NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Introduction
Part of this book is dedicated to the large amount of experimental data presently available on the biochemical and molecular aspects of telomerase. Therefore no details on this enzymatic function will be mentioned in this Chapter. However it should be pointed out that telomerase is able to restore telomeric repetitive sequences, that are lost during cell division. Therefore, it provides one of the most efficient mechanisms to ensure a sort of “cell immortality”, characteristic of stem cells and tumors.
Telomerase activity has been detected in more than 85% of solid tumors and leukemias whereas it is absent in terminally differentiated cells,1 except for selected cellular components of the immune system and other tissues with high cell turnover.2 Therefore, telomerase activity has been considered a widespread and relatively selective tumor cell marker that can be used for detecting the presence of viable neoplastic cells.3
Based on the difference in telomerase activity between normal and tumor cells, a number of potential clinical applications relative to telomerase in cancer treatment has been proposed. It could be a tool for detecting cancer cells in various organs, or a useful indicator for prognosis of some type of tumors, as suggested for neuroblastoma,4 or breast cancer.5 Moreover, since the endless proliferative potential of primary and metastatic tumor cells is ensured by this enzymatic function, telomerase appears to be a logical target for developing cancer therapies (reviewed in refs. 3, 6,7).
Another relatively recent application of tumor-associated telomerase activity concerns its use as a tumor marker for in vitro sensitivity assays to establish the effect of antineoplastic treatments on cancer cells.8–12 The final goal would be to set-up tests able to predict the therapeutic response of patients to antitumor agents, before or shortly after the onset of treatment. Actually, the idea of developing in vitro assays to measure the sensitivity of a tumor of each single patient to chemotherapy, radiotherapy or immunotherapy, stems from the observation that tumor responses to therapy are subjected to wide individual variability, even in the case of neoplasias of the same histological type. Spontaneous or induced resistance to anticancer drugs, to radiation, or to immuno-mediated mechanisms remains one of the major obstacles for the success of therapy. Therefore, monitoring susceptibility to treatment prior and during therapy appears to be of great importance for ameliorating the chances of patient's response and possibly survival.
In this Chapter of the book we are presenting the current state of research on the evaluation of residual telomerase activity of in vitro cultured tumor cells after exposure to antitumor agents or to immune effector cells.
This concept and the related assay have been developed in our laboratory for in vitro chemosensitivity testing. The classical “Telomeric Repeat Amplification Protocol” (TRAP13) to evaluate telomerase activity has been adopted. Therefore the assay has been called “Residual-TRAP” assay, hereafter referred to as R-TRAP. The theoretical basis of the assay was furnished mainly by two simple considerations: (a) in a mixture of normal and tumor cells (i.e., in most cases of tumor biopsies or surgery specimens), telomerase-positive cells represent mainly the neoplastic component of the entire cell population; (b) when a tumor cell dies, it loses telomerase function, although no exhaustive information is presently available on the precise temporal relationship between cell death (either for apoptotic or necrotic mechanism) and disappearance of the enzymatic activity. Actually, Akiyama et al14 pointed out that telomerase activity disappears in apoptotic cells when plasma membrane damage is present. However, tumor cells at the initial stage of the apoptotic process, before exhibiting plasma membrane alteration, still maintain high telomerase activity. It follows that monitoring residual telomerase activity could be considered a sort of classical “viable cell count” performed after drug treatment, being the viability criteria based on membrane integrity.
Scope of this report is to present and discuss the main results relative to two areas in which R-TRAP provided successful results in experimental models, although no clinical validation was still obtained. They are the chemosensitivity assay where the feasibility and the reproducibility of the R-TRAP has already been demonstrated8–12 and the more recent immunosensitivity assay. This technique, also developed in our laboratory, appears to be able to evaluate the cytotoxic activity of immune effector cells against target tumor cells (Faraoni et al, in preparation).
A possible linkage between telomerase and radiosensitivity of tumors is presented in the Chapter by Tej Pandita.
The in Vitro Chemosensitivity Assays and the Possible Role of R-TRAP
A relatively large number of antineoplastic drugs are now available in clinical oncology. In the majority of human neoplasias, the mutator phenotype associated with malignant cells, generates an exceptionally high number of different clones in tumor cell population. This provides one of the main mechanisms underlying the wide unpredictability of response to drug treatment. Therefore, an in vitro chemosensitivity assay, predicting the percentage of tumor cells killed by a panel of antitumor compounds, will provide the unique opportunity to identify the most active agents. Meanwhile, it could avoid the use of drugs that, being ineffective, would provoke useless toxicity for the patient.
