NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Cells show diverse appearances and sizes, ranging from some 30 nanometers up to several meters in length. Besides the classical prokaryotic and eukaryotic cells, there are also very bizarre cells such as the highly reduced symbiotic mitosomes which lack DNA. Other examples of extremely small cells in the nanometre range are the mycosomes and nanobacteria. On the other hand, there are huge eukaryotic cells, the size of which can reach up to several meters. Most of these are multinucleate (coenocytic) due to mitotic divisions not having been followed by cytokinesis. Moreover, cells at all levels of cellular complexity show an inherent tendency to form cell-cell channels. The most conspicuous example is the plant ‘supercell’ where all the cells of the plant body are permanently connected via plasmodesmata. In the last year, the first reports of similar cell-cell channels between animal cells have been published. Moreover, fungal cells fuse together into supracellular mycelia, even exchanging their motile nuclei. This phenomenon is also known for plant cells. Intriguingly, transcellularly moving fungal nuclei communicate with their mating partners via pheromone-like signaling mechanisms.
Already in 1892 Julius Sachs was aware of most of the problems associated with the Cell Theory, which in fact survive until the present. Sachs proposed the Energide concept, postulating that it is the nucleus and its protoplasm which represent the vital unit of living matter within a supracellular construction, while the cell periphery is only a secondary structure generated by the active Energide for its shelter and protection. Recently, we elaborated the Cell Body concept which explains how and why the nucleus and the microtubular cytoskeleton have become merged together to build a coherent and universal unit of eukaryotic life which is autonomous and can synthesize the rest of the cell. However, there are several problems with the term Cell Body as it is sometimes used in other meanings. Here we show that the Energide concept of Sachs can be united with the Cell Body concept. Moreover, we agree with Julius Sachs that the term Energide better invokes the unique properties of this universal unit of supracellular living matter endowed with the vital energy.
Introduction
Current biological thinking is dominated by cells. All living things are defined as either being composed of cells or are cells themselves—membraneous compartments filled with organic molecules in a way that allows them to be autonomous self-replicating units of life. The cell concept originates from Robert Hooke's seminal observations of plant tissue in 1665 when he saw small chambers (in fact, dead and empty cells of cork tissue) which he called ‘cells’.1 These were later, during the next 200 years (when living cells were being examined), shown to contain a nucleus and all the other now familiar organelles. In 1838, Matthais Schleiden and Theodor Schwann proposed that both plants and animals are built of cells; Rudolf Virchow announced his famous dictum ‘omnis cellula e cellula’ in 1858.2 Virchow also wrote that: ‘every animal appears as the sum of vital units, each of which bears in itself the complete characteristics of life’.3 After the cellular nature of brain tissue was confirmed,4 cells became synonymous with life. Cell Theory has been ever since the most influential dogma of contemporary biology.2-7
The Permanent Crisis Surrounding Cell Theory
There are fundamental problems with the Cell Theory. In fact, many of these problems started to be apparent soon after Schleiden and Schwann proposed the cellular basis of plants and animals.8,9 First of all, multinuclear cells were found. Next, prokaryotic cells were discovered which lacked organelles, and their simple architecture was taken as evidence for their ancient or archaic nature. Nowadays, however, several genome-based data have emerged which suggest just the opposite — that the contemporary prokaryotic cells are not so ancient and primitive after all.10,11 More importantly, the eukaryotic cell is defined as a structurally coherent and physically autonomous entity being completely separated from its surroundings by a plasma membrane. However, there are numerous examples, listed in the next sections of this chapter as well as in other chapters of this volume, of cells which are interlinked via cell-cell channels (fig. 1), cells which contain many nuclei either due to nuclear divisions not followed by cell divisions, or which are generated by fusions between cells.
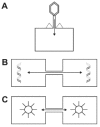
Figure 1
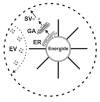
Cell-cell channels in viruses, bacteria, organelles, eukaryotic cells. In eukaryotic cells, whole Energides can move from cell-to-cell via microtubule-based motility.
Permanent cell-cell channels are typical of plant tissues. Similar supracellular constructions which allow the exchange of organelles occur also in the animals kingdom (see other chapters in this volume).12,13 Furthermore, classical cell organelles such as mitochondria and plastids have finally been demasked, after many years of discussion, as being cells too. They are highly reduced descendents of once free-living cells. Besides these smaller organelles, nuclei can also be considered to represent vestiges of ancient endosymbionts.12,14-20 In fact, the nucleus emerges as the first endosymbiont to have invaded the ancient host cell.15 Recently, we have reviewed numerous data in favour of the Cell Body concept which considers not the cell but rather the nucleus with its associated microtubules, the so-called Cell Body, to represent the smallest unit of life in the construction of multicellular or, more exactly, supracellular, organisms.12,21 However, by 1892, Julius Sachs (fig. 2) had already discussed these problems from the perspective of giant cells, such as multinucleate coenocytes and syncytia, which did not obviously conform with the Cell Theory. In order to resolve this problem, he proposed the ‘Energide’ concept.6,22,23

Figure 2
Julius Sachs
Below, we discuss the Energide concept of Sachs in relation to our Cell Body concept and we conclude that both concepts can easily be combined, resulting in an ultimate concept that explains the smallest autonomous unit of supracellular eukaryotic life (summarized in BOX 1 and fig. 3). As the Cell Body is not as suitable term, from now on we will use ‘Energide’ instead of ‘Cell Body’ for this universal unit of eukaryotic life.
More Problems with Cell Theory: What Is a Cell?
Obviously, the definition of a cell is very vague. If the cell should be the smallest unit of life showing all the attributes of living matter, how can Cell Theory accommodate the fact that cellular organelles are, or have been, cells also? Even recently evolved eukaryotic cells have been internalized and enslaved by a recent endosymbiotic event and now act as chloroplasts.24,25 There are other obvious examples of ‘cells within cells’ such as bacterial and yeast spores, as well as pollen tubes and embryo sacs in plants.9,21 Moreover, as Sachs noted more than a hundred years ago, the structural organization of multinucleate cells of coenocytic algae, for instance, those of Ventricaria ventricosa,26 cannot be explained by the Cell Theory.9,21-23
Cell-merging is one of the most important factors shaping the evolution of more complex eukaryotic cells.15 Not only are these fusions examples of secondary endosymbiotic events, but the enslaved cells accomplish dramatic reductions which do not permit their recognition as cells anymore.9 Hydrogenosomes are endosymbiotic organelles related to mitochondria, and some of them, like those of Trichomonas and Neocallimastix, lack any DNA.27 Moreover, another type of mitochondria-derived relict organelles of human pathogen, Entamoeba histolytica, the so-called mitosomes, have sizes below 500 nm and lack any DNA.28 Nevertheless, these mini-cells can reproduce themselves as their numbers are maintained at about 250 for one host cell.29 Both mitosomes and hydrogenosomes provide us with convincing evidences that DNA can be completely removed from one partner cell during an endosymbiotic partnership. Further examples of exotic cellular organisms are mycosomes (0.1—1.0 mm in diameter) isolated from plant plastids and which can give rise to yeast-like cells.30 Even smaller are nanobacteria which are about 30-100 nm in diameter. These tiny spheres are predicted to cause several human diseases.31 The bacterium Spiroplasma is one of the smallest of cells, comparable in size to the diameter of a bacterial flagellar bundle. Despite its extremely small size, it is equipped with a quite complex cytoskeleton.32 On the other hand, there are some viruses which are larger than the smallest bacteria.33 Even more, there are large eukaryotic cells which are visible to the naked eye. Avian egg cells are the largest cells by volume organized by a single nucleus. Plant pollen cells grow several centimeters in length to deliver sperm cells to ovules deeply buried within flower tissues, and multinucleate laticifers may extend the whole length of a Euphorbia plant. The marine algae, Caulerpa and Acetabularia, are single cells which can attain up to several meters in length (in the case of Caulerpa). They are filled with numerous nuclei, each organizing its own microtubular array and cytoplasmic domain. One of the most dramatic examples of huge cells is the mammalian placental syncytiotrophoblast whose surface area can reach up to ten square meters.34
In the classical cell concept, the limiting membrane plays the dominant role whereas the internal parts are not as important and vary from having no DNA, such as in the above-mentioned mitosomes and some hydrogenosomes, to having just a few DNA molecules encoding several proteins within highly reduced organelles, up to the extremely complex cellular interiors encompassing hundreds of nuclei and enormous numbers of other organelles (also cells) with a cytoplasm. In contrast to this diffuse and vague situation with cells, the structure and morphology of the Energide (Cell Body) units are essentially the same throughout the eukaryotic superkingdom (see below), implying that they are much more useful for defining the basic vital units of supracellular eukaryotic life. As we discuss below, cell-cell channels are inherent for all these cell types, starting with the prokaryotic cells and symbiotic cellular organelles, up to eukaryotic cells covering all kingdoms including fungi, plants and animals. If we really need a basic unit of supracellular eukaryotic life, then we must define a new one.
