu and Arg residues forming ion pair clusters which contribute to thermal stability of the thermophilic AsnRS. Such type of stabilization has not been reported for T. thermophilus AspRS and LysRS, but for other thermophilic enzymes.28
Despite strong resemblances of their modular organization, superposition of the catalytic domains of class IIb aaRS reveals angular shifts of the Nt domain which can reach up to 18.8°.16,27,29 (fig. 1).
Contacts between the subunit interfaces. Association of the subunits involves four types of interactions which are mostly preserved in AspRS and LysRS. (i) On the top of the interface, contacts between the Ct domains involve residues from motif 1 distributed along the interfaces of H3 α-helices and the following A1 strands. These interactions, essentially polar, involve Asp, Glu and Arg residues but are reinforced by stacking of the His142 residues from each subunit. (ii) On the bottom of the dimer interface, β-strands I1 and I2 from each monomer form an antiparallel β-sheet. (iii) The internal parts of the interfaces are associated by hydrophobic interactions involving Ile, Leu and Val residues from strands A1, I1 and I2, and Phe 207 of motif 2 from each subunit. (iv) Finally, association is strengthened by inter-subunit contacts between the Ct domain from one subunit and the Nt and hinge domains from the other subunit.
Interaction of Asparaginyl-tRNA Synthetase with ATP•Mg2+, Asparaginyl-Adenylate and its Analog
Subunit interactions link intimately the dimer interface, in particular the conserved motif 1, with the ATP-binding site. ATP interacts as in the other class II aaRSs with the expected contribution of the conserved residues from motifs 2 and 3 (Table 1). Interactions of aa residues with the adenine ring and the γ-phosphate stabilize ATP in the bent-conformation, essential for aa activation by class II aaRSs. Three Mg2+ ions are associated with the triphosphate group. Mg2+1 binds to α and β phosphates and to Asp352 and Glu361, two residues conserved in class II aaRSs (Table 1). Mg2+ 2 and 3 interact with phosphates β and γ, one on each side from phosphoester bond.
Table 1
Amino acid groups of AspRS and AsnRS from T. thermophilus contacting the chemical groups of aa∼AMP.
Cocrystallization of AsnRS with ATP•Mg2+ and Asn leads to formation of Asn∼AMP bound in the active site in an elongated conformation with Mg2+1. The protein residues contacting the substrate groups were identified with the 5'-O-(N-asparaginylsulfamoyl) adenosine analog. The adenosine moiety interacts as in ATP (Table 1 and fig. 2A). The α-amino group of Asn is positioned by Glu164, Ser185 and Gln187, whereas the carbonyle and the amide groups make hydrogen bonds with Arg368 and Glu225, respectively (fig. 2A).
Comparison of the 3D structures of free AsnRS with that complexed with Asn∼AMP shows that binding of the activated aa promotes domain movements in the protein resulting in changes of the enzyme conformation and a more precise delimitation of the catalytic site. Motif 2 loop and the loop preceding β-sheet I1, become more ordered. The first moves in a concerted way with the Ct peptide towards the active site, whereas the latter swings over the catalytic site closing its access and sequestering the activated Asn. The novel interactions shift the two helices H10 and H11 towards the active site and promote rotation by 2.5° of the Nt β-barrel relative to the catalytic domain. This conformation is stabilized by inter-subunit interactions between residues of helix H10 of one subunit and the Nt and hinge domains from the other subunit. This sequence of events, induced by formation of aa∼AMP, was also reported for the other subclass IIb aaRSs.27,29,30
Exposure of motif 2 loop on the enzyme surface and its contribution in stabilization of the complex with the small ligands are supported by experimental evidences. (i) Limited trypsic digestion of E. coli AsnRS generates a stable peptide of Mr 26,000 starting with residue His243 located in this loop.12 (ii) Escherichia coli mutant HO202, in which Pro231 from motif 2 (residue 439 from alignment) is replaced by Leu, exhibits a temperature-dependent increase of the KM for Asn and ATP in ATP-PPi exchange without significant alteration of the kcat.9 The reversible effect of this mutation indicates that Pro231 is not directly involved in substrate binding but more likely in stabilization of the complex and suggests that this residue participates in the positioning of the loop formed by motif 2.
Comparison of Asparaginyl-tRNA Synthetases from Various Phylae and Similarities with Aspartyl-tRNA Synthetases
Figure 3 shows multiple sequence alignments of AsnRSs from all domains of life. Two representatives from each phylum are presented, but the consensus sequence derives from alignment of the 46 known AsnRSs. The modular organization described for the T. thermophilus enzyme is conserved in all AsnRSs. The Nt-, hinge-, and Ct domains, include respectively residues 1 to 272, 273 to 321 and 322 to 921 from consensus sequence. Thirty-four residues are strictly conserved, respectively 4, 1 and 29 in the Nt-, hinge- and catalytic domains. About 122 residues are conserved in 80% of AsnRSs. Most conserved residues are involved in substrate binding or in intra/inter-domain interactions. Thirty of the conserved or semi-conserved residues are also conserved in AspRSs. Table 1 and Figures 2 and 3 summarize the role of the conserved residues in ATP and aa∼AMP binding as revealed by comparison of the 3D structures of AsnRS and AspRS complexed with the ligands.16,30 The role of these residues in AsnRS has, until now, not been investigated by site-directed mutagenesis. The role of only two of these residues has been defined. In yeast strain YHRO19 auxotroph for Asn, which permitted cloning of the AsnRS gene by complementation, Gly479 from motif 3 of AsnRS is substituted by a Ser residue. It was shown that replacement of this conserved residue, adjacent to residues essential for adenylate binding, increases 10 fold KM for Asn.31 Further, substitution in E. coli AsnRS of Tyr426, a residue at the beginning of motif 3, by Ser increases KM for ATP, suggesting implication of this residue in ATP binding.32
With the exception of the appended Nt extension common to all eukaryotic aaRSs (108 to 116 residues in eukaryotic AsnRSs), the alignment does not reveal phylum-specific insertions or extensions in AsnRSs. This contrasts with AspRSs that exhibit species-specific structural signatures.33 The Nt extension, larger in AsnRSs than in AspRSs, contains 9 to 19% of Lys residues. However, Lys-rich sequences were also found in the appended domain of other eukaryotic aaRSs. Experimental evidences have shown implication of the Nt extension in formation of a high molecular weight complex involving AspRS and 8 other aaRSs in metazoans.34 However, AsnRS is found only in free form, excluding such role for its extension. Moreover, presence of an Nt extension in AspRS and AsnRS from yeast, lacking aaRS complexes, argues further for another role for this domain. Modelization and NMR investigations have shown organization of the Lys-rich sequence of yeast and mammalian AspRSs in an amphiphilic α-helix.35,36 Recent results suggest involvement of this fold, probably conserved in eukaryotic AsnRSs, in tRNA binding and in stabilization of the complex with the enzyme. It has been shown that in yeast AspRS, the Lys-rich sequence (residues 32xSKxxLKKxxK42 boxed in the consensus sequence in fig. 3) together with other residues from Nt domain, binds tRNA by the minor groove of the anticodon stem, and increases its affinity for the enzyme.37 Since the consensus sequence is partly conserved in LysRSs and in AsnRSs (fig. 3), this domain may be implicated in selection and recognition of tRNA by aaRSs of subclass IIb. Variable position of this sequence in the Nt extension of the three enzymes might be related to different orientation of the anticodon-binding domain with respect to the catalytic domain (fig. 1).16
Structural Basis for the Discrimination between Asp and Asn by Asparaginyl-tRNA Synthetase
Superposition of AsnRS and AspRS catalytic sites complexed with their homologous aa∼AMP shows that most residues contacting ATP and aa are identical,16,30 and reveals how discrimination between Asp and Asn can occur (fig.2A,B). Specific recognition is directed by a distinct structural context in the two aaRSs related to 3 systematic differences found in all AspRSs and AsnRSs16. (i) A Lys residue conserved in AspRS (Lys204 in T. thermophilus AspRS) is replaced by a small or an uncharged residue in AsnRS (Ala 190 in T. thermophilus AsnRS). (ii) This residue is followed by Gln in AspRS and Glu in AsnRS (Gln205 and Glu191 in T. thermophilus AspRS and AsnRS). (iii) An invariant Asp in motif 2 of AspRS is substituted by Glu in AsnRS (Asp239 and Glu225 in T. thermophilus AspRS and AsnRS).