In vitro chemosensitivity assays are largely used for identifying the agents of choice to be used in infectious diseases. In spite of the difficulties encountered in the interpretation of the results, these tests often provide valuable suggestions for adopting efficacious antibacterial regimens. However, a similar approach is by far more difficult in the case of human tumor cells. Complex pharmacokinetic and pharmacodynamic mechanisms underlie drug resistance to antineoplastic compounds. Therefore the in vivo conditions are not easily reproducible in vitro. Moreover, a series of technical factors are encountered with cultures of primary tumor cells.
In the last years various in vitro chemosensitivity tests have been proposed for predicting tumor cell response to anticancer agents, with very limited clinical success. These tests require a relatively large number of tumor cells collected from each patient. Therefore, only a limited number of prospective randomized studies could have been performed, so that no clear correlation between the results of in vitro assays and clinical responses to cancer chemotherapy has been detected (reviewed in refs. 15,16).
However, other problems must be considered. For example, among assays based on cell viability, the number of cells able to exclude a supravital stain (e.g., trypan blue) has been mainly used as a measure of the number of cells surviving after drug treatment: In this case, damaged cells are stained. In the differential staining cytotoxicity (DiSC) assay, a further stain with haematoxylin-eosin is needed to distinguish tumor cells from non-malignant cells present in tumor biopsies.17 The test is time-consuming and requires extensive work. Simpler to perform is the sulforhodamine B (SRB) assay.18 In this test the stain SRB binds the basic amino acids of cellular macromolecules giving an estimate of the total cellular protein content of each microculture well. Unfortunately, it does not discriminate debris—already present in the sample—from whole cells. Probably, this is one of the main reasons why the results of the assay did not provide a satisfactory correlation with clinical response.15
Radioactive incorporation of nucleic acid precursors, that provides a measure of total cellular proliferation within cultured tumor samples, has also been used to test in vitro the effect of antitumor agents. However, in many cases, tumor specimens are composed of a variable percentage of tumor cells admixed with normal cells and debris. It follows that non-malignant cells present in tumor biopsies are responsible of part of the radioactive incorporation of the microcultures. Other problems include the lack of incorporation of the radioactive precursor by cells with a long cell cycle, or by cells in which cell cycle is arrested by a damage inflicted during the isolation process.
A number of chemosensitivity assays based on drug-induced alteration of cell metabolism, independent from nucleic acid synthesis, has been also developed. Among them, particular attention has been dedicated to the MTT test, which is based on the ability of viable cells to reduce a tetrazolium salt [3-(4,5-dimethylthiazol-2-yl)2,5-diphenyl-tetrazolium bromide] (MTT), to a colored formazan product.19,20 In particular, the reaction, based on dehydrogenase activity, is thought to take place in intact mitochondria. However, feasibility of the assay depends on the metabolic rate characteristic of each cell type. Therefore this test can be applied to a limited number of tumors that meet these metabolic requirements.
ATP bioluminescence assay is a more recent test that requires a limited number of cells. But again, this test does not discriminate between ATP from tumor and normal cells present in the biopsy.21,22
The classical human tumor cloning assay, developed by Hamburger and Salmon,23 is based on the evaluation of the effects of drugs on the generation of tumor cell clones in soft agar. This method is considered one of the most reliable chemosensitivity assays. Using this technique numerous retrospective studies have been published (reviewed from 1978 to 1990, by Von Hoff24), showing a significant correlation between the results of the in vitro assay and the patient's clinical response. He estimated a total positive predictive value of 69% and a negative predictive value of 91%. Interestingly, the human tumor cloning assay is the only chemosensitivity assay that can distinguish malignant cells from the others, since only tumor cells grow in soft agar. A serious problem that represents the main drawback to this test is that few tumors can proliferate adequately in vitro. Moreover, the number of colonies, which are generated in agar, is usually in the range of 0.001 to 1% of the number of cells that have been plated. Therefore, 5–8 million cells are required for each drug tested. Hence, only 31–41% of the primary tumor samples can be evaluated, either for insufficient number of available cells, or for inadequate tumor cell proliferation.25,26 Another disadvantage with the clonogenic assay concerns the time required to obtain a reliable answer. Actually, 20 days or more are needed before suggestions on the drug(s) to be used in a given patient could be available.