Julius Sachs: Energide as the Basic Unit of Eukaryotic Life Endowed with Vital Energy
Julius Sachs was well aware of all these problems of Cell Theory 110 years ago. He proposed the Energide concept in which it was not the cell, but rather the nucleus with a surrounding sphere of protoplasmic influence (‘Wirkungssphäre’) that defined the autonomous vital unit bearing all the attributes of life.22,23 Energide does not encompass the plasma membrane (‘Zellhaut’) and cell wall which, in contemporary cells, are postulated to be of a secondary nature produced by the active Energide to protect itself from the hostile environment.23 Importantly, while the Energide contains the ‘vital energy’ (‘Lebenskraft’) and divides by a template-based duplication followed by fission, the cell periphery cannot multiply in this way and grows rather via fusion of vesicles generated by the secretory activities of the Energide (‘…von ihm selbst erzeugten Gehäuse’). For Julius Sachs, the cell is only a chamber (‘Zellstoffkammer’) which can harbour one, several, or many Energides. Recently, we discussed numerous modern data confirming most of the predictions made by Julius Sachs more than 100 years ago.9,21 In order to convey his line of arguments to contemporary cell biologists as well as to a broader community of scientists, we translate from German into English the most important parts of his text.
Julius Sachs, Flora 1892; 75:57-67.
Pages 57, 59:
‘Unter einer Energide denke ich mir einen einzelnen Zellkern mit dem von ihm beherrschten Protoplasma, so zwar, dass ein Kern und das ihn umgebende Protoplasma als ein Ganzes zu denken sind und dieses Ganze ist eine Organische Einheit, sowohl im morphologischen wie im physiologischem Sinne. Den Namen Energide wähle ich, um damit die Haupteigenschaft diese Gebildes zu bezeichnen: dass es nämlich innere Thatkraft, oder wenn man will: Lebenskraft besitzt.’‘ ...die einzelne Energide für sich frei leben kann, ohne von einer Zellhaut oder Zelle umgeben zu sein...’‘Zum Begriff der Energide gehört also die Zellhaut nicht; die Sache liegt vielmehr so, dass jede einzelne Energide sich mit einer Zellhaut umgeben kann, oder aber mehrere Energiden zusammen bilden eine Zellhaut ...’
‘Energide is represented by a nucleus associated with its protoplasm in such a way that the nucleus and surrounding protoplasm form an organic unit, both from the morphological and physiological perspectives. The name Energide is chosen to stress the major property of this structure: endowment with the inner power for action or, if one wants: endowment with a vital power.’ ‘…an individual Energide is capable of living freely, without being enclosed within the cell skin or cell chamber…’ ‘The term Energide does not encompass the cell skin; the case is more that each individual Energide is able to enclose itself by a cell skin, or that several Energides together can enclose themselves with one single cell skin …’
Page 60:
‘…dagegen beginnt die Wissenschaft von den lebendingen Dingen mit einem Wort, welches vor mehr als 200 Jahren infolge eines Irrthums entstanden und dann beibehalten worden ist: dem Wort Zelle.’
‘Bekantlich ist das Wort Zelle als Terminus technicus der Botanik nur historisch zu verstehen, insofern Robert Hooke 1667 die innere Configuration des Korkes und der Holzkohle eine zellige, im Sinn einer Bienenwabe, nannte. Auch die Zootomie hat später dieses unglückliches Wort aufgegriffen und für die Elementartheile des thierischen Organismus verwendet, obgleich es dort noch weinger Sinn hatte, als bei denn Pflanzen. — In der 40er Jahren erkannten die Botaniker, dass das Wesentliche der Pflanzenzelle nich ihr Gehäuse, sondern ihr Inhalt, wie wir jetzt sagen, das Protoplasma mit dem Kern ist, und so unterschied man Zelle und Zellinhalt.’
‘…the science which deals with living matter commenced with a controversial word, originating as a mistake more than 200 years ago, and then maintained up to the present day: it is the word Cell.’
‘It is well-known that the word Cell should be understood only from a historical perspective as Robert Hooke referred to the inner structures of cork as being cellular because they closely resembled hexagonal wax cells of honeycomb. Later, zoology also accepted this unfortunate term, despite this word being even more controversial when applied to animals. In the forties [i.e., 1840s], botanists realized that the true living matter is not the shell, but that it resides inside the Cell, and is called protoplasm and nucleus. One should therefore be careful in distinguishing the Cell's interior from its external boundary’.
Energide versus Cell Periphery
Julius Sachs stressed that although the Energide can undergo autonomous template-based growth followed by binary fission, its boundary structures (‘Gehäuse’ or ‘Zellstoffkammer’) can divide only after the accomplishment of Energide-based growth and Energide-instructed division. Importantly, Energide division does not need to be followed by cell periphery division, and this results in the formation of coenocytes. Even more importantly, the opposite situation, that cell divison could occur without a preceding mitosis, was never recorded. Moreover, ‘cells’ lacking their Energides, so-called cytoblasts, cannot divide. On the other hand, Energides can be released from ruptured coenocytic algae, whereupon each Energide regenerates both its plasma membrane and cell wall to form a new complete cell.9 The Energide is the primary unit of supracellular life as it can escape from its cellular boundaries and, later, it can autonomously regenerate these boundary structures. Cell periphery structures can not undergo sustained growth and binary fission without being supported by an Energide.
Recently, we have discovered that cytokinesis in plant cells is driven by the fusion of endosomes which are enclosed by the plasma membrane-derived membrane and enriched with cell wall pectins.35,36 These endosomes are recruited to the division site by the Energide (Cell Body) microtubules.36 In animal cells, too, Energide microtubules specify cytokinesis both spatially and temporally.37 Similarly, sporulating yeast cells generate their plasma membrane de novo from early endosomes enriched with PI(3)P.38 Also, in this situation, the Energide and its microtubules play the essential role of enclosing the cell with a plasma membrane.39 Last, but not least, de novo formation of the plasma membrane was demonstrated during the fragmentation of coenocytic algae, releasing ‘naked’ Energides which then enveloped themselves with boundary structures, thus regenerating new cells.9 All these examples, from throughout the eukaryotic superkingdom, convincingly document the secondary nature of the boundary structures, including the plasma membrane and cell wall/extracellular matrix. This is evident also from the mode of their growth and division. In contrast to the Energide, cell periphery grows and divides by fusion of vesicles.
Adaptability versus Complexity in Cellular Evolution
The formation of eukaryotic cells represented a great leap forward in cellular complexity.40 The Darwinian selection-based evolution towards increased fitness, which acted even on ancient proto-cells early in the development of life,41,42 cannot explain the dramatic increase in cellular complexity.40 In fact, both viruses and prokaryotic cells are far better adapted to harsh environments, and this is the reason why they have been so successful up to the present time. They have been selected by their environment to be efficient mutators and replicators. In several aspects, these simple cells are not primitive at all, and they clearly dominate the biosphere of the Earth. Darwinian evolution is strong in explaining how their adaptability comes about, driven by variation, competition, and selection. However, Darwinian adaptive evolution is weak in explaining satisfactorily why cells should proceed from the less complex towards the more complex forms of cellular organization. It may be a question of selection creating the most efficient combinations of molecular modules at the subcellular level,43 whereas at the cellular level evolution is a question of finding the best combination of symbiotic associations.