Interaction of AspRS with Asp is based on the electrostatic complementarity between the negatively charged β-carboxylate group of Asp and the positively charged side-chains of Arg and Lys residues (Arg483 and Lys204 in T. thermophilus AspRS), which are positioned by the carboxylate groups of Glu241 and Asp239 (fig. 2B). This structural context disfavours interaction of the carboxyamide group of Asn which would lead to an unfavorable head-on interaction with the Lys and Arg residues. Conversely, AsnRS lacks the Lys which is substituted by a small side-chained residue (Ala 190), but displays the conserved Arg (Arg368) in the same configuration. However, the unfavorable head-on interaction of the carboxyamide of Asn with the Arg residue is avoided by a 60° rotation of the C-Cα bound which turns the Asn side chain more into the catalytic site. As a consequence, the carbonyle and the amido groups of Asn are hydrogen bounded to Arg368 and the adjacent Glu225 (fig. 2A). The latter residue plays the dual role of recognition of Asn and rejection of the negatively charged β-carboxylate group of Asp. The rotation of the side chain of Asn implies a displacement of its α-amino group which is precisely positioned by three hydrogen bonds with conserved Ser185, Gln187 and Glu164 residues (fig. 2A).
In AspRS, Lys204 fulfils the reverse dual role of Glu225 in AsnRS, by interacting with the negatively charged β-carboxylate of Asp and discriminating against Asn. This residue is positioned by Asp239, shorter than its equivalent Glu225 in AsnRS, and is thus too distant from Asp side chain to provoke electrostatic repulsion (fig. 2B). The residues which in AsnRS orient Asn side chain, are conserved in AspRS (Ser199, Gln201, and Glu177), but subtle changes in the position of their side-chains induce a different orientation of the α-amino group of Asp. In conclusion, a limited number of residues differ in the catalytic sites of the two enzymes and are directly implicated in selection of the homologous aa, but other residues participate additionally in correct orientation of the substrates side-chains and are indirectly involved in their selection.
Organellar Asparaginyl-tRNA Synthetases
Mitochondrial and cytosolic AsnRSs from yeast are encoded by distinct nuclear genes, like for 17 other aaRSs in this organism.38 As a consequence, disruption of the gene encoding the mitochondrial enzyme induces a ‘petite’ phenotype but does not affect cell viability. The 14 Nt residues of the mitochondrial yeast enzyme constitute the targeting signal that is removed by proteolytic cleavage upon import.38
The genome of A. thaliana encodes 4 AsnRSs, one of which contains an Nt peptide of 71 residues with overlapping mitochondrial (51 aa) and chloroplastic (63 aa) targeting sequences.39,40 Green fluorescent protein (GPF) fusion experiments demonstrated import of this AsnRS in chloroplasts and mitochondria. Phylogenetic analyses show that the 4 AsnRSs are clearly distinct from other eukaryotic cytosolic AsnRSs, suggesting that they evolved by repeated duplication of a gene transferred from an ancestral plastid. They constitute an example of capture of an organellar aaRS by the cytosolic protein synthesis machinery.
Involvement of Asparaginyl-tRNA Synthetases in Immune Reactions
AsnRS from the human filarial parasite Brugia malayi, which causes lymphatic filariasis, was shown to be a protective immunodominant antigen.41,42 It was later shown that sera from patients with interstitial lung disease and inflammatory arthritis contain antibodies which very specifically inhibit AsnRS and precipitate the synthetase complexed with tRNA.43,44 This was the sixth example of a human aaRS involved in autoimmune reactions. Analysis of the effect of the autoantibodies on AsnRS deletion mutants allowed identification of two distinct epitopes.45 A first one located in the Nt domain (residues 1-221) was identified by Western-blot and is not involved in enzyme inactivation. A second one localized in the Ct domain (residues 108-548), probably in the catalytic β-barrel and/or in the hinge region, is involved in enzyme inactivation. The antibodies are without effect on aa activation but inactivate tRNA charging and, surprisingly, increase affinity of the enzyme for tRNA. Thus, enzyme inactivation might be provoked by blocking either dissociation of the charged tRNA, or the conformational changes promoting formation of the competent enzyme•tRNA complex. This effect contrasts with that described for other aminoacylation systems where inactivation by autoantibodies was related to inhibition of aa activation or to precipitation of the cognate tRNA. The physiological processes involved in production of autoimmune phenomena are not understood. It has been proposed that autoantibodies arose through an immune response to foreign antigens such as infectious agents that share, by molecular mimicry, common structures with host proteins.45
Structure of tRNAAsn and Recognition by Asparaginyl-tRNA Synthetase
Structural and Functional Peculiarities of tRNAAsn
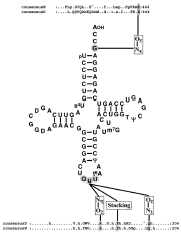
The various organisms contain one tRNAAsn with anticodon GUU able to recognize the AAU and AAC codons encoding Asn. Sixteen tRNAAsn of various origins are sequenced.46 tRNAAsn belongs to class 1 tRNAs characterized by a small variable-loop containing 5 or exceptionnally, in organellar tRNAAsn, 4 nucleotides. Prokaryotic and eukaryotic tRNAAsn contain mostly respectively 76 and 77 nucleotides, because of variations in their D-loops containing 8 or 9 nucleotides. However, the D-loop of Methanobacterium thermoautotrophicum tRNAAsn contains 10 nucleotides and that of mitochondrial tRNAAsn displays 3 (e.g., in bronchiostoma) to 11 (in albinoni coerulea). The D-loops of eubacterial tRNAAsn contain two dihydrouridines (D) whereas archaebacterial tRNAAsn are deprived of this modification. In contrast, D-loops of eukaryotic tRNAAsn contain four modified U: four D are present in yeast tRNAAsn and three D in addition to 3-(3-amino-3-carboxypropyl)-Uridine (acp3U) in mammalian tRNAAsn. The latter modified U is also present in the D-loop of M. thermoautotrophicum tRNAAsn. D is also found on position 47 of the variable-loop of eukaryotic tRNAAsn and pseudouridine (F) in positions 27 and 28 of the anticodon-stem in mammalian tRNAAsn. In contrast to other tRNA species, no atypical secondary structures were found in mitochondrial tRNAAsn.
Two unusual modifications are found in tRNAAsn: queuine at position 34, the first nucleotide from anticodon, in eukaryotic and most eubacterial tRNAAsn and N-((9-β-Dribofuraanosylpurine- 6-yl)carbamoyl)threonine (t6A) at position 37 in almost all tRNAAsn. These modifications can, however, also be present in other tRNA species. Queuine, a modified guanine, is inserted by exchange with G34 by a tRNA-guanine transglycosylase.47 Certain eubacterial subgroups such as Thermus and all archaea are unable to synthesize queuine. In these species, tRNAAsn contains G34. tRNAAsn from T. thermophilus like the other tRNAs sequenced from this organism contain 3 invariant post-transcriptional modifications: Gm18, s2T54 and m1A58 (see ref. 33 and refs. therein). It has been shown that these modifications are not involved in the functionality of the tRNAs, but they reinforce their thermal stability. 10,48 Finally T. themophilus tRNAAsn contains m2G at positions 6 and 10. These modifications were until now not found in other prokaryotic tRNAAsn. In contrast m2G10 and m22G26 are present in eukaryotic tRNAAsn and additionally m1G9 in mammalian tRNAAsn.
Cross-reactions with tRNAAsn and AsnRSs of various origins reveal species specificity in tRNA asparaginylation. Human and B. malayi AsnRSs charge well mammalian and yeast tRNAAsn but poorly E. coli tRNAAsn,31,42,44 whereas AsnRS from T. thermophilus aspartylates efficiently T. thermophilus and E. coli tRNAAsn but only poorly yeast tRNAAsn. 10 It has been suggested that species specificity of AsnRSs is related to the additional base inserted in position 21 of the D-loop in eukaryotic tRNAAsn.44
Identity of tRNAAsn
The nucleotides defining Asn identity and the possible implication of post-transcriptional modifications in tRNA recognition by AsnRS have been investigated in prokaryotic systems.49,50
Eubacterial tRNAAsn all begin with 5'pU, preventing efficient transcription with T7 RNA polymerase. However, the tRNAAsn transcript from Thermus thermophilus could be obtained by self-cleavage of a large transcript in which tRNAAsn was extended at the 5' end by a hammerhead ribozyme. This unmodified tRNAAsn is charged only 6-fold less efficiently by AsnRS than the modified tRNAAsn isolated from T. thermophilus.10 Thus, the post-transcriptional modifications, are not crucial for asparaginylation, although they exert a limited effect on its charging efficiency.