The analysis of the status of the art on cancer chemosensitivity assays in 1995, prompted us to consider the possibility that tumor-associated telomerase activity could represent the starting point for new technical approaches. Keeping in mind the characteristics of telomerase, it was anticipated that part of the problems previously discussed could have been solved, adopting a chemosensitivity assay based on the evaluation of telomerase activity. In particular, it was considered that: (a) it is possible to detect telomerase activity testing a very limited number of neoplastic cells. Therefore, large amount of tumor cells are not required for the R-TRAP; (b) the presence of contaminating, telomerase-negative normal cells in the sample does not interfere with the assay. Actually, evaluation of telomerase activity allows to distinguish tumor from normal cells without previous separation; (c) prolonged tumor cell culture is not required; (d) proliferation of tumor cells is not necessary because the marker (i.e., telomerase activity) is constitutively associated with the neoplasia, and no cellular uptake of biochemically active precursors is required.
On the other hand, it must be pointed out that cancer chemosensitivity assays based on R-TRAP, rely on residual telomerase activity remaining after drug-induced cell killing. Therefore, it was recognized that the antitumor agents under investigation must be devoid of suppressive effects on telomerase activity itself. Moreover, these assays must be reasonably quantitative and reproducible to provide reliable information on drug sensitivity of individual tumors.
Experimental Approaches
Piatyszek et al demonstrated that the TRAP assay is able to detect as low as 10 neoplastic cells.13 In a context of differentiated cell population, this level of sensitivity is extremely important, for the following reasons: (a) the experiments must be conducted on few primary cancer cells present in tumor biopsies; (b) in most cases, time-dependent reduction of the number of viable neoplastic cells and, consequently of the TRAP signal, takes place in tissue culture. This occurs not only as a result of drug-induced cytotoxicity, but also as a consequence of spontaneous decline of cell viability in untreated controls.10 Therefore, for the majority of tumor specimens, the time required to evaluate the killing effect of antitumor agents cannot be longer than 1–3 days; (c) differential decline of telomerase activity in untreated controls or in drug-treated samples, following exposure to antitumor agents in vitro, must be evaluated quantitatively. Therefore, detection techniques of living tumor cells must be sensitive enough to discriminate between non-treated cells, and cells subjected to treatment with different drugs, in order to select the agent of choice.
In order to avoid misinterpretation of the data concerning the effect of drugs on telomerase activity reported and discussed in this Chapter, the following terms “telomerase activity per viable cell” (TA/cell), and “telomerase activity of total cultured cells” (TA/culture), will be used throughout the text.
Telomerase activity relative to each viable tumor cell (i.e., TA/cell) is calculated by dividing the telomerase activity value (expressed in arbitrary units resulting from the sum of absorbance of each ladder band obtained, for example, by densitometric analysis of classical TRAP assay blots) detected in a given amount of cell extract, by the number of “viable” cells present in the same volume of the extract. In general, viability criteria are based on a rather crude technique, that considers a cell “viable” when it excludes the uptake of certain dyes (e.g., trypan blue). However, this seems to be an universally accepted parameter, that has been considered valid in most of the published studies.
Telomerase activity relative to the total cells present in a culture (i.e., TA/culture) is calculated taking into account a limited aliquot only of the entire cell culture, for obvious practical reasons. An established volume of cultured cell suspension is centrifuged and the pellet processed for TRAP assay, independently of the number of cells present in the pellet. Telomerase activity values are expressed in the same terms that have been previously mentioned for TA/cell. It is reasonable to consider TA/culture a sort of viable tumor cell count in control or treated samples, when treatment or culture conditions do not alter TA/cell. In all cases, in order to determine the effects of antitumor agents on TA/cell or TA/culture, telomerase activity of controls must be compared with that of treated samples.
Telomerase activity related to chemosensitivity assays is not always computed according to cell number criteria. In some instances it is expressed on the basis of various biochemical parameters (e.g., enzyme activity per weight unit of protein, DNA or RNA content of cell extract). In this case, TA/cell or TA/culture are not easily applicable, since no strict and fully reliable relationship between viable cell count and protein or nucleic acid content can be demonstrated, especially in drug-treated cells.