In fact, we know several examples where complex cells undergo ‘regressive’ evolution towards a less complex type of cell if they are enslaved by other cells or exposed to extremely challenging environments. Recent advances in the study of symbiogenesis and its role in cellular evolution allowed a breakthrough in our understanding of this paradigm. It was primarily Lynn Margulis15,45-48 who revived this old concept, after more than 70 years of oblivion.44 It appears that the most powerful source of innovation with regard to cellular complexity is the merging of individual cells.15 Of course, this cellular merging is a by-product of cellular competition. But this time it is not the adaptive competition in the sense of Darwinian adaptive evolution, but it rather is predative competition9 in the sense of Margulisian symbiotic evolution which is based on serial endosymbiosis.15,48 In other words, adaptive evolution towards increased fitness is driven by the conflict of organisms within a harsh selectionist environment, whereas predative evolution that drives towards increased cellular complexity is powered by nutritive conflicts between heterotrophic unicellular organisms entertaining a predator/prey based life style. Sometimes, a balance of ‘forces’ is reached between predator and prey cells allowing the development of stable symbiotic relationships between the host and its guest. This ultimately leads to the generation of more complex cells.15,48
Recent analysis of genomes and their genes have revealed that the last universal common ancestor of eukaryotic cells was formed by cell-cell fusion.14,49 This universal ancestor enslaved endosymbiotic cells (organelles) via predatory phagocytosis.9,15-20,45-48,50-52 Thus there are two processes which pave the way towards more complex eukaryotic cells and which disobey Darwinian evolution: lateral gene transfer and endosymbiosis.14,49 Both these processes played a crucial role in evolution of complex eukaryotic cells endowed with nuclei and Energides.9,15
Recently, even viruses have come into consideration as having invented the first nucleus in ancient protocells. Viruses have the inherent ability to invade cells and there are several intriguing similarities between nuclei and viruses.52 Nuclei and viruses lack the protein synthesis machinery and lipid-producing pathways; both transcribe DNA but do not translate RNA, and they contain linear chromosomes equipped with primitive telomeres rather than the circular chromosomes found in bacteria, mitochondria, and plastids.52 Furthermore, viruses are well-known for their ability to induce cell-cell fusion,53 a phenomenon which was essential for the early events required for the formation of ancient eukaryotic cells.14
Duality of Eukaryotic Cells
Eukaryotic cells are characterized by a distinct duality at the structural level (Energide and cell periphery), at the level of genome organization (eubacterial and archaebacterial features), cytoskeleton (actin and tubulin), membrane flow (exocytosis and endocytosis), and cell division (mitosis and cytokinesis).9 These features strongly suggest that the most ancient eukaryotic cell was generated from at least two cellular organisms fusing together and generating the ancient nucleus by transformation of the host cell via recruitment of all the host DNA.9 Recent analysis of diverse genomes has revealed that the eukaryotic genome was, in fact, generated from a fusion of at least two prokaryotic-like genomes, an event which massively speeded-up horizontal gene transfer.14,49 Due to this, all host DNA was transferred into the guest cell which was then transformed into a eukaryotic nucleus.9,15 Moreover, some mitochondria-related hydrogenosomes have also lost all of their DNA.27 This new twist in the evolutionary origin of the contemporary eukaryotic cell is in accordance with the symbiotic origin of eukaryotic nuclei,9,14-20 and it can also explain the absence of DNA from the plasma membrane. Very convincing examples of the complete loss of DNA from endosymbiotic cells are mitosomes.28 In fact, any DNA introduced into a eukaryotic cell is transported into the nucleus and integrated into the genome, allowing for the easy transformability of eukaryotic cells.
As mentioned above, endosymbiosis and lateral gene transfer are not only the most important processes shaping evolution of complex eukaryotic cells, but also these are phenomena which disobey Darwinian evolution.49 Thus, the mechanisms shaping the evolution of eukaryotic cells are extremely important for understanding biological evolution in a much broader perspective. Evolution of complex eukaryotic cells provides us with an important paradigm for the elusive nature of living matter and why it should evolve from low to high complexity. The next big issue to be solved by cell biology is the topic of this book — cells at all levels of complexity are characterized by cell-cell channels. Why and what for? This feature must be very ancient and of extreme importance because it is evident in all contemporary cellular organisms, at all levels of cellular complexity. It seems that both RNA and DNA molecules could be relevant in this respect. Importantly, viewing nuclear pores as cell-cell channels not only supports the endosymbiotic origin of nuclei but also explains the perplexing similarity between nuclear pores and plasmodesmata.54
Cell-Cell Channels: Supracellularity Is Found at All Levels of Cellular Organization
As is apparent throughout this book, cells which have been created by division also seem to have an inherent tendency to fuse into supracellular assemblies. The first breakthrough in this respect was the early discovery of plasmodesmata by Eduard Tangl in 1867.55 It is perhaps typical of biology that this discovery was not fully accepted until some 100 years later when electron microscopy allowed the submicroscopic resolution of these cell-cell channels.55 Much more recently, cell-cell channels of animal cells have been discovered.13,56-58 Hopefully it will not take a further 100 years to have this discovery accepted (or definitely rejected) by mainstream biology.
Cell-Cell Channels in Viruses, Prokaryotes and in Eukaryotic Organelles: Something Special about Cellular End-Poles
Cell-cell channels can be found at all levels of cellular complexity, suggesting that this feature is inherent to cellular organization. Bacterial viruses inject phage DNA into host cells via a channel which is still not characterized in detail (fig.1A).59 This ability of viruses suggests that they should be considered as highly reduced cells specialized for a unique form of cellular parasitism. Bacteria also develop a wide spectrum of transcellular injection machines for cell-cell delivery of their virulence factors into host cells (fig. 1B).60 Interestingly, the nongrowing cellular end-poles assemble these injection organelles which export DNA to other prokaryotic or eukaryotic cells.61-63 In Streptomyces colonies, pore-like structures assemble at end-poles for the conjugative spread of plasmids throughout the colony.64 These cells interacting via their end-poles are also equipped with pores secreting slime and motile hair-like filaments known as pili.65-68 Could this polarisation of cell-cell interactions at the end-poles have implications for the siting of plant plasmodesmata?
Dynamic pili protruding from the bacterial end-poles can rapidly extend and then retract, exerting pulling and pushing forces.66 These might contribute to the alignment of cells into filamentous rows65,67 resembling the cell files of plant tissues.69 In the cyanobacteria, Phormidium and Anabaena, junctional pore complexes are similarly formed at the end-poles of cells which are also arranged into cell files.70 These end-poles are traversed by cell-cell channels which allow direct transport of metabolites as well as communication between heterocysts specialized for nitrogen fixation and other cells accomplishing photosynthesis.6 Similarly, end-poles/cross-walls of plant cells are abundantly traversed by plasmodesmata.69 Intriguingly, prokaryotic cell-cell channels can be targeted by movement proteins of plant viruses which then modify these prokaryotic cell-cell channels in a manner known from virus-affected plant plasmodesmata.71 This finding strongly suggest that prokaryotic and eukaryotic cell-cell channels are evolutionarily related. Intriguingly in this respect, nongrowing end-poles traversed by cell-cell channels, play a central role in polarity of both bacteria and plant cells.69 Furthermore, viruses are well-known to induce cell-cell channels and cell-cell fusions in eukaryotic cells.53
In conformity with free-living bacteria, bacteria-like descendent organelles of eukaryotic cells, such as plastids, mitochondria, and peroxisomes, are all known to fuse together, an event which necessitates the existence of fusion channels. Plastids generate stroma-filled long tubules, so-called stromules. These are highly dynamic structures interconnecting adjacent plastids and allowing an exchange of molecules as large 560 kDa.72-78 Mitochondria are also well-known to undergo fusion and fission processes.79-82 The fusion machinery for plastids are not known yet, but there are data accumulating on the fusion machineries of mitochondria83 and peroxisomes.84 It is obvious that these ‘cell-cell’ fusion mechanisms differ from those which drive membrane fusion in exocytic and endocytic pathways.85
The outer leaflet of the nuclear envelope extrudes endoplasmic reticulum (ER) whereas the inner leaflet is anchored via the nuclear lamina, to the chromatin complex. ER pervades the whole cytoplasmic space, budding as a Golgi Apparatus (GA) and secretory vesicles (fig. 3), as well as interacting with other organelles.86-88 In plants, ER elements extend from cell-to-cell across the plasmodesmata,54-55 but this is likely to be a secondary event following from the primary contact and fusion of adjacent cell peripheries. Microinjection of 3 kDa dye into the lumen of ER enables visualization of dye movement into adjacent cells where it first appears within the nuclei.89 Obviously, Energides from adjacent plant cells are now able to interact via the physical continuity of their ER membranes.