Because of the difficulty to obtain tRNAAsn transcripts, Asn identity has not been investigated by analysis of the charging capacity of variants created by in vitro transcription of the mutated gene. Also, in vivo approaches, using variants of either initiator tRNA transplantated with the Asn anticodon or tRNAAsn suppressor failed. Indeed, transplantation of the Asn anticodon into tRNAfMet did not lead to initiation of protein synthesis from an Asn codon.49 Further, conversion of tRNAAsn into amber or opal suppressors, through change of the anticodon, inactivates the tRNA, and it was not possible to restaure its aminoacylation capacity by compensatory mutations in the anticodon stem and loop like for tRNAAsp.51 In fact, random mutagenesis in yeast tRNAAsn(CUA) creates variants charged by GlnRS.52 Acquisition of Gln specificity by the tRNAAsn amber suppressor containing already several Gln identity elements is promoted by creation of mutations triggering disruption of the first base-pair from acceptor stem, essential for tRNA glutaminylation. Nevertheless, these studies showed that the anticodon of tRNAAsn is an essential element in Asn identity. Additionnal informations about implication of the anticodon nucleotides and other elements from the tRNA in Asn identity were obtained by analysis of variants of E. coli tRNALys(UUU) transcribed in vitro and E. coli tRNAAsn(GUU) expressed in vivo.49,50
The primordial role of the first base from anticodon (G34) was revealed by switch of the specificity of a tRNALys transcript transplantated with both the anticodon and the discriminator base of tRNAAsn (GUU and G73 instead of UUU and A73). This chimeric tRNALys(GUU) becomes well charged with Asn.50 Since tRNALys(UUU) with a G73 does not accept Asn, G34 plays a crucial role in Asn specificity. However, the discriminatory G is also important, since the presence of any other nucleotide on this position decreases asparaginylation efficiency of this tRNA.
The contribution of the anticodon nucleotides to Asn identity was more extensively investigated by in vivo analysis of the charging capacity of tRNAAsn variants.49 Charged and uncharged tRNA were fractionated by PAGE and quantified by Northern blot. Single base changes of U35 to C35 or U36 to C36 in tRNAAsn abolish its charging capacity whereas substitution of G34 by C34 converting the anticodon GUU to CUU confers Lys accepting capacity to the mutated tRNA. Finally, the chimeric tRNAAsn (CUU) with A73 is deprived of any Asn accepting capacity and switches, in vivo, to Lys specificity. Thus, the anticodon nucleotides G34, U35 and U36 as well as the discriminator G73 are crucial for asparaginylation (fig.4). In addition to its functional role, G73 plays also a pivotal role by maintaining the overall structure of tRNAAsn since presence of C73 or U73 provokes in vivo instability. These nucleotides are probably the major but not the sole elements determining Asn identity, since the chimeric tRNAfMet with transplantated Asn anticodon and G73 does not initiate synthesis of a polypeptide chain from an Asn initiation codon.49
Discrimination by Asparaginyl-tRNA Synthetase of tRNAAsn against tRNAAsp and tRNALys
The strongest identity elements of tRNAAsn, namely the anticodon nucleotides and the discriminator base are partly conserved in tRNAAsp53-56 and tRNALys.57-60 Then, how does AsnRS discriminate tRNAAsn from tRNAAsp and tRNALys? Among the nucleotides involved in identity of tRNAAsn and tRNAAsp only nucleotides 36, (respectively C and U) differs. Since substitution in E. coli tRNAAsn of U36 by C36 drastically decreases charging efficiency,50 C36 prevents recognition of tRNAAsp by AsnRS. In contrast, U36 in tRNAAsn by itself probably does not prevent aspartylation by AspRS since substitution in tRNAAsp of C36 by U36 only affects moderately tRNA aspartylation.56 Specific asparaginylation of tRNAAsn may be reinforced by the postranscriptionnal modification t6A preventing aspartylation.
Among the nucleotides involved in identity of tRNAAsn and tRNALys(UUU) only nucleotide 34, (respectively G and hypermodified U) differs in cytosolic eukaryotic tRNAs, whereas both, nucleotide 34 and the discriminator base (respectively G73 and A73), differ in prokaryotic tRNAs. So, in eukaryotic tRNALys(UUU) the modified U34 is probably the sole nucleotide which prevents asparaginylation, whereas in prokaryotic tRNALys(UUU) the discriminator base A73 may also prevent asparaginylation.
Finally it has been suggested that strong contribution of U35 and U36 to Asn identity determines the distribution of the two distantly related groups of class I LysRS which recognize either both U35 and U36 or U36 alone, in organisms possessing or lacking AsnRS.61
Elements of Asparaginyl-tRNA Synthetase Involved in Recognition of tRNAAsn
The aa residues of AspRS contacting the anticodon and the discriminator base of tRNAAsp were identified in the 3D structure of the complex from yeast.25,62 Implication of most of these residues in efficient tRNA charging was confirmed by site-directed mutagenesis of the enzyme.63 Alignments show conservation in AsnRS of the residues from AspRS contacting U35 and G73 of tRNAAsp.54,64 Since these nucleotides are conserved in tRNAAsn and contribute also to efficient asparaginylation they are probably contacted by the conserved residues from AsnRS (fig. 4).
The Indirect Pathway of tRNA Asparginylation
Characterization, General Properties and Occurrence
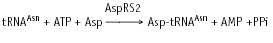
Biochemical and genomic data reveal the existence in various prokaryotes of a non-conventional pathway of tRNA asparaginylation. This pathway was first discovered in Haloferax volcani.21,65 It was shown that in this archaea, deprived of AsnRS, Asn-tRNA is formed by amidation of Asp bound on tRNA. This suggested indirect synthesis of Asn-tRNAAsn by a pathway resembling that previously described for formation of Gln-tRNAGln.23 Further informations about this pathway, were obtained by analysis of Asn-tRNAAsn synthesis in T. thermophilus. This thermophilic eubacterium possesses two AspRS (AspRS1 and AspRS2).64 Alignments show that AspRS1 resembles structurally eubacterial AspRS, and AspRS2, of significantly shorter size, resembles archaeal AspRS.33 AspRS1 is discriminating and aspartylates only tRNAAsp, whereas AspRS2, non-discriminating, aspartylates tRNAAsn in addition to tRNAAsp, with similar efficiencies (reaction 1).22 Asp-tRNAAsn formed by AspRS2 is then converted into Asn-tRNAAsn by an tRNA-dependent aspartyl-tRNAAsn amidotransferase (Asp-AdT) in the presence of an amide group donor (Asn, Gln or ammonia) and ATP (reaction 2, fig. 5).
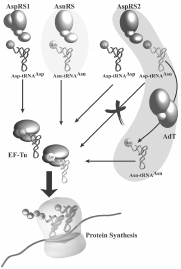
Figure 5
Interrelation between tRNA aspartylation and asparaginylation in T. thermophilus. The direct and indirect routes of Asn-tRNA formation are shown respectively on light and dark grey.
Reaction 1:

This pathway resembles the indirect route of Gln-tRNAGln formation discovered in B. megaterium23 and later described in other Gram + eubacteria, the Gram-eubacterium Rhizobium meliloti, cyanobacteria, archaea, and most organelles.66-68 In these organisms deprived of GlnRS, Gln-tRNAGln is formed by amidation of Glu mischarged on tRNAGln by a GluRS able to attach Glu on tRNAGln as efficiently than on tRNAGlu. Biochemical and genomic investigations reveal that absence of GlnRS is widespread among eubacteria and ubiquitous in archaea, whereas absence of AsnRS is widespread in archaea but exceptionnal in eubacteria69 (Table 2). Therefore, prokaryotes use the indirect pathway much more frequently for glutaminylation than for asparaginylation of tRNA69. In contrast, eukaryotes form Gln-tRNAGln and Asn-tRNAAsn directly by GlnRS and AsnRS. Organelles possess AsnRS and use the direct pathway for tRNA asparaginylation. However, they are deprived of GlnRS and glutaminylate tRNA by the indirect pathway; the sole known exception is presence of GlnRS in mitochondria from Leishmania tarentolae.70
Table 2
Prokaryotes using the direct or/and the indirect route for formation of Asn-tRNA.
The indirect pathway involves formation of Asp-tRNAAsn whose participation in protein synthesis would be lethal. It was shown that Asp-tRNAAsn is not vehiculated to the ribosome because it cannot bind to the elongation factor Tu (fig. 5).22 Absence of binding of mischarged Glu-tRNAGln, involved in organellar tRNA glutaminylation, to elongation factor Tu from chloroplasts from Pisum sativum has also been reported.71 Therefore, the partners of the translation machinery adapted their function to the formation of the mischarged tRNA to improve accuracy. The structural basis for the rejection, by the elongation factor Tu, of mischarged aa-tRNA carrying a physiological function remains still unknown.
Coexistence of Direct and Indirect Pathways of tRNA Asparaginylation
Most species using the indirect route for tRNA asparaginylation and all glutaminylating indirectly tRNA are deprived of respectively AsnRS or GlnRS and thus are unable to form directly the homologous aa-tRNA. However, T. thermophilus and Deinococcus radiodurans can asparaginylate tRNA directly and indirectly, since AsnRS coexists with the partners of the indirect pathway.22,72 Duplication of the pathways of Asn-tRNA synthesis could be rationalyzed by biochemical and genetic analyses showing absence of asparagine synthetase (AS) in these organisms. Under Asn starvation, tRNA asparaginylation occurs by formation of Asn on tRNAAsn whereas, when the organism is supplied with free Asn, Asn-tRNAAsn is formed directly by AsnRS. Further, in vitro analysis of the efficiencies of the two pathways shows faster asparaginylation of tRNA directly by AsnRS than indirectly.22 Thus, the direct pathway is conserved, despite absence of its autonomy, since it provides the organism more efficiently with asparaginylated tRNA than the indirect pathway. However, the latter pathway is essential for survival of the organism since it constitutes the unique route of Asn-tRNAAsn formation in the absence of free Asn (fig. 5).