The initial approaches to set-up the R-TRAP were performed with human tumor cell lines.8,10 The sensitivity of the TRAP assay and the semiquantitative relationship between cell number and ladder band intensity were measured using cell extracts corresponding to graded numbers of cells. A 50% reduction of tumor cells was easily detected by a substantial decrease of the ladder band intensity.9,10 With most tumor cell lines (i.e., MCF-7, A2780, Jurkat, Daudi) an overnight exposure of the gel showed a telomeric ladder in cell extracts corresponding to 1–3 cells. The sum of ladder band intensities, determined by densitometric analysis, expressed in terms of conventional numbers, provided a quantitative estimate of telomerase activity. The regression line obtained by plotting the values of densitometric analysis vs the log of cell number showed a direct correlation in the range of 15 to 103 tumor cells.9 However, the system reached a “saturation state” with loss of linearity when larger number of cells were used. Since a wide range of telomerase activity is associated with different tumors, in some cases it is difficult to run R-TRAP assays using a number of cells that gives a signal included in the linearity range of detection. Actually, in samples obtained from biopsies, the number of tumor cells cannot be easily established. However, we have performed several TRAP analysis on primary tumors showing that compared to tumor cell lines a larger number of cells are required to detect telomerase activity. In these conditions, the increment of enzymatic activity was linear in a rather wide range (i.e., from 103 to 2 × 104 cells10).
Figure 1 illustrates an outline of the R-TRAP method, that can be performed with tumor cell lines or tumor specimens. In the experimental design illustrated in the figure, TA/culture is the parameter studied by the R-TRAP assay.
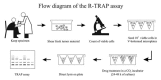
Figure 1
Schematic view of the major steps involved in the R-TRAP assay. See text for details.
Fresh tumor material is taken just after surgery, discarding fat and necrotic tissue as much as possible. Alternatively, fresh tumor material can be obtained from other clinical sources. The material is then sheared in order to obtain a homogeneous single cell suspension. When blast cells of leukemic patients are studied, no processing of the sample is generally required. Viable cell count is performed and cells (0.52 × 104 cells depending on tumor type) are seeded in V-bottomed microplates and treated with graded concentrations of the drug. Drug levels are calculated according to the plasma peak concentration (PPC) of the agents at the dose utilized in clinical protocols. After 24–48 h, or even, in selected cases, at 72 h, the plates are centrifuged and the medium is taken off. If necessary, the plates can be stored at 80°C until use. Thereafter, cells are harvested from each well and extracted with the same amount of lysis buffer. The extraction is done on ice, directly in the culture plate. For each sample equal volumes of cell extracts (4–8 ml) are analyzed for residual telomerase activity using the TRAP assay. The estimate number of control cells that will be processed in each sample for TRAP assay, must be adjusted according to the TRAP signal intensity. Actually, the number of cells obtained from fresh tumor biopsies must be generally much higher than that of the majority of cultured standard tumor lines. In any case, the percentage of drug-induced inhibition is always calculated on the basis of the TRAP signal intensity of the relative untreated control.
Several classes of chemotherapeutic agents have been used by our and other groups to check whether the results of the R-TRAP assay were similar to those obtainable with other more classical in vitro tests (e.g., MTT assay, viable cell count, colony formation, etc.). Satisfactory results were obtained after 24–48 h of treatment with: (a) anthracyclines (daunorubicin, doxorubicin, epirubicin); (b) natural products, such as Vinca alkaloids (vincristine, vinorelbine), epipodophyllotoxins (etoposide) and taxol; (c) antimetabolites (5-fluorouracil); (d) alkylating agents, such as nitrosureas (bis-chloroethyl-nitrosurea), temozolomide and maphosphamide (the in vitro active analogue of cyclophosphamide); (e) platinum compounds (cisplatin and carboplatin) (refs. 8–12,27 and unpublished data).
All these antitumor agents were also tested by adding drugs to the cell extracts, just before telomerase reaction, to exclude false-positive results originated from direct inhibition of the enzyme, not related to cell death. In all cases, no direct suppressive effects on telomerase activity by the agents under investigation were detected, when the drugs were used at clinically attainable concentrations.