Nuclear Pores as Prototypic Cell-Cell Channels
If the endosymbiotic origin of nuclei can be definitely confirmed, then nuclear pores also can be viewed as specialized cell-cell channels resembling plant plasmodesmatal cell-cell channels.54 These are optimized for effective cell-cell transport of proteins, as well as RNA and DNA molecules. Intriguingly, nuclear pores are highly selective with respect to the macromolecules which they transport. Particles as large as ribosomes can pass through nuclear pores whereas much smaller molecules may be actively excluded.90, 91 Similarly, plant plasmodesmata support transport of certain large proteins whereas the small auxin molecule, for example, is excluded from direct cell-cell transport.92 Recently, both plasmodesmata93-95 and nuclear pores96,97 were revealed to be closely associated with molecules driving endocytosis. Viruses, RNA and DNA molecules all are internalized into endosomes which then deliver them into either the ER or the nuclear pores.98,99 Many cell periphery and endocytic proteins use this endosomal route for their transport into and out of the nucleus.100-102 Interestingly in this respect, endosomes are the only type of organelle which invades and travels through cell-cell channels of animal cells.13 Furthermore, the acrosomal compartment which drives the cell-cell fusion between sperm cells and oocytes, has endosomal features; and it is also enriched with synaptic molecules which recycle via secretory endosomes.103-104
Cell-Cell Channels in Filamentous Fungi, Plants and Animals
Cell-cell channels are found in all multicellular organisms, including filamentous fungi, plants and animals. As their presence, structure, and functions are extensively discussed in several chapters of this volume, we limit ourselves to just some basic information. Fungal hyphae are tip-growing tubules which inherently fuse together. As a result, complex mycelial networks are formed to allow the exchange of large masses of protoplasm, organelles and, importantly, the highly motile Energides.105-109 All cells of plant bodies are well-known to be interconnected via plasmodesmata, 55,71,87 and if they are not formed as an accompaniment of cytokinesis, they can form later as so-called secondary plasmodesmata. But plant tissues also form much larger cell-cell channels, known as cytoplasmic channels or intercellular bridges.110,111 These macro-channels allow transfer not only of large organelles but also of nuclei (Energides). This spectacular process was documented both in situ110,112 and in vivo.113 In addition, plasmodesmata can be transformed into the massive pores of sieve plates, thus allowing the mass flow of metabolites along phloem elements.114
There are numerous examples of cell-cell channels in animals. Well studied are fusomes and intercellular bridges of germinal cells of numerous taxa. They form as a result of incomplete cytokinesis. 115-118 Fusomes develop from spherical vesicular compartments into large cell-cell channels which not only nurse developing oocytes but also determine their polarity due to the anchoring of the Energide microtubules.119 Similar to plasmodesmata of plant cells, fusomes provide direct Energide-Energide connectivity via ER elements.120 Recently, exciting observations have revealed thin (50-200 nm) nanotubules between cultured PC12 rat neural cells and kidney cells.13 These actin-based cell-cell channels allow transcellular transport of myosin-associated endosomes. Similar nanotubes were found also at immunological synapses where exchange of GPI-anchored proteins and class I MHC protein was noted.56,57 Even larger (100 nm-5 μm in width, and up to 100 μm in length) cell-cell tubules were reported between DU 145 human prostate cancer cells. These are positive for alpha-tubulin and transport very large membrane vesicles (up to 3 μm in diameter).58
Cell-Cell Channels and the Energide: Implications for Cell Theory
Microtubules radiating from the Energide/Cell Body represent an ideal instrument by which to delimit the cytoplasmic, cellular boundary. Moreover, this limit is somehow controlled by the DNA mass, or amount, stored within the nucleus.9 The dynamic instability of microtubules allows the Energide to measure continuously the size and shape of its ‘sphere of influence’ (cell). Also, the microtubules collectively serve as a highly effective tool for Energide motility within the confines of its cell.9,122 But it is mysterious how DNA, irrespective its coding information, can determine both the size of the nucleus (Energide) and the size of its cell. Concerning the DNA amount/size of the nucleus correlation, the skeletal DNA hypothesis has been proposed121 while the Energide/Cell Body concept can explain the tight correlation between DNA amount and cell size.9,123
Energide/Cell Body Microtubules Act as a Tool to Prevent their Fusion, to Obtain Information Which Is Acquired and Processed at the Plasma Membrane, to Explore the Cellular Space and to Invade Adjacent Cells, as Well as to Repair Old and Generate New Cell Periphery
What features are so special about the Energide (Cell Body) that confer upon it properties which have allowed the Energide to act as the vital unit of supracellular eukaryotic organisation involving constructions composed of millions of Energides? We argue that this feature is due to the unique association between the DNA-based nuclei and the tubulin-based microtubular cytoskeleton.9 Julius Sachs obviously could not incorporate microtubules into his Energide concept. So, by merging the Energide concept with the Cell Body concept,9,21 we should be able to unravel the full scope of the unique properties of the Sachsian Energide.
First of all, whereas cells show an extremely wide range of sizes and structural organizations, Energides are basically constant structures, always consisting of nucleus sheathed within perinuclear radiating microtubules. These are instrumental for numerous properties of a vital Energide. Energide microtubules are essential for preventing accidental nuclear fusions which would easily happen either during cytokinesis or after cell-cell fusions.9 In fact, depolymerization of Energide microtubules during mitosis results in the immediate fusion of daughter Energides and the subsequent formation of a polyploid nucleus.124 This inherent tendency of adjoining nuclei to fuse together, if they are not ensheathed with microtubules or cell periphery boundaries, is a very strong argument for cellular nature and endosymbiotic origin of nuclei. Energide-Energide fusions can be observed in some ephemeral plant tissues like endosperm.21,123-125 This is because radiating microtubules from adjacent Energides, which interact via plus ends of their microtubules so that they usually keep their distance from each other, under some circumstances can fail to be effective organizers of their cytoplasmic ‘spheres’ of influence.9,21
Next, Energides of syncytial and coenocytic cells are regularly distributed in their cytoplasmic space. This is also due to the physical interactions between plus ends of microtubules from the adjacent Energides.9,21 The claim upon protoplasmic space, the volume of which is directly related to the amount of DNA stored within a given nucleus, leads to the phenomenon known as the nucleo-cytoplasmic ratio.9,123 However, also important is the way in which DNA is packed within the nuclei: highly condensed generative nuclei of pollen grains form small Energides whereas decondensed large vegetative nuclei form large Energides, despite the fact that the DNA amount in both these nuclei can be the same.123 The mechanisms behind these phenomena include both nucleus-stored molecules having MTOC properties123 and the Energide's ability to generate longer microtubules when more MTOC molecules are available.9,21 Microtubules initiated at the nuclear surface, and having their dynamic plus ends interacting with the plasma membrane, are ideally suited for the transfer towards the Energide nucleus of information acquired and processed at the cell periphery and plasma membrane. Importantly, Energide-associated dynamic microtubules can act as vehicles allowing the movement of whole Energides/Cell Bodies through cellular space.122
Finally, these microtubules can gain control over the distribution of cell periphery-derived endosomes filled with internalized cell wall material and enclosed by a membrane, which are ideally suited for cell periphery repair or for rapid generation of a new cell periphery.35,36,39 In plants, Energides are actively recruited to cell periphery sites which have been compromised by pathogen attack or which accomplish rapid and highly polarized cell growth via tip-growth.126,127 Similar tipward localization of Energides is typical also of filamentous fungi.109,128,129
Energide/Cell Body Can Use Cell-Cell Channels to Travel across Cellular Boundaries
As briefly mentioned above, Energides (Cell Bodies) not only explore the confines of their own cells122 but can also move to an adjacent cell if the connecting channels are sufficiently large. This phenomenon, which is clearly incompatible with the current version of Cell Theory, is the final proof for the autonomous primary nature of the Energide. In plants, there are numerous ultrastructural observations of this process, known also as cytomixis.110,112 However, these findings have been criticized as representing nothing more than aberrant cytokinesis where daughter nuclei have been squeezed thtough a maturing cell plate. But in vivo studies provided the final proof for a genuine cytomixic cell-cell movement of nuclei (Energides).105,106,109,113 Moreover, other well studied examples of nuclear transfer involve a structure called the conjusome, a cell-cell channel specialized for conjugation in Tetrahymena.130,131
A rather exotic cell-cell transfer of nuclei occurs between the parasitic alga, Choreocolax, and cells of its red-alga host, Polysiphonia.132 This resembles the situation in fusing fungal hyphae in which genetically different Energides co-exist and are exchanged.107,108,133,134 Intriguingly, the fungal Energides can recognize their respective mating Energide partners.135 This mysterious process is accomplished via a pheromone/receptor system operating at the level of Energides (Cell Bodies) of the opposite mating types.136,137 This pheromone-like communication between individual Energides is a further very strong argument for the cellular and endosymbiotic nature of nuclei. When a spindle pole body is inserted into the nuclear envelope, it acts as an MTOC and interacts with the DNA. This DNA-spindle pole body complex was proposed to be responsible for the recognition of self and nonself at the level of individual Energides.138
Energide microtubules are extremely sensitive to mechanical treatments. Thus, it is not surprising to find an extremely low (typically between 0-4%) success rate in experiments of animal cloning which have involved nuclear transfer.139-141 We can safely envision that if more care were taken over the intactness of the whole Energide during this process,the success rate (as judged by the frequency of developing embryos) would increase considerably and would help optimize the cloning process. To achieve this, one needs to be aware that the whole intact Energide (Cell Body), including its ensheathing microtubules, must be transferred into the recipient cell, not just the nucleus alone.