The Partners of the Indirect Pathway of tRNA Asparaginylation
The Aspartyl-tRNA Synthetase of Dual Specificity
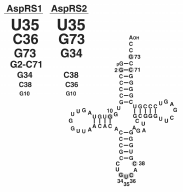
The first step of indirect formation of Asn-tRNAAsn consists in aspartylation of tRNAAsn by an AspRS of dual specificity. Until now, only AspRSs present in archaea and in Deinococcaceae, an eubacterial group, were found able to aspartylate in vitro tRNAAsn in addition to tRNAAsp.22,72 These AspRSs, of archaeal-type, are the smallest AspRSs. They differ structurally from the eubacterial and eukaryotic AspRSs by the absence of the additional domains present in AspRSs from other origins. It is thus tempting to speculate that only archaeal-like AspRSs are non discriminating and eubacterial and eukaryal AspRSs exert strict specificity as a consequence of their distinct structural properties. Interestingly, archaeal-type AspRSs differ from other AspRSs by the size of the loop located in the anticodon binding domain (residues 173 to 186 from alignment).33 This loop comprises 14 to 16 residues in eubacterial and eukaryal AspRSs but only 5 to 9 in archaeal AspRSs. The 3D structure of yeast AspRS complexed to tRNAAsp has shown involvement of this loop in recognition of C36 from tRNAAsp anticodon through backbone contacts.25,62 Since anticodons of tRNAAsp and tRNAAsn differ only by this nucleotide (respectively C and U), it has been proposed that the strict or relaxed specificity of AspRSs in tRNA recognition is determined by the size of this loop.73 The large loop of eubacterial and eukaryal AspRSs would determine strict specificity by discriminating C against U, whereas the small loop of archaeal AspRSs deprived of the discrimination ability, would determine relaxed specificity. However, the eubacterial AspRSs from Helicobacter pylori and Chlamydia trachomatis which possess a large loop, display a relaxed specificity since, in these organisms deprived of AsnRS, Asn-tRNAAsn may be formed by the indirect pathway. Thus, strict and relaxed specificities of AspRSs are not solely confined to the size of this loop. The structural peculiarities conferring strict or relaxed specificity to AspRSs will likely be revealed by comparison of the 3D structures of AspRSs exerting distinct specificities but of the same structural type (e.g., AspRS1 and AspRS2 from T. thermophilus) complexed to tRNA.
tRNAAsp and tRNAAsn Recognitions by Aspartyl-tRNA Synthetase of Relaxed Specificity
Since AspRS1 and AsnRS discriminate tRNAAsp from tRNAAsn and AspRS2 does not, one may predict that AspRS1 and AsnRS recognize peculiar elements in the cognate tRNA, whereas AspRS2 recognizes elements common to both tRNAs. The elements of tRNAAsp from T. thermophilus involved in recognition by the two thermophilic AspRSs are presented in Figure 6.56 With only one exception, the same nucleotides determine aspartylation by AspRS1 and AspRS2. However, they differ by their contribution. The 3 major determinants for charging by AspRS1 are U35 and C36 from anticodon followed by the discriminator base G73. This triad is followed by the G2-C71 pair from acceptor stem, the first base from anticodon G34 and finally by C38. The hierarchy of contribution of these elements differs for charging by AspRS2. Contribution of U35 preceds immediately that of the discriminator base G73 which is followed by G34, C38 and finally C36. Therefore, specific charging and selection of the cognate tRNA by AspRS1 and AsnRS is very likely promoted by nucleotide 36 of the anticodon. This base distinguishes tRNAAsp from tRNAAsn (respectively C and U) and is essential for charging by the homologous aaRS. Relaxed specificity of AspRS2 is due to the minimized contribution of nucleotide 36 in tRNA recognition.
The tRNA-Dependent Amidotransferase
The Asp-tRNAAsn amidotransferase (Asp-AdT) converting Asp mischarged on tRNAAsn into Asn was first isolated from T. thermophilus in an αβ heterodimeric form.22 Analysis of the genome from T. thermophilus revealed ORF encoding orthologs of GatA, GatB and GatC subunits of the heterotrimeric Glu-tRNAGln amidotransferase (Glu-AdT) from B. subtilis.74,75 Comparison of the Nt sequences showed identity of the α and β subunits of Asp-AdT with GatA and GatB of Glu-AdT, suggesting that both constitute the same enzyme, probably of trimeric aβ? (GatABC) structure.
When the genes encoding GatA, GatB and GatC subunits of T. thermophilus Asp-AdT were cloned in an operon for expression in E. coli, a fully active trimeric enzyme converting Asp-tRNAAsn into Asn-tRNAAsn was expressed with a subunit stoichiometry of 1/1/1. This enzyme also converts, in vitro, Glu mischarged on B. subtilis tRNAGln into Gln.75 Dual specificity was further observed for Glu-AdT of B. subtilis capable to convert, in vitro, Asp mischarged on tRNAAsn from T. thermophilus or D. radiodurans into Asn.75 Finally, orthologs of this enzyme identified in D. radiodurans, Acidothiobacillus ferrooxidans, Chlamydia trachomatis and in the archaeon M. thermoautotrophicum also display dual specificity in vitro.70,76-77 Thus the trimeric amidotransferase called Asp/Glu-AdT or AdT can ensure in vitro formation of Asn-tRNAAsn and Gln-tRNAGln. However, the cellular function of this enzyme is dictated by the biochemical and genetic background of the host. In the absence of AsnRS (e.g., in H. volcanii) the AdT supplies the organism with Asn-tRNAAsn, in the absence of GlnRS (e.g., in B. subtilis) with Gln-tRNAGln and in the absence of GlnRS and AsnRS (e.g., in A. ferrooxidans and C. trachomatis), with both aa-tRNA.
Despite identical functional properties, important divergences are observed in organization of the AdT genes in the various species. In B. subtilis, A. ferrooxidans and C. trachomatis the genes are organized in an operon (gatCAB), contrasting with their dispersion in the genomes from T. thermophilus, D. radiodurans and archaea.74-78
Alignment of AdT polypeptide chains shows two interesting features in GatA. (i) The signature sequence of amidases rich in Gly, Ser and Ala (residues 179-210 from consensus sequence), involved in the metabolic pathways of various amino acids (Arg, Pro, Phe, Trp) in procaryotes (e.g., Rhodococcus, Brevibacterium and Pseudomonas species) (fig. 7).74,77 Inhibition of AdT by boronic acid suggests implication of a Ser residue in catalysis.79,80 (ii) A P-loop (residues 142-148) which is a structural domain involved in ATP and GTP binding. These features indicate that GatA binds ATP and catalyzes transamidation, and agree with the ability of B. subtilis GatA, when expressed together with GatC, to promote ATP-independent deamidation of Gln.74 GatB, structurally related to Pet112, an essential mitochondrial protein, is probably involved in tRNA recognition. Finally GatC might be involved in folding and/or stability of GatA, since in vivo expression of active GatA requires coexpression of GatC.74
Catalytic Properties of tRNA-Dependent Amidotransferase
The enzyme only catalyzes amidation of Asp and Glu mischarged, respectively, on tRNAAsn and tRNAGln. Asp and Glu, in free form or attached on their cognate tRNA are not substrates. 21,22,74 AdT from B. subtilis discriminates well Glu-tRNAGln from Glu-tRNAGlu but less well Asp-tRNAAsn from Asp-tRNAAsp which, in contrast, are well discriminated by T. thermophilus AdT.21 Thus, AdT probably adapted their specificity to the physiological necessity to form only the homologous aa-tRNA.
Amidation requires as amide donor, Asn, Gln or ammonium ions, and is accompagnied by hydrolysis of ATP into ADP and Pi in the presence of Mg2+.10,81 Efficiency of the various donors varies with the origin of the enzyme. Gln is the most efficient for AdT from B. subtilis, and Asn for that from T. thermophilus.10,74 Ammonium ions are poor substrates because of their low affinity for the enzyme.10 Kinetic investigations of B. subtilis and Streptococcus pyogenes AdT suggest that transamidation of Glu-tRNAGln comprises three chemical events.81,82
- Hydrolysis of the amide donor Gln or Asn forming NH4 + sequestered by the enzyme.
- ATP-dependent activation of accepting Glu attached on tRNA, by formation of an enzyme-bound carboxyl-phosphate intermediate.
- Transfer of the amide group on the activated carboxyle to form Gln-tRNAGln.
Transamidation of Asp-tRNAAsn into Asn-tRNAAsn> probably involves the same chronology of events.