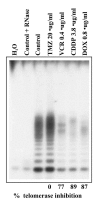
Figure 2 illustrates the results of a representative experiment in which a clone of the Jurkat leukemia line was treated with a panel of drugs, i.e., temozolomide (TMZ), vincristine (VCR), cisplatin (CDDP) and doxorubicin (DOX) at concentrations corresponding to their PPC.21 After 48 h of drug treatment, no difference in telomerase activity was found between control and TMZ-treated cells, indicating no change of TA/culture of treated cells with respect to that of controls. Moreover, comparable increase in the number of viable cells and similar TA/cell values were found in both groups. The cytotoxic effects of TMZ are mediated principally by methylation of the DNA at the O6 atom of guanine (O6-G).28 The DNA repair enzyme O6-methylguanine-DNA methyltransferase (MGMT) specifically transfers the alkyl group from the O6-G to a cysteine residue present in the MGMT itself, thus restoring the integrity of the DNA.29 Since Jurkat cells express high levels of MGMT, they are highly resistant to TMZ, in line with the results of the R-TRAP assay.28 On the other hand, Figure 2 shows also that a marked reduction of telomerase activity (TA/culture) occurred 48 h after treatment with VCR (77%), CDDP (89%) and DOX (87%). Cell counts and MTT tests run in parallel with R-TRAP assays, point out that drug-induced decline of telomerase activity was consistently correlated with reduction of the number of viable cells present in the cultures exposed to the antitumor agents. This finding provides strong support to the hypothesis that the mechanism underlying drug-mediated telomerase impairment is essentially the result of cell killing.
The presence of a substantial number of normal cells might influence the results of chemosensitivity assay. When the MTT assay is used, as demostrated by Kaspers et al,30 the contamination of leukemia samples with cells from normal peripheral blood can interfere with the results of the chemosensitivity assay. These authors demonstrated that normal blood lymphocyte are significantly more resistant to a large number of antineoplastic drugs than leukaemic cells from children with acute lymphoblastic leukaemia. In case of samples with less than 80% leukaemic cells, they suggest not to use MTT assay or other tests that evaluate drug-mediated killing of the total cell population. They suggest to consider rather assays in which a distinction between non-malignant and leukaemic cells can be made.
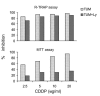
A series of experiments provide support to the use of R-TRAP assay also in those cases in which a high number of contaminating normal cells is present in the samples to be tested. Several R-TRAP assays were performed with leukemic Jurkat cells, alone or admixed with telomerase-negative normal cells, such as B lymphocytes27 or endometrial cells9 at the ratio of 1:9. Neoplastic cells were exposed to TMZ, CDDP and DOX, and R-TRAP was performed after 24, 48 and 72 h of culture. In all cases, no substantial differences in TA/culture pattern were found when cells where treated alone or admixed with telomerase-negative cells. This important finding is confirmed by the results illustrated in Figure 3, that show a direct comparison of in vitro drug sensitivity between ovarian cancer cells alone (TUM) or admixed with normal B lymphocytes (Ly) using both R-TRAP and MTT assays. The R-TRAP assay demonstrated comparable telomerase reduction after CDDP treatment of tumor cells cultured alone or in presence of Ly. In contrast, when chemosensitivity was evaluated on the basis of the MTT assay, the results in the presence of normal cells were entirely unreliable. Actually, the percentage of growth inhibition detectable in samples containing normal + neoplastic cells was at least 2–3-fold lower than that observed in samples containing tumor cells alone. This can be explained considering that Ly are only marginally affected by CDDP treatment. Therefore, the difference in the percentage of drug inhibition should be attributed to the capability of the MTT assay to detect metabolism of the CDDP-resistant normal B cells.
On the whole, these experiments indicate that R-TRAP assay can provide valuable information on chemosensitivity of cancer cells, even in the presence of considerable amounts of differentiated telomerase-negative normal cells.
Reproducibility of the R-TRAP assay was tested using ovarian cells obtained from a primary carcinoma, that were divided into aliquots and frozen in liquid nitrogen. Thereafter, several R-TRAP assays were performed at different days. Analysis of the control samples indicated a high intra-experiment reproducibility of the assay, with a coefficient of variation (CV) of 13.9% [CV was calculated by dividing the-standard deviation by the mean value of signal intensity (measured in terms of pixels) and multiplied by 100]. Moreover, the CV was of 11.6% when tests were performed with duplicate samples and the mean values of two telomeric ladders were used.10
In addition, inter-experiment reproducibility of the R-TRAP was calculated using the percentage of telomerase inhibition at 72 h, in cultured ovarian cancer cells exposed to graded concentrations of CDDP (0.6, 1.2, 2.5, 5 mg/ml). Figure 4 (left panel) shows one representative gel out of six. Each test was done in duplicate, and the percentage of telomerase inhibition respect to untreated controls was calculated using the mean signal intensity value obtained from two lane scan. Thereafter, CV was calculated taking into account the percent inhibition values relative to each concentration of CDDP. In these experiments the percentage of inhibition was found to be directly correlated with the concentration of CDDP. Interestedly, the CV values were always satisfactory, and decreased