Impacts of Cell-Cell Channels on a Hypothetical Scenario of Eukaryotic Cell Evolution
From the currently available data, two major phases in the evolution of complex eukaryotic cells can be envisioned. An initial ‘communal’ phase142,143 dominated by repeated fusion events between proto-cells allowed the early cells to increase their amounts of DNA. As a vestige of this primordial phase, the cell periphery of contemporary cells still shows an inherent tendency to fuse together thereby forming cell-cell channels. Some ancient proto-cells were more effective in this fusion process than others, which resulted in their larger cell size. These large cells suffered from osmotic imbalance resulting in the rupture of the early plasma membrane.144,145 The frequent bursting of these proto-cells forced them to develop a means of rapid repair of the cell periphery as well as an ability to assemble more robust peripheries by enveloping the plasma membrane with a cell wall. These events heralded the second ‘predatory’ phase which, in fact, continues up to the present time. A convincing example of this is, the mutual enslavement of eukaryotic algae thereby generating eukaryote-eukaryote chimaeras.24,25
The onset of the ‘predatory’ phase was preceded by the generation of the most ancient eukaryotic cell. Now it is clear that this last common ancestor of eukaryotic cells was a complete cells equipped with nucleus, cytoskeleton, mitochondria, endoplasmic reticulum, Golgi apparatus, and vesicle trafficking. 146,147 Merging of cells was accomplished presumably with the invasion of a large and soft-periphery host cell, still suffering from the frequent bursting events, by a more advanced smaller guest cell already equipped with a more rigid-periphery.9 These ancient nucleated proto-cells then went on to ‘invent’ actin-based phagocytosis which allowed them not only to initiate an active predatory lifestyle but also to accomplish a very efficient cell periphery repair process using recycling secretory endosomes filled with extracellular matrix components and enclosed by a plasma membrane-derived membrane.35,36 Similarly, a phagocytosis-like process is known to be accomplished in the bacterium Bacillus subtilis during the formation of spores.148,149 The cellular prey internalized by these proto-cells active in phagocytosis, and perhaps not fully equipped with all the enzymes needed to digest prey, often survived and developed into organelles such as peroxisomes, mitochondria, and plastids which are now features of eukaryotic cells.9 During evolution, lateral gene transfer resulted in loss of DNA from developing organelles (DNA-losing ancient guest cells) and their accumulation within nuclei (DNA-gaining ancient guest cells). In some situations, all DNA can be lost: for example, mitosomes are remnants of mitochondrion-related organelles that lack any detectable DNA.28 Furthermore, some hydrogenosomes also lost all of their DNA. Similarly, all DNA from the host cell (the cell contributing the plasma membrane) was transferred into the guest cell (the cell which became the nucleus), and it might well be that peroxisomes and centrosomes also represent highly reduced guest cells, having lost all DNA via lateral gene transfer.150 For instance, the cell wall of plant cells appears to be descended from endosymbiotic cyanobacteria which, during their evolutionary transformation into plastids, have donated their cell wall genes to the guest cell.151
Conclusions and Outlook
The most important message of this chapter (and the whole book) is that cell-cell channels are an inherent feature of cells at all levels of complexity, from relatively simple bacteria up to large and complex plant and animal cells. In fact, plants are supracellular organisms because most of their cells (Energides) are interconnected by plasmodesmatal channels.55,87 This might turn out to be true for animals. Conclusive evidence would be if the sensitive and apparently short-lived nanotubes13,56-58 were found in intact tissues. There are some intriguing generalities with respect to cell-cell channels. First, viruses and molecules of RNA and DNA seem to have the ability to induce cell-cell channels. Both viruses and bacteria—and here for plant cells, consider Agrobacterium tumefaciens152—can induce cell-cell channels, enabling them to invade host cells and then to pirate the nuclear pores and thereby gain access to the DNA replication machinery within the host nuclei. After their replication, they escape from the nucleus and return to the cytoplasm; they then either kill the cell to gain access to other cells (animals) thereby inducing new cell-cell channels, or they gate the existing channels to allow cell-cell spread of virus or bacterium throughout the whole organism (plants). Besides viruses and bacteria, small RNA molecules also can spread from cell-to-cell, and this ability is used for transcellular and global silencing of specific genes. This transfer of small RNAs across cell boundaries is an ancient feature, allowing application of the RNA interference (RNAi) technology for both animals and plants.153-155 The original discovery of RNAi was in C. elegans156 where the transport of RNAs across cellular borders is dependent on the transmembrane protein SID1.
Darwinian adaptive evolution that tends towards increased fitness is still the major force with respect to the adaptation of cells and other more complex units of life, like multicellular organisms, to the ever-changing ambient environment. However, it operates in parallel with the Margulisian symbiotic evolution which generates an increased complexity of evolving systems such as eukaryotic cells and their vital units of supracellular life, which are here called Energides. This is true both for the evolution of complex eukaryotic cells, as well as for their assembly into multi-cellular or, better, supra-cellular, organisms. Cell-cell fusion emerges as important factor determining animal cell behavior. Cell-cell channels are inherent not only to plants87 but also to animals.13,56-58 Both unicellular15 as well as supracellular organisms87 should be considered as complex ecosystems composed of numerous modified cellular micro-organisms. Finally, in contrast to cells which are widely heterogeneous in their size (ranging from 30 nanometres up to several metres), architecture, and mechanism of their divisions; Energides are extremely conservative in all these respects. This makes the Energide an ideal candidate for the universal unit of supracellular eukaryotic life. Appreciation of Energides as vital units endowed with all attributes of life is critical for our understanding of still enigmatic processes and phenomena of contemporary cell biology, such as transformation, plasticity, and cloning of eukaryotic cells.
References
- 1.
- Hooke R. Of the schematisme or texture of cork, and of the cells and pores of some other such frothy bodies. Micrographia, Observation 18. London. 1665:112–116.
- 2.
- Harris H. The Birth of the Cell. New Haven: Yale University Press. 1999
- 3.
- Lodish H, Baltimore D, Berk A. et al. Molecular Cell Biology, 3rd ed. New York: W.H. Freeman and Company. 1995
- 4.
- Mazzarello P. A unifying concept: the history of cell theory. Nat Cell Biol. 1999;1:E13–E15. [PubMed: 10559875]
- 5.
- Alberts B, Bray D, Hopkin K. et al. Essential Cell Biology. 2nd ed. New York: Taylor & Francis Group: Garland Science. 2004
- 6.
- Kleinig H, Sitte P. Zellbiologie. Stuttgart, New York: Gustav Fischer Verlag. 1984
- 7.
- Pollard TD, Earnshaw WC. Cell Biology. Philadelphia, London, New York, St. Louis, Sydney, Toronto: Saunders, Elsevier Science. 2002
- 8.
- Richmond ML.T.H. Huxley's criticism of German Cell Theory: an epigenetic and phsiological interpretation of cell structure. J Hist Biol. 2000;33:247–289. [PubMed: 11640226]
- 9.
- Baluska F, Volkmann D, Barlow PW. Eukaryotic cells and their Cell Bodies: Cell Theory revisited. Ann Bot. 2004;94:9–32. [PMC free article: PMC4242365] [PubMed: 15155376]
- 10.
- Forterre P, Philippe H. Where is the root of the universal tree of life. BioEssays. 1999;21:871–879. [PubMed: 10497338]
- 11.
- Poole A, Jeffares D, Penny D. Early evolution: prokaryotes, the new kids on the block. BioEssays. 1999;21:880–889. [PubMed: 10497339]
- 12.
- Baluska F, Hlavacka A, Volkmann D. et al. Getting connected: actin-based cell-to-cell channel in plants and animal. Trends Cell Biol. 2004;14:404–408. [PubMed: 15308205]
- 13.
- Rustom A, Saffrich R, Markovic I. et al. Nanotubular highways for intercellular organelle transport. Science. 2004;303:1007–1110. [PubMed: 14963329]
- 14.
- Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. [PubMed: 15356622]
- 15.
- Margulis L. Serial endosymbiotic theory (SET) and composite individuality. Transition from bacterial to eukaryotic genomes. Microbiol Today. 2004;31:172–174.
- 16.
- Margulis L, Dolan MF, Guerrero R. The chimeric eukaryote: origin of the nucleus from karyomastigont in amitochondriate protist. Proc Natl Acad Sci USA. 2000;97:6954–6999. [PMC free article: PMC34369] [PubMed: 10860956]
- 17.
- Horiike T, Hamada K, Kanaya S. et al. Origin of eukaryotic cell nuclei by symbiosis of Archaea in Bacteria is revealed by homology-hit analysis. Nat Cell Biol. 2001;3:210–214. [PubMed: 11175755]
- 18.