Deamidation of the donor occurs in the absence of the other substrates, but Glu-tRNAGln increases kcat 10-fold and Glu-tRNAGln with ATP 70-fold.82 Formation of the carboxyl-Pi was evidenced by retention of enzyme bound [32P]Pi-Glu-tRNAGln on nitrocellulose disks, when B. subtilis AdT was incubated with Glu-tRNAGln and γ-[32P]ATP in the absence of amide group donor.81 Activation of the γ-carboxylate of Glu was demonstrated by conversion, under appropriate conditions, of [32P]Pi-Glu-tRNAGln into pyrrolidone-tRNAGln or γ-glutamylhydroxamate-tRNAGln.81 Transfer of the amide group on the activated carboxyl is accompagnied by stoichiometric formation of Gln-tRNAGln and release of [32P]Pi.82 Gln-tRNAGln formation is rate-limiting in the overall transamidation (kcat 1.18, 1.09 and 0.6 s-1 respectively for glutaminase, ATP hydrolysis and transamidase activities).82
Kinetic analysis of the partial steps in presence of an analog of ATP unable to stimulate efficiently transamidation, shows that this substrate provokes allosteric effects which trigger tight coupling in ligand binding and stabilize the AdT•Glu-tRNAGln complex.82 A similar conclusion emerges from study by glycerol gradient sedimentation of AdT from Chlamydomonas reinhardtii in the absence or presence of substrates. The enzyme forms a stable complex with Glu-tRNAGln only when ATP is present.83
Functional and Structural Interrelations between tRNA-Dependent and tRNA-Independent Amidotransferases and between Amidotransferases and Aminoacyl-tRNA Synthetases
The transamidation catalyzed by AdT resembles mechanistically amidation of free Glu into Gln by glutamine synthetase (GS) since both form a γ carboxyl-Pi intermediate. In contrast, conversion of free Asp into Asn by AS involves formation of a β carboxyl-AMP intermediate. Interestingly aaRS activate their homologous aa by formation of a a carboxyl-AMP. Comparison of the 3D structures of E. coli AS A and B with yeast AspRS reveals a strong resemblance of the catalytic center, in particular conservation of essential aa residues and presence in AS of the ATP-binding motif of class II aaRS.84,85 The most stricking difference between both structures are related to activation of distinct carboxylate groups of Asn. This observation indicates that AspRS, AsnRS and AS are closely related in evolution. In contrast, binding of ATP by AdT and AS respectively by a P-loop and a motif conserved in class II aaRSs, and presence of the signature motif of amidases in AdTs but not in ASs (fig. 7), argue for a distinct origin of both enzymes and suggest that AdTs and aaRSs are not phylogenetically interconnected. Finally, GS which binds ATP by a non canonical motif and differs structurally from AS and aaRS is evolutionary distant from these enzymes.86
Interestingly, AsnRS and AS are related by regulation processes. In L. bulgaricus synthesis of both enzymes is coregulated since their genes are organized in an operon depending on a unique leader region located upstream of AS gene87. Evidences have been brougth that transcription of both genes is regulated by a common antitermination mechanism and would be triggered by increase of the uncharged tRNAAsn in the cell caused by insufficient endogenous Asn.
Glu/Asp-AdT does not constitute the unique tRNA-dependent amidotransferase. In archaea, deprived of GlnRS, tRNA glutaminylation is achieved by a tRNA-dependent amidotransferase converting Glu mischarged on tRNAGln into Gln. This heterodimeric enzyme, called Glu-AdT, exhibits, in contrast to Glu/Asp-AdT, a strict specificity and is absent in eubacteria.77 However, both enzymes are present in archaea deprived of GlnRS and AsnRS and form respectively Asn-tRNAAsn and Gln-tRNAGln. Presence of a specific Glu-AdT, in addition to Asp/Glu-AdT of dual specificity, which leads to the redundancy of Glu-AdT activity, remains unexplained.
The Evolutionary History of Asparaginyl-tRNA Synthetase
Synthesis of Asn-tRNAAsn and Gln-tRNAGln, in contrast to the other aa-tRNAs, can be achieved by two distinct pathways, a direct and an indirect one (fig. 5) involving different enzymes which in most cases, are mutually exclusive. The exceptions are illustrated by the Thermus-Deinococcus group where, for metabolic reasons, both pathways coexist,22,72 and by archaea which are all depleted of GlnRS and use an AdT unique to their genre for Gln-tRNA production.76 The predominance of AdT over AsnRS and GlnRS in procaryotes versus eukaryotes suggests that AsnRS and GlnRS were not present in the latest common ancestor but appeared after the split into the three distinct phylae.
Two Amide Aminoacyl-tRNAs and Two Different Evolutionary Histories
Origin of GlnRS has been confidently established by several phylogenic studies which show that this enzyme arose in eukarya from a duplicate of the gene of GluRS that acquired this new specificity and was transferred to eubacteria by horizontal gene transfer.88,89 The lack of GlnRS in the archaeal domain and in complete subgroups of eubacteria supports the idea that GlnRS specificity was one of the latest to emerge. This hypothesis is also supported by the theory of coevolution of the genetic code and biosynthesis of the aa,90 which postulates that, initially Gln codons encoded Glu as the result of tRNAGln charging with Glu by the ancestral GluRS from which originated GlnRS latter in evolution.
Similarly, in primitive systems, Asn codons were likely encoded by Asp as the result of aspartylation of tRNAAsn by the primordial AspRS. The remarkable extent of structural homologies between AspRSs and AsnRSs lead to the suggestion that AsnRS arose from AspRS by gene duplication and subsequent acquisition of AsnRS specificity by one of the copy. So far, three attempts were made to decipher evolution of the Asp/Asn-tRNA synthetase family and to pinpoint the origin of AsnRS.91-93 They vary by the pool of AsnRS and AspRS sequences processed and by the phylogenic approaches applied. Each strategy identifies a different kingdom for the origin of AsnRS. Three evolutionary models are currently in competition, which were already proposed by the first and most extensive study (fig. 8).91 Although conflicting about the origin of AsnRS, these studies agree with emergence of AsnRS prior to GlnRS and evolution of both along different paths. Closer examination of the methods and strategies used to build these phylogenic trees shows that either a limited number of sequences and/or only the catalytic domain of these enzymes were subjected to analysis. We therefore reinvestigated the filiation of AsnRS by using an extended pool of AspRS and AsnRS sequences encompassing both the catalytic and anticodon-binding domains. The tree (fig. 9) was constructed by the Neighbor Joining method from a bootstrap analysis (1000 replicates). This tree shows that AsnRS like AspRS are organized into three clades following the organismal phylogeny. However, bacterial AsnRSs are splitted into two groups: a large one encompassing most of the bacterial AsnRSs and the mitochondrial species (Bact. N1), and a small one constituted of 6 species of firmicutes and T. thermophilus (Bact. N2) and arising from the archaeal AsnRS branch. Although AsnRSs seem to be closer to the eukaryal/archaeal branch of AspRSs in terms of evolution distances, there is no clear indication that these enzymes arose from this subgroup of AspRSs, lesser that eukaryal or archaeal AspRSs have originated them.
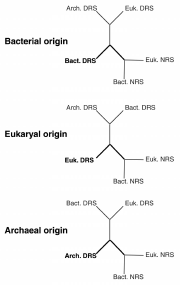
Figure 8
The three possible evolutionary trees of Asp/AsnRS family. Asp- and AsnRS are abbreviated by DRS and NRS. Suffixes Bact., Arch. and Euk. refer to the bacterial, archaeal or eukaryotic origins of these aaRSs.
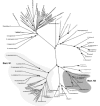
Figure 9
Phylogenic tree of Asp/AsnRS sequences. The tree was deduced by Neighbor Joining analysis from a bootstrap of 1000 replicates based on 53 and 46 AspRS and AsnRS sequences aligned by clustal X.
AsnRS are present in all domains and their phylogenic distribution, with the exception of Bact. N2 species, follows that of AspRS. Topology of AsnRS trees suggests that either AsnRS appeared before separation of the phylae or that early after split of the phylae, the bacterial and the archaeal/eukaryal ancestors independently evolved AsnRS leading to a wide spread of this enzyme among the various organisms, especially among bacteria. Another hypothesis explaining the lack of likelihood between Bact. N1 AsnRS and their eukaryal/archaeal counterparts would be that AsnRS emerged in the latter domain after its separation from bacteria and was immediately transferred into bacteria (from the Bact. N1 type) early thereafter, and evolved independently since then. The small group of Bact. N2 AsnRS did not, however, follow this evolutionary scheme. Their position, arising from the archaeal branch, suggests that their presence in the 7 bacterial species results from a more recent horizontal gene transfer from archaea. Although, existence of an archaeal-type AspRS has already been described, its acquisition has also been correlated to a functional advantage (formation of Asp-tRNAAsn, substrate of the AdT) for the bacterial host. So far there is no apparent advantage, nor explanation, for the existence of an archaeal-like AsnRS in some bacterial species.
From Aspartate to Asparagine, the Emergence and Evolution Path of a New tRNA Charging Specificity
Based on biochemical and structural data available on Asn-tRNA formation and the consensual phylogenetic arguments produced by studies on AsnRS, we can try to retrace the steps leading to emergence of AsnRS. This evolutionary scenario starts with the assumption that an aaRS displaying the highest degree of specificity for both the aa and its corresponding tRNA is the ultimate stage in the evolution of aa-tRNA synthesis. Conversely, aaRS with relaxed specificities, and formation of aa-tRNA requiring mutiple steps, are hallmarks of lesser evolved enzymes and pathways.
It is admitted that Asn was one of the latest aa introduced in the cell's repertoire for protein synthesis.90 In the latest common ancestor, before apparition of Asn, codons specifying Asn (AAU and AAC) were encoded by Asp as the result of the capacity of the ancestral AspRS to acylate tRNAAsp and tRNAAsn with Asp. The first step leading to actual incorporation of Asn into proteins, at codons specifying Asn, was apparition of AdT able to convert Asp mischarged on tRNAAsn into Asn. At some point in the evolution, AsnRS appeared by duplication of the AspRS gene and acquisition by one copy, of the capability to attach Asn on tRNAAsn. The only restriction to this event relies in the ability of the cell to produce free Asn since existence of an autonomous direct pathway of Asn-tRNA synthesis requires the coexistence of AsnRS and AS. Therefore, acquisition and stabilization of an AsnRS was very likely related to apparition of AS A or B. In most cases, acquisition of the autonomous direct route of Asn-tRNA formation led to loss of the indirect route. However, if AsnRS or AS is missing, organismal synthesis of Asn-tRNA is performed by Asp-AdT (Table 2). In most cases Asp-AdT supplies the absence of AsnRS, or of both AsnRS and AS, but exceptionally only the absence of AS. In these organisms, AsnRS which should be dispensable, has been conserved because it forms Asn-tRNA more efficiently than AdT. However, this selective advantage is only expressed under certain environmental conditions namely when Asn is present.
Conclusion and Perspectives
Surprisingly, distribution of the direct and indirect pathways of Asn-tRNA formation does not follow organismal phylogeny (Table 2). One striking example is illustrated by the γ proteobacteria E. coli and P. aeruginosa. They share more than a phylogenic ressemblance and have been grouped together also because of similar biochemical backgrounds. However, there is no rational reason as to why one uses the direct route of Asn-tRNA formation and the other one the indirect route. Likewise, there is no explanation as to why inside the same organismal subgroups some species developped or acquired an autonomous direct route of tRNA asparaginylation and others only part of this pathway (either AsnRS or AS; Table 2). In contrast, the direct pathway of tRNA glutaminylation is always autonomous, since in all organisms studied so far, GS is present, and provides Gln if GlnRS was acquired. Both enzymes, however, did not appeared concomittantly. GS was probably acquired by the lattest common ancestor, whereas phylogeny of GlnRS unquestionably shows that this enzyme appeared in eukaryotes and was transferred only to a limited number of eubacterial species. In contrast, the presence in various prokaryotes of AsnRS but not of AS or vice-versa agrees with emergence of both enzymes not in the latest common ancestor, but early after the split of the phylae. Therefore, synthesis of free Gln was established before emergence of GlnRS whereas AsnRS emerged before formation of free Asn or more likely at the same moment in evolution.
The functional and structural properties of AdT able to replace either components of the protein synthesis (GlnRS or AsnRS) or metabolic enzymes (AS) suggest that this enzyme is very likely primitive. Furthermore, this enzyme, forming the two amidated aa on the homologous tRNA, connected tRNA asparaginylation and glutaminylation in primitive systems catalyzed surprisingly by aaRSs of distincts classes. However, the important functional and structural divergences between the amidases involved in Asn and Gln formation (AS, GS and AdT) indicate distinct origins of each. Presence in AdT of signatures motifs of amidases and a P-loop, both absent in AS and GS, suggests a common origin of this enzyme and the amidases involved in metabolic conversion of carboxylates into amides. AdT might have constituted the primitive enzyme synthesizing Asn and Gln and forming Asn-tRNAAsn and Gln-tRNAGln. This enzyme however, did not originated the modern enzymes forming Asn and Gln (AS and GS) and amide aa-tRNA (AsnRS and GlnRS). Nevertheless, the non-discriminating AspRS and GluRS of the indirect pathways generated the modern and specific AsnRS and GlnRS.
A more extensive knowledge of the distribution of the direct and indirect pathways of asparaginylation in the living kingdom will allow to explore their origin and their phylogenetic interrelation. A more precise knowledge of the properties of the various partners of both pathways will allow to obtain a better insigth of each, and to better understand their structural and functional interconnexion and their relation with other physiological processes. The structural and functional similarities between AspRS and AS and between AspRS and AsnRS offers the possibility to explore the evolutionary rules used by nature to evolve biochemical processes and to create new activities by conversion of preexisting enzymes.
Acknowledgements
The authors thank R. Giegé (IBMC, Strasbourg) for critical reading of the manuscript and S. Cusack (EMBL Grenoble) for the coordinates of the 3D structures of T. thermophilus AsnRS and LysRS. The project of analysis of the indirect pathway of tRNA asparaginylation is supported by grants from Université Louis Pasteur, from Centre National de la Recherche Scientifique et Technique and from Association de la Recherche contre le Cancer. HR is recipient of a grant from Ministère de l'Enseignement Supérieur et de la Recherche.
References
- 1.
- Hedgcoth C, Ravel J, Shive W. The separation of the aspartyl- and asparaginyl-tRNA synthetases of Lactobacillus arabinosus. Biochem Biophys Res Commun. 1963;13:495–499.
- 2.
- Nass G, Stoffler G. Molecular weight distribution of the aminoacyl-tRNA synthetases of Escherichia coli by gel-filtration. Mol Gen Genet. 1967;100:378–382. [PubMed: 4869079]
- 3.
- Muench KH, Safille PA. Transfer ribonucleic acids in Escherichia coli. Multiplicity and variation. Biochemistry. 1968;7:2799–2808. [PubMed: 4299084]
- 4.
- Barnett WE, Brown DH, Epler JL. Mitochondrial-specific aminoacyl-RNA synthetases. Proc Natl Acad Sci USA. 1967;57:1775–81. [PMC free article: PMC224546] [PubMed: 5231411]
- 5.
- Davies MR, Marshall RD. Partial purification of L-asparaginyl-tRNA synthetase from rabbit liver. Biochem Biophys Res Commun. 1972;47:1386–1395. [PubMed: 5040239]
- 6.
- Grosjean H, Charlier J, Darte C. et al. Purification, characterization and mechanism of action of several aminoacyl-tRNA synthetases from Bacillus stearothermophilus. Experientia Suppl. 1976;26:347–362. [PubMed: 939278]
- 7.
- Samuelson T, Lundvik L. Purification and some properties of asparagine, lysine, serine and valine: tRNA ligases from Bacillus stearothermophilus. J Biol Chem. 1978;253:7033–7039. [PubMed: 690138]
- 8.
- Andrulis IL, Chiang CS, Arfin SM. et al. Biochemical characterization of a mutant asparaginyl-tRNA synthetase from Chinese hamster ovary cells. J Biol Chem. 1978;253:58–62. [PubMed: 618867]
- 9.
- Madern D, Anselme J, Härtlein M. Asparaginyl-tRNA synthetase from Escherichia coli temperature-sensitive strain HO202. FEBS Lett. 1992;299:85–89. [PubMed: 1544480]
- 10.
- Roy H, Becker HD, Kern D. Unpublished results.
- 11.
- Davies MR, Marshall RD. Enhancement of the activity of asparaginyl-tRNA synthetase by an activator from rabbit liver. Biochim Biophys Acta. 1975;390:94–104. [PubMed: 164948]
- 12.
- Anselme J, Härtlein M. Asparaginyl-tRNA synthetase from Escherichia coli has significant sequence homologies with yeast aspartyl-tRNA synthetase. Gene. 1989;84:481–485. [PubMed: 2693216]
- 13.
- Eriani G, Dirheimer G, Gangloff J. Aspartyl-tRNA synthetase from Escherichia coli: cloning and characterisation of the gene, homologies of its translated amino acid sequence with asparaginyl- and lysyl-tRNA synthetases. Nucleic Acids Res. 1990;18:7109–7118. [PMC free article: PMC332776] [PubMed: 2129559]
- 14.
- Levêque F, Plateau P, Dessen P. et al. Homology of lysS and lysU, the two Escherichia coli genes encoding distinct lysyl-tRNA synthetases. Nucl Acids Res. 1990;8:305–312. [PMC free article: PMC330268] [PubMed: 2183178]
- 15.