- Horiike T, Hamada K, Shinozawa T. Origin of eukaryotic cell nuclei by symbiosis of Archaea in Bacteria supported by the newly clarified origin of functional genes. Genes Genet Syst. 2002;77:369–376. [PubMed: 12441648]
- 19.
- Dolan MF, Melnitsky H, Margulis L. et al. Motility proteins and the origin of the nucleus. Anat Rec. 2002;268:290–301. [PubMed: 12382325]
- 20.
- Hartman H, Fedorov A. The origin of the eukaryotic cell: a genomic investigation. Proc Natl Acad Sci USA. 2002;99:1420–1425. [PMC free article: PMC122206] [PubMed: 11805300]
- 21.
- Baluska F, Volkmann D, Barlow PW. Cell bodies in a cage. Nature. 2004;428:371. [PubMed: 15042068]
- 22.
- Sachs J. Beiträge zur Zellentheorie. Energiden und Zellen. Flora. 1892;75:57–67.
- 23.
- Sachs J. Weitere Betrachtungen über Energiden und Zellen. Flora. 1892;81:405–434.
- 24.
- Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae). Philos Trans R Soc Lond B Biol Sci. 2003;358:109–133. [PMC free article: PMC1693104] [PubMed: 12594921]
- 25.
- Keeling PJ. Diversity and evolutionary history of plastids and their hosts. Am J Bot. 2004;91:1481–1493. [PubMed: 21652304]
- 26.
- Shepherd VA, Beilby MJ, Bisson MA. When is a cell not a cell? A theory relating coenocytic structure to the unusual electrophysiology of Ventricaria ventricosa (Valonia ventricosa). Protoplasma. 2004;223:79–91. [PubMed: 15221513]
- 27.
- Embley TM, van der Giezen M, Horner DS. et al. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life. 2003;55:387–395. [PubMed: 14584589]
- 28.
- Leon-Avila G, Tovar J. Mitosomes of Entamoeba histolytica are abundant mitochondrion-related remnant organelles that lack a detectable organellar genome. Microbiology. 2004;150:1245–1250. [PubMed: 15133087]
- 29.
- Tovar J, Leon-Avila G, Sanchez LB. et al. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. [PubMed: 14614504]
- 30.
- Atsatt PR. Fungus propagules in plastids: the mycosome hypothesis. Int Microbiol. 2003;6:17–26. [PubMed: 12730709]
- 31.
- Maniloff J, Nealson KH, Psenner R. et al. Nanobacteria: size limits and evidence. Science. 1997;276:1773–1776.
- 32.
- Kürner J, Frangakis AS, Baumeister W. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science. 2005;307:436–438. [PubMed: 15662018]
- 33.
- Raoult D, Audic S, Robert C. et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. [PubMed: 15486256]
- 34.
- Benirschke K. Remarkable placenta. Clin Anat. 1997;11:194–205. [PubMed: 9579593]
- 35.
- Baluska F, Liners F, Hlavacka A. et al. Cell wall pectins and xyloglucans are internalized into dividing root cells and accumulate within cell plates during cytokinesis. Protoplasma. 2005 In press [PubMed: 16228896]
- 36.
- Dhonuksche P. Visualizing microtubule dynamics and membrane trafficking in live and dividing plant cells. Ph.D. Thesis, University of Amsterdam. 2005
- 37.
- Burgess DR, Chang F. Site selection for the cleavage furrow at cytokinesis. Trends Cell Biol. 2005;15:156–165. [PubMed: 15752979]
- 38.
- Onishi M, Koga T, Morita R. et al. Role of phosphatidylinositol 3-phosphate in formation of forespore membrane in Schizosaccharomyces pombe. Yeast. 2003;20:193–206. [PubMed: 12557273]
- 39.
- Shimoda C. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J Cell Sci. 2004;117:389–396. [PubMed: 14702385]
- 40.
- de DuveC. The birth of complex cells. Scient Amer. 1996;274(4):38–45. [PubMed: 8907651]
- 41.
- de DuveC. The onset of selection. Nature. 2005;433:581–582. [PubMed: 15703726]
- 42.
- Ingber DE. The origin of cellular life. BioEssays. 2002;22:1160–1170. [PubMed: 11084632]
- 43.
- Hartwell LH, Hopfield JJ, Leibler S. et al. From molecular to modular cell biology. Nature. 1999;402(suppl):C47–C52. [PubMed: 10591225]
- 44.
- Altman R. Die Elementarorganismen und Ihre Beziehungen zur den Zellen. Leipzig: Verlag von Veit. 1890
- 45.
- Margulis L. Origin of Eukaryotic Cells. Yale University Press: New Haven. 1970
- 46.
- Margulis L. Symbiosis in Cell Evolution. Life and Its Environment on the Early Earth. San Francisco: W. H. Freeman. 1981
- 47.
- Margulis L. Symbiosis in Cell Evolution. San Francisco: W. H. Freeman. 1993
- 48.
- Margulis L, Sagan D. Acquiring Genomes: a Theory of the Origin of Species. New York: Basic Books. 2002
- 49.
- Martin W, Embley TM. Early evolution comes full circle. Science. 2004;431:134–137. [PubMed: 15356611]
- 50.
- López-García P, Moreira D. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci. 1999;24:88–93. [PubMed: 10203753]
- 51.
- Moreira D, López-García P. Symbiosis between methanogenic Archaea and Proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J Mol Evol. 1998;47:517–530. [PubMed: 9797402]
- 52.
- Pennisi E. The birth of the nucleus. Science. 2004;305:766–768. [PubMed: 15297641]
- 53.
- Hernandez LD, Hoffman LR, Wolfsberg TG. et al. Virus-cell and cell-cell fusion. Annu Rev Dev Biol. 1996;12:627–661. [PubMed: 8970739]
- 54.
- Lee J-Y, Yoo B-C, Lucas WJ. Parallels between nuclear-pore and plasmodesmal trafficking of information molecules. Planta. 2000;210:177–187. [PubMed: 10664123]
- 55.
- Carr DJ. Historical perspectives on plasmodesmataIn: Gunning BES, Robards AW, eds.Intercellular Communication in Plants: Studies on PlasmodesmataBerlin, Heidelberg, New York: Springer Verlag,1976291–295.
- 56.
- önfelt B, Nedvetzki S, Yanagi K. et al. Membrane nanotubes connect immune cells. J Immunol. 2004;173:1511–1513. [PubMed: 15265877]
- 57.
- önfelt B, Davis DM. Can membrane nanotubes facilitate communication between immune cells? Biochem Soc Trans. 2004;32:676–678. [PubMed: 15493985]
- 58.
- Vidulescu C, Clejan S, O'Connor KC. Vesicle traffic through intercellular bridges in DU 145 human prostate cancer cells. J Cell Mol Med. 2004;8:388–396. [PMC free article: PMC6740136] [PubMed: 15491514]
- 59.
- Errington J, Bath J, Wu LJ. DNA transport in bacteria. Nat Rev Mol Cell Biol. 2001;2:538–544. [PubMed: 11433368]
- 60.
- Gauthier A, Thomas NA, Finlay BR. Bacterial injection machines. J Biol Chem. 2003;278:25273–25276. [PubMed: 12759358]
- 61.
- Kumar RB, Das A. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol Microbiol. 2002;43:1523–1532. [PubMed: 11952902]
- 62.
- Judd PK, Kumar RB, Das A. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol Microbiol. 2005;55:115–124. [PubMed: 15612921]
- 63.
- Rohde M, Püls J, Buhrdorf R. et al. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49:219–234. [PubMed: 12823823]
- 64.
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol Molec Biol Rev. 2003;67:277–301. [PMC free article: PMC156469] [PubMed: 12794193]
- 65.
- Kaiser D. Coupling cell movement to multicellular development in myxobacteria. Nat Rev Microbiol. 2003;1:45–54. [PubMed: 15040179]
- 66.
- Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA. 2001;98:6901–6904. [PMC free article: PMC34450] [PubMed: 11381130]
- 67.
- Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci USA. 1998;95:3054–3058. [PMC free article: PMC19693] [PubMed: 9501214]
- 68.
- Wolgemuth C, Hoiczyk E, Kaiser D. et al. How myxobacteria glide. Curr Biol. 2002;12:369–377. [PubMed: 11882287]
- 69.
- Baluska F, Wojtaszek P, Volkmann D. et al. The architecture of polarized cell growth: the unique status of elongating plant cells. BioEssays. 2003;25:569–576. [PubMed: 12766946]
- 70.
- Hoiczyk E, Baumeister W. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr Biol. 1998;8:1161–1168. [PubMed: 9799733]
- 71.