- Seignovert L, Härtlein M, Leberman R. Asparaginyl-tRNA synthetase from Thermus thermophilus HB8. Sequence of the gene and crystallization of the enzyme expressed in Escherichia coli. Eur J Biochem. 1996;239:501–508. [PubMed: 8706760]
- 16.
- Berthet-Colominas C, Seignovert L, Härtlein M. et al. The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid. EMBO J. 1998;17:2947–2960. [PMC free article: PMC1170635] [PubMed: 9582288]
- 17.
- Eriani G, Delarue M, Poch O. et al. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. [PubMed: 2203971]
- 18.
- Cusack S. Eleven down and nine to go. Nat Struct Biol. 1995;2:824–831. [PubMed: 7552701]
- 19.
- Sprinzl M, Cramer F. Site of aminoacylation of tRNAs from Escherichia coli with respect to the 2'- or 3'-hydroxyl group of the terminal adenosine. Proc Natl Acad Sci USA. 1975;72:3049–3053. [PMC free article: PMC432916] [PubMed: 1103137]
- 20.
- Fraser TH, Rich A. Amino acids are not all initially attached to the same position on transfer RNA molecules. Proc Natl Acad Sci USA. 1975;72:3044–3048. [PMC free article: PMC432915] [PubMed: 1103136]
- 21.
- Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. [PubMed: 8757127]
- 22.
- Becker HD, Kern D. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc Natl Acad Sci USA. 1998;95:12832–12837. [PMC free article: PMC23616] [PubMed: 9789000]
- 23.
- Wilcox M, Nirenberg M. Transfer RNA as a cofactor of protein. Proc Natl Acad Sci USA. 1968;61:229–236. [PMC free article: PMC285927] [PubMed: 4972364]
- 24.
- Giegé R, Lorber B, Théobald-Dietrich A. Crytallogenesis of biological macromolecules: facts and perspectives. Acta Crystallogr. 1994;D50:339–350. [PubMed: 15299382]
- 25.
- Ruff M, Krishnaswamy S, Boeglin M. et al. Class II aminoacyl transfer-RNA synthetases: Crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science. 1991;252:1682–1689. [PubMed: 2047877]
- 26.
- Briand C, Poterszman A, Eiler S. et al. An intermediate step in the recognition of tRNA(Asp) by aspartyl-tRNA synthetase. J Mol Biol. 2000;299:1051–1060. [PubMed: 10843857]
- 27.
- Cusack S, Yaremchuk A, Tukalo M. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 1996;15:6321–6334. [PMC free article: PMC452455] [PubMed: 8947055]
- 28.
- Yip KS, Stillman TJ, Britton KL. et al. The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures. Structure. 1995;3:1147–1158. [PubMed: 8591026]
- 29.
- Delarue M, Poterszman A, Nikonov S. et al. Crystal structure of a prokaryotic aspartyl-tRNA synthetase. EMBO J. 1994;13:3219–3229. [PMC free article: PMC395218] [PubMed: 8045252]
- 30.
- Poterszman A, Delarue M, Thierry J-C. et al. Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase. J Mol Biol. 1994;244:158–167. [PubMed: 7966328]
- 31.
- Landrieu I, Vandenbol M, Leberman R. et al. Identification of YHRO19 in Saccharomyces cerevisiae chromosome VIII as the gene for the cytosolic asparaginyl-tRNA synthetase. Yeast. 1998;14:527–533. [PubMed: 9605503]
- 32.
- Anselme J, Härtlein M. Tyr-426 of the Escherichia coli asparaginyl-tRNA synthetase, an amino acid in a C-terminal conserved motif, is involved in ATP binding. FEBS Lett. 1991;280:163–166. [PubMed: 2009959]
- 33.
- Becker HD, Roy H, Moulinier L. et al. Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase. Biochem. 2000;39:3216–3230. [PubMed: 10727213]
- 34.
- Agou F, Mirande M. Aspartyl-tRNA synthetase from rat: in vitro functional analysis of its assembly into the multisynthetase complex. Eur J Biochem. 1997;243:259–267. [PubMed: 9030747]
- 35.
- Lorber B, Mejdoub H, Reinbolt J. et al. Properties of N-terminal truncated yeast aspartyl-tRNA synthetase and structural characteristics of the cleaved domain. Eur J Biochem. 1988;174:155–161. [PubMed: 3286258]
- 36.
- Agou F, Yang Y, Gesquiere JC. et al. Polyanion-induced alpha-helical structure of a synthetic 23-residue peptide representing the lysine-rich segment of the N-terminal extension of yeast cytoplasmic aspartyl-tRNA synthetase. Biochem. 1995;34:569–576. [PubMed: 7819251]
- 37.
- Frugier M, Moulinier L, Giegé R. A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J. 2000;19:2371–2380. [PMC free article: PMC384352] [PubMed: 10811628]
- 38.
- Landrieu I, Vandenbol M, Härtlein M. et al. Mitochondrial asparaginyl-tRNA synthetase is encoded by the yeast nuclear gene YCR24c. Eur J Biochem. 1997;243:268–273. [PubMed: 9030748]
- 39.
- Peeters NM, Chapron A, Giritch A. et al. Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J Mol Evol. 2000;50:413–423. [PubMed: 10824085]
- 40.
- Aubourg S, Cheron A, Kreis M. et al. Structure and expression of an asparaginyl-tRNA synthetase gene located on chromosome IV of Arabidopsis thaliana and adjacent to a novel gene of 15 exons. Biochim Biophys Acta. 1998;1398:225–231. [PubMed: 9655910]
- 41.
- Nilsen TW, Maroney PA, Goodwin RG. et al. Cloning and characterization of a potentially protective antigen in lymphatic filariasis. Proc Natl Acad Sci USA. 1988;85:3604–3607. [PMC free article: PMC280262] [PubMed: 3368467]
- 42.
- Kron M, Marquard K, Härtlein M. et al. An immunodominant antigen of Brugia malayi is an asparaginyl-tRNA synthetase. FEBS Lett. 1995;374:122–124. [PubMed: 7589498]
- 43.
- Hirakata M, Suwa A, Nagai S. et al. Anti-KS: Identification of autoantibodies to asparaginyl-tRNA synthetase associated with interstitial lung disease. J Immunol. 1999;162:2315–2320. [PubMed: 9973509]
- 44.
- Beaulande M, Tarbouriech N, Härtlein M. Human cytosolic asparaginyl-tRNA synthetase: cDNA sequence, functional expression in Escherichia coli and characterization as human autoantigen. Nucleic Acids Res. 1998;26:521–524. [PMC free article: PMC147268] [PubMed: 9421509]
- 45.
- Beaulande M, Kron M, Härtlein M. Human anti-asparaginyl-tRNA synthetase autoantibodies (anti KS) increase the affinity of the enzyme for its tRNA substrate. FEBS Lett. 2001;494:170–174. [PubMed: 11311235]
- 46.
- Sprinzl M, Horn D, Brown M. et al. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26: 148–153. [PMC free article: PMC147216] [PubMed: 9399820]
- 47.
- Gradler U, Gerber HD, Goodenough-Lashua DM. et al. A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase. J Mol Biol. 2001;306:455–467. [PubMed: 11178905]
- 48.
- Horie N, Hara-Yokoyama M, Yokoyama S. et al. Two tRNAIle1 species from an extreme thermophile, Thermus thermophilus HB8: Effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochem. 1985;24:5711–5715. [PubMed: 3853464]
- 49.
- Li S, Pelka H, Schulman L. The anticodon and the discriminator base are important for aminoacylation of Escherichia coli tRNAAsn. J Biol Chem. 1993;268:18335–18339. [PubMed: 8349709]
- 50.
- Shimizu M, Asahara H, Tamura K. et al. The role of anticodon bases and the discriminator nucleotide in the recognition of some E. coli tRNAs by their aminoacyl-tRNA synthetases. J Mol Evol. 1992;35:436–443. [PubMed: 1487827]
- 51.
- Normanly J, Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. [PubMed: 2673006]
- 52.
- Martin F, Eriani G, Reinbolt J. et al. Genetic selection for active E. coli amber tRNAAsn exclusively led to glutamine inserting suppressors. Nucleic Acids Res. 1995;23:779–784. [PMC free article: PMC306759] [PubMed: 7708493]
- 53.
- Pütz J, Puglisi JD, Florentz C. et al. Identity elements for specific aminoacylation of yeast tRNAAsp by cognate aspartyl-tRNA synthetase. Science. 1991;252:1696–1699. [PubMed: 2047878]
- 54.
- Becker HD, Giegé R, Kern D. Identity of prokaryotic and eukaryotic tRNAAsp for aminoacylation by aspartyl-tRNA synthetase from Thermus thermophilus. Biochem. 1996;35:7447–7458. [PubMed: 8652522]
- 55.
- Giegé R, Florentz C, Kern D. et al. Aspartate identity of transfer RNAs. Biochimie. 1996;78:605–623. [PubMed: 8955904]
- 56.
- Becker HD. Mise en évidence et propriétés de deux aspartyl-tRNA synthétases dans Thermus thermophilus. Thesis of the University Louis Pasteur, Strasbourg. 1998
- 57.
- Mc ClainWH, Foss K, Jenkins RA. et al. Nucleotides that determine Escherichia coli tRNAArg and tRNALys acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc Natl Acad Sci USA. 1990;87:9260–9264. [PMC free article: PMC55144] [PubMed: 2251270]
- 58.
- Normanly J, Kleina LG, Masson JM. et al. Construction of Escherichia coli amber suppressor tRNA genes. Determination of tRNA specificity. J Mol Biol. 1990;213:719–726. [PubMed: 2141650]
- 59.
- Tamura K, Himeno H, Asahara H. et al. In vitro study of E. coli tRNAArg and tRNALys. Nucleic Acids Res. 1992;20:2335–2339. [PMC free article: PMC312350] [PubMed: 1375736]
- 60.
- Shiba K, Stello T, Motegi H. et al. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescue Escherichia coli double-defective mutant. J Biol Chem. 1997;272:22809–22816. [PubMed: 9278442]
- 61.
- Söll D, Becker H, Plateau P. et al. Context-dependent anticodon recognition by class I lysyl-tRNA synthetases. Proc Natl Acad Sci USA. 2000;97:14224–14228. [PMC free article: PMC18899] [PubMed: 11121028]
- 62.
- Cavarelli J, Rees B, Ruff M. et al. Yeast tRNAAsp recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993;362:181–184. [PubMed: 8450889]
- 63.
- Eriani G, Gangloff J. Yeast aspartyl-tRNA synthetase residues interacting with tRNAAsp identity bases connectively contribute to tRNAAsp binding in the ground and transition-state complex and discrimination against non-cognate tRNAs. J Mol Biol. 1999;291:761–773. [PubMed: 10452887]
- 64.
- Becker HD, Reinbolt J, Kreutzer R. et al. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochem. 1997;36:8785–8797. [PubMed: 9220965]
- 65.
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984;259:9461–9471. [PubMed: 6746655]
- 66.
- Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln 1 in vitro. J Bacteriol. 1986;165:88–93. [PMC free article: PMC214374] [PubMed: 3079749]
- 67.
- Gagnon Y, Lacoste L, Champagne N. et al. Widespread use of the Glu-tRNAGln transamidation pathway among bacteria. J Biol Chem. 1996;271:14856–14863. [PubMed: 8662929]
- 68.
- Schön A, Kannangara G, Gough S. et al. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. [PubMed: 3340166]
- 69.
- Kim SI, Nalaskowska M, Germont J-E. et al. Asn-tRNA in Lactobacillus bulgaricus is formed by asparaginylation of tRNA and not by transamidation of Asp-tRNA. Nucleic Acids Res. 1996;24:2649–2651. [PMC free article: PMC146019] [PubMed: 8758990]
- 70.
- Nabholz C, Hauser R, Schneider A. Leishmania tarentolae contains distinct cytosolic and mitochondrial glutaminyl-tRNA synthetase activities. Proc Natl Acad Sci USA. 1997;94:7903–7980. [PMC free article: PMC21527] [PubMed: 9223285]
- 71.
- Stanzel M, Schön A, Sprinz lM. Discrimination against misacylayed tRNA by chloroplast Elongation Factor Tu. Eur J Biochem. 1994;219:435–439. [PubMed: 8307009]
- 72.
- Curnow AW, Tumbula DL, Pelaschier JT. et al. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc Natl Acad Sci USA. 1998;95:12838–12843. [PMC free article: PMC23620] [PubMed: 9789001]
- 73.
- Schmitt E, Moulinier L, Fujiwara S. et al. Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation. EMBO J. 1998;17:5227–5237. [PMC free article: PMC1170850] [PubMed: 9724658]
- 74.
- Curnow AW, Hong K-W, Yuan R. et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. [PMC free article: PMC23611] [PubMed: 9342321]
- 75.
- Becker HD, Min B, Jacobi C. et al. The heterotrimeric Thermus thermophilus Asp-tRNAAsn amidotransferase can also generate Gln-tRNAGln. FEBS Lett. 2000;476:140–144. [PubMed: 10913601]
- 76.
- Salazar JC, Zuniga R, Raczniak G. et al. A dual-specific Glu-tRNA(Gln) and Asp-tRNA(Asn) amidotransferase is involved in decoding glutamine and asparagine codons in Acidithiobacillus ferrooxidans. FEBS Lett. 2001;500:129–131. [PubMed: 11445070]
- 77.
- Tumbula DL, Becker HD, Chang W-Z. et al. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. [PubMed: 10993083]
- 78.
- Raczniak G, Becker HD, Söll D. A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J Biol Chem. 2001;276:45862–45867. [PubMed: 11585842]
- 79.
- Kobayashi M, Fujiwara Y, Goda M. et al. Identification of active sites in amidase: evolutionary relationship between amide bond- and peptide bond-cleaving enzymes. Proc Natl Acad Sci USA. 1997;94:11986–11991. [PMC free article: PMC23678] [PubMed: 9342349]
- 80.
- Decicco CP, Nelson DJ, Luo Y. et al. Glutamyl-γ-boronate inhibitors of bacterial Glu-tRNAGln amidotransferase. Bioorg Med Chem Lett. 2001;11:2561–2564. [PubMed: 11549469]
- 81.
- Wilcox M. γ-Glutamyl phosphate attached to glutamine-specific tRNA. A precursor of glutaminyl-tRNA in Bacillus subtilis. Eur J Biochem. 1969;11:405–412. [PubMed: 4983848]
- 82.
- Horiuchi KY, Harpel MR, Shen L. et al. Mechanistic studies of reaction coupling in Glu-tRNAGln amidotransferase. Biochem. 2001;40:6450–6457. [PubMed: 11371208]
- 83.
- Jahn D, Kim YC, Ishino Y. et al. Purification and functional characterization of the Glu-tRNA(Gln) amidotransferase from Chlamydomonas reinhardtii. J Biol Chem. 1990;265:8059–8064. [PubMed: 1970821]
- 84.
- Nakatsu T, Kato H, Oda J. Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetases. Nat Struct Biol. 1998;5:15–19. [PubMed: 9437423]
- 85.
- Larsen TM, Boehlein SK, Schuster SM. et al. Three-dimensional structure of Escherichia coli asparagine synthetase B: A short journey from substrate to product. Biochem. 1999;38:16146–16157. [PubMed: 10587437]
- 86.
- Russell RB, Sternberg MJE. Two new examples of protein structural similarities within the structure-function twilight zone. Protein Eng. 1997;10:333–338. [PubMed: 9194157]
- 87.
- Kim SI, Germond J-E, Pridmore D. et al. Lactobacillus bulgaricus asparagine synthetase and asparaginyl-tRNA synthetase: coregulation by transcription antitermination. J Bacteriol. 1996;178:2459–2461. [PMC free article: PMC177964] [PubMed: 8636057]
- 88.
- Lamour V, Quevillon S, Diriong S. et al. Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc Natl Acad Sci USA. 1994;91:8670–8674. [PMC free article: PMC44668] [PubMed: 8078941]
- 89.
- Brown JR, Doolittle WF. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J Mol Evol. 1994;49:485–495. [PubMed: 10486006]
- 90.
- Wong JT. A co-evolution theory of the genetic code. Proc Natl Acad Sci USA. 1975;72:1909–1912. [PMC free article: PMC432657] [PubMed: 1057181]
- 91.
- Shiba K, Motegi H, Yoshida M. et al. Human asparaginyl-tRNA synthetase: molecular cloning and the inference of the evolutionary history of Asx-tRNA synthetase family. Nucleic Acids Res. 1998;26:5045–5051. [PMC free article: PMC147956] [PubMed: 9801298]
- 92.
- Wolf YI, Aravind L, Grishin NV. et al. Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed: 10447505]
- 93.
- Woese CR, Olsen GJ, Ibba M. et al. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236. [PMC free article: PMC98992] [PubMed: 10704480]
- Asparaginyl-tRNA Synthetases - Madame Curie Bioscience DatabaseAsparaginyl-tRNA Synthetases - Madame Curie Bioscience Database
- Therapies of Nonsense-Associated Diseases - Madame Curie Bioscience DatabaseTherapies of Nonsense-Associated Diseases - Madame Curie Bioscience Database
- Coronin: The Double-Edged Sword of Actin Dynamics - Madame Curie Bioscience Data...Coronin: The Double-Edged Sword of Actin Dynamics - Madame Curie Bioscience Database
- PARP and the Release of Apoptosis-Inducing Factor from Mitochondria - Madame Cur...PARP and the Release of Apoptosis-Inducing Factor from Mitochondria - Madame Curie Bioscience Database
- THE RNA INFRASTRUCTURE: An Introduction to ncRNA Networks - Madame Curie Bioscie...THE RNA INFRASTRUCTURE: An Introduction to ncRNA Networks - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...