- Heinlein M, Wood MR, Thiel T. et al. Targeting and modification of prokaryotic cell-cell junctions by tobacco mosaic virus cell-to-cell movement protein. Plant J. 1998;14:345–351. [PubMed: 9628027]
- 72.
- Menzel D. An interconnected plastidom in Acetabularia: implications for the mechanism of chloroplast motility. Protoplasma. 1994;179:166–171.
- 73.
- Köhler RH, Cao J, Zipfel WR. et al. Exchange of protein molecules through connections between higher plant plastids. Science. 1997;276:2039–2042. [PubMed: 9197266]
- 74.
- Köhler RH, Schwille P, Webb WW. et al. Active protein transport through plastid tubules: velocity quantified by fluorescence correlation spectroscopy. J Cell Sci. 2000;113:3921–3930. [PubMed: 11058079]
- 75.
- Kwok EY, Hanson MR. Plastids and stromules interact with the nucleus and cell membranes in vascular strands. Plant Cell Rep. 2004;23:188–195. [PubMed: 15252692]
- 76.
- Kwok EY, Hanson MR. GFP-labeled Rubisco and aspartate aminotransferase are present in plastid stromules and traffic between plastids. J Exp Bot. 2004;55:595–604. [PubMed: 14754918]
- 77.
- Natesan SKA, Sullivan JA, Gray JC. Stromules: a characteristic cell-specific feature of plastid morphology. J Exp Bot. 2005;56:787–797. [PubMed: 15699062]
- 78.
- Gunning BES. Plastid stromules: video microscopy of their outgrowth, retraction, tensioning, anchoring, branching, bridging and tip growth. Protoplasma. 2005;225:33–42. [PubMed: 15868211]
- 79.
- Bereiter-Hahn J, Vöth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. [PubMed: 8204911]
- 80.
- van GestelK, Verbelen J-P. Giant mitochondria are a response to low oxygen pressure in cells of tobacco (Nicotiana tabacum L.). J Exp Bot. 2002;53:1215–1218. [PubMed: 11971932]
- 81.
- Logan DC. Mitochondrial dynamics. New Phytol. 2003;160:463–478. [PubMed: 33873653]
- 82.
- Westermann B. Merging mitochondria matters. Cellular role and molecular machinery of mitochondrial fusion. EMBO Rep. 2002;3:527–531. [PMC free article: PMC1084147] [PubMed: 12052774]
- 83.
- Mozdy AD, Shaw JM. A fuzzy mitochondrial fusion apparatus comes into focus. Nat Rev Mol Cell Biol. 2003;4:468478. [PubMed: 12778126]
- 84.
- Boukh-Viner T, Guo T, Alexandrian A. et al. Dynamic ergosterol- and ceramide-rich domains in the peroxisomal membrane serve as an organizing platform for peroxisome fusion. J Cell Biol. 2005;168:761–773. [PMC free article: PMC2171827] [PubMed: 15738267]
- 85.
- Jahn R, Lang T, Südhof TC. Membrane fusion. Cell. 2003;112:519–533. [PubMed: 12600315]
- 86.
- Staehelin LA. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 1991;11:1151–1165. [PubMed: 9225461]
- 87.
- Gamalei YuV. Supercellular plant organization. Russ J Plant Physiol. 1997;44:706–730.
- 88.
- Holthuis JC, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol. 2005;6:209–220. [PubMed: 15738987]
- 89.
- Faulkner C, Brandom J, Maule A. et al. Plasmodesmata 2004. Surfing the symplasm. Plant Physiol. 2005;137:607–610. [PMC free article: PMC1065361] [PubMed: 15710689]
- 90.
- Fahrenkrog B, Köser J, Aebi U. The nuclear pore complex: a jack of all trades? Trends Biochem Sci. 2005;29:175–182. [PubMed: 15082311]
- 91.
- Timney BL, Rout MP. Robbing from the pore. Nat Cell Biol. 2004;6:177–179. [PubMed: 15039784]
- 92.
- Sheldrake AR. Effects of osmotic stress on polar auxin transport in Avena mesocotyl sections. Planta. 1979;145:113–117. [PubMed: 24317665]
- 93.
- Baluska F, Samaj J, Hlavacka A. et al. Myosin VIII and F-actin enriched plasmodesmata in maize root inner cortex cells accomplish fluid-phase endocytosis via an actomyosin-dependent process. J Exp Bot. 2004;55:463–473. [PubMed: 14739268]
- 94.
- Haupt S, Cowan GH, Ziegler A. et al. Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell. 2005;17:164–181. [PMC free article: PMC544497] [PubMed: 15608333]
- 95.
- Oparka KJ. Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 2004;9:33–41. [PubMed: 14729217]
- 96.
- Devos D, Dokudovskaya S, Alber F. et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PloS Biol. 2004;2(12):e380. [PMC free article: PMC524472] [PubMed: 15523559]
- 97.
- Antonin W, Mattaj IW. Nuclear pore complexes: round the bend? Nat Cell Biol. 2005;7:10–12. [PubMed: 15632943]
- 98.
- Guyader M, Kiyokawa E, Abrami L. et al. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. J Virol. 2002;76:10356–10364. [PMC free article: PMC136590] [PubMed: 12239312]
- 99.
- Manunta M, Tan PH, Sagoo P. et al. Gene delivery by dendrimers operates via a cholesterol dependent pathway. Nucl Acids Res. 2004;32:2730–2739. [PMC free article: PMC419601] [PubMed: 15148360]
- 100.
- Vecchi M, Polo S, Poupon V. et al. Nucleocytoplasmic shuttling of endocytic proteins. J Cell Biol. 2001;153:1511–1517. [PMC free article: PMC2150719] [PubMed: 11425879]
- 101.
- Benmerah A, Scott M, Poupon V. et al. Nuclear function for plasma membrane-associated proteins? Traffic. 2003;4:503–511. [PubMed: 12839493]
- 102.
- Benmerah A. Endocytosis: signalling from endocytic membranes to the nucleus. Curr Biol. 2004;14:R314–R316. [PubMed: 15084302]
- 103.
- Ramalho-Santos J, Schatten G, Moreno RD. Control of membrane fusion during spermiogenesis and the acrosome reaction. Biol Reprod. 2002;67:1043–1051. [PubMed: 12297516]
- 104.
- Redecker P, Kreutz MR, Bockmann J. et al. Brain synaptic junctional proteins at the acrosome of rat testicular germ cells. J Histochem Cytochem. 2003;51:809–819. [PubMed: 12754292]
- 105.
- Giovannetti M, Fortuna P, Citernesi AS. et al. The occurrence of anastomosis formation and nuclear exchange in intact arbuscular mycorrhizal networks. New Phytol. 2001;151:717–724. [PubMed: 33853252]
- 106.
- Giovannetti M, Sbrana C, Avio L. Patterns of below-ground plant interconnections established by means of arbuscular mycorrhizal networks. New Phytol. 2004;164:175–181. [PubMed: 33873483]
- 107.
- Glass NL, Kaneko I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell. 2003;2:1–8. [PMC free article: PMC141178] [PubMed: 12582117]
- 108.
- Glass NL, Rasmussen C, Roca MG. et al. Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 2004;12:135–141. [PubMed: 15001190]
- 109.
- Xiang X, Fischer R. Nuclear migration and positioning in filamentous fungi. Fung Gen Biol. 2004;41:411–419. [PubMed: 14998524]
- 110.
- Wang XY, Yu CH, Li X. et al. Ultrastructural aspects and possible origin of cytoplasmic channels providing intercellular connection in vegetative tissues of anthers. Russ J Plant Physiol. 2004;51:97–106.
- 111.
- Guo G-Q, Zheng G-C. Hypotheses for the functions of intercellular bridges in male germ cell development and its cellular mechanisms. J Theor Biol. 2004;229:139–146. [PubMed: 15178192]
- 112.
- Guzicka M, Wozny A. Cytomixis in shoot apex of Norway spruce (Picea abies L. Karst.). Trees. 2005;18:722–724.
- 113.
- Zhang WC, Yan WM, Lou CH. Intercellular movement of protoplasm in vivo in developing endosperm of wheat caryopses. Protoplasma. 1990;153:193–203.
- 114.
- van BelA. The phloem, a miracle of ingenuity. Plant Cell Environm. 2003;26:125–149.
- 115.
- Telfer WH. Development and physiology of the oocyte-nurse cell syncytium. Adv Insect Physiol. 1975;11:223–319.
- 116.
- Spradling A. Germline cysts: communes that work. Cell. 1993;72:649–651. [PubMed: 8453660]
- 117.
- Robinson DN, Cooley L. Stable intercellular bridges in development: the cytoskeleton lining the tunnel. Trends Cell Biol. 1996;6:474–479. [PubMed: 15157506]
- 118.
- Kramerova IA, Kramerov AA. Mucinoprotein is a universal constituent of stable intercellular bridges in Drosophila melanogaster germ line and somatic cells. Dev Dyn. 1999;216:349–360. [PubMed: 10633855]
- 119.
- Haynh J-R, St JohnstonD. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr Biol. 2004;14:R438–R449. [PubMed: 15182695]
- 120.
- Snapp EL, Iida T, Frescas D. et al. The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol Biol Cell. 2004;15:4512–4521. [PMC free article: PMC519145] [PubMed: 15292454]
- 121.
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann Bot. 2005;95:147–175. [PMC free article: PMC4246715] [PubMed: 15596464]
- 122.
- Baluska F, Volkmann D, Barlow PW. Motile plant cell body: a ‘bug’ within a ‘cage’ Trends Plant Sci. 2001;6:104–111. [PubMed: 11239608]
- 123.
- Baluska F, Volkmann D, Barlow PW. Nuclear components with microtubule organizing properties in multicellular eukaryotes: functional and evolutionary considerations. Int Rev Cytol. 1997;175:91–135. [PubMed: 9203357]
- 124.
- Baroux C, Fransz P, Grossniklaus U. Nuclear fusions contribute to polyploidization of the gigantic nuclei in the chalazal endosperm of Arabidopsis. Planta. 2004;220:38–46. [PubMed: 15248065]
- 125.
- Guitton AE, Page DR, Chambrier P. et al. Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development. 2004;131:2971–2981. [PubMed: 15151989]
- 126.
- Baluska F, Volkmann D, Barlow PW. Actin-based domains of the ‘cell periphery complex’ and their associations with polarized ‘cell bodies’ in higher plants. Plant Biol. 2000;2:253–267.
- 127.
- Ketelaar T, Faivre-Moskalenko C, Esseling JJ. et al. Positioning of nuclei in Arabidopsis root hairs: an actin-regulated process of tip growth. Plant Cell. 2002;14:2941–2955. [PMC free article: PMC152738] [PubMed: 12417712]
- 128.
- Freitag M, Hickey PC, Raju NB. et al. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 2004;41:897–910. [PubMed: 15341912]
- 129.
- Martin R, Walther A, Wendland J. Deletion of the dynein heavy-chain gene DYN1 leads to aberrant nuclear positioning and defective hyphal development in Candida albicans. Eukaryot Cell. 2004;3:1574–1588. [PMC free article: PMC539012] [PubMed: 15590831]
- 130.
- Orias JD, Hamilton EP, Orias E. A microtubule meshwork associated with gametic pronucleus transfer across a cell-cell junction. Science. 1983;222:181–184. [PubMed: 6623070]
- 131.
- Janetopoulos C, Cole E, Smothers JF. et al. The conjusome: a novel structure in Tetrahymena found only during sexual reorganization. J Cell Sci. 1999;112:1003–1011. [PubMed: 10198282]
- 132.
- Goff LJ, Coleman AW. Transfer of nuclei from a parasite to its host. Proc Natl Acad Sci USA. 1984;81:5420–5424. [PMC free article: PMC391716] [PubMed: 16593507]
- 133.
- Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Molec Biol Rev. 2000;64:489–502. [PMC free article: PMC99001] [PubMed: 10974123]
- 134.
- Kuhn G, Hijri M, Sanders IR. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature. 2001;414:745–748. [PubMed: 11742398]
- 135.
- Shiu PKT, Glass NL. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr Opin Microbiol. 2000;3:183–188. [PubMed: 10744990]
- 136.
- Schuurs TA, Dalstra HJP, Scheer LML. et al. Positioning of nuclei in the secondary mycelium of Schizophyllum commune in relation to differential gene expression. Fung Genet Biol. 1998;23:150–161. [PubMed: 9578628]
- 137.
- Debuchy R. Internuclear recognition: a possible connection between Euascomycetes and Homobasidiomycetes. Fung Genet Biol. 1999;27:218–223. [PubMed: 10441447]
- 138.
- Thompson-Coffe C, Zickler D. How the cytoskeleton recognizes and sorts nuclei of opposite mating type during the sexual cycle in filamentous ascomycetes. Dev Biol. 1994;165:257–271. [PubMed: 8088443]
- 139.
- Wilmut I, Beaujean N, de Sousa PA. et al. Somatic cell nuclear transfer. Nature. 2002;419:583–586. [PubMed: 12374931]
- 140.
- Gurdon JB, Byrne JA, Simonsson S. Nuclear reprogramming and stem cell creation. Proc Natl Acad Sci USA. 2003;100:11819–11822. [PMC free article: PMC304092] [PubMed: 12920185]
- 141.
- Fujita N, Wade PA. Nuclear transfer: epigenetics pay a visit. Nat Cell Biol. 2004;6:912–922. [PubMed: 15459719]
- 142.
- Woese CR. On the evolution of cells. Proc Natl Acad Sci USA. 2002;99:8742–8747. [PMC free article: PMC124369] [PubMed: 12077305]
- 143.
- Woese CR. A new biology for a new century. Microbiol Mol Biol Rev. 2004;68:173–186. [PMC free article: PMC419918] [PubMed: 15187180]
- 144.
- Koch AL. Development and diversification of the Last Universal Ancestor. J Theor Biol. 1994;168:269–280. [PubMed: 8072294]
- 145.
- Koch AL. The bacterium's way for safe enlargement and division. Appl Environm Microbiol. 2000;66:3657–3663. [PMC free article: PMC92203] [PubMed: 10966373]
- 146.
- Simpson AGB, Roger AJ. The real ‘kingdoms’ of eukaryotes. Curr Biol. 2004;14:R693–R696. [PubMed: 15341755]
- 147.
- Walsh DA, Doolittle WF. The real ‘domains’ of life. Curr Biol. 2005;15:R237–R240. [PubMed: 15823519]
- 148.
- Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 1999;96:14553–14559. [PMC free article: PMC24474] [PubMed: 10588743]
- 149.
- Abanes-De MelloA, Sun Y-L, Aung S. et al. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. [PMC free article: PMC187501] [PubMed: 12502745]
- 150.
- Timmis JN, Ayliffe MA, Huang CY. et al. Endosymbiotic gene transfer: organelles genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. [PubMed: 14735123]
- 151.
- Hoiczyk E, Hansel A. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriol. 2000;182:1191–1199. [PMC free article: PMC94402] [PubMed: 10671437]
- 152.
- Valentine L. Agrobacterium tumefaciens and the plant: the David and Goliath of modern genetics. Plant Physiol. 2003;133:948–955. [PMC free article: PMC1540339] [PubMed: 14612581]
- 153.
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. [PubMed: 12110901]
- 154.
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. [PubMed: 15372043]
- 155.
- McManus MT. Small RNAs and immunity. Immunity. 2004;21:7. [PubMed: 15589164]
- 156.
- Fire A, Xu S, Montgomery MK. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. [PubMed: 9486653]
- Introduction
- The Permanent Crisis Surrounding Cell Theory
- More Problems with Cell Theory: What Is a Cell?
- Julius Sachs: Energide as the Basic Unit of Eukaryotic Life Endowed with Vital Energy
- Energide versus Cell Periphery
- Adaptability versus Complexity in Cellular Evolution
- Duality of Eukaryotic Cells
- Cell-Cell Channels: Supracellularity Is Found at All Levels of Cellular Organization
- Cell-Cell Channels in Filamentous Fungi, Plants and Animals
- Cell-Cell Channels and the Energide: Implications for Cell Theory
- Conclusions and Outlook
- References
- Cell-Cell Channels and Their Implications for Cell Theory - Madame Curie Bioscie...Cell-Cell Channels and Their Implications for Cell Theory - Madame Curie Bioscience Database
- Antibiotic Resistance in Bacteria Caused by Modified Nucleosides in 23S Ribosoma...Antibiotic Resistance in Bacteria Caused by Modified Nucleosides in 23S Ribosomal RNA - Madame Curie Bioscience Database
- Proteolytic Degradation of Αβ by Neprilysin and Other Peptidases - Madame Curie ...Proteolytic Degradation of Αβ by Neprilysin and Other Peptidases - Madame Curie Bioscience Database
- The Role of IL-10 in Autoimmune Pathology - Madame Curie Bioscience DatabaseThe Role of IL-10 in Autoimmune Pathology - Madame Curie Bioscience Database
- Mating Cell-Cell Channels in Conjugating Bacteria - Madame Curie Bioscience Data...Mating Cell-Cell Channels in Conjugating Bacteria - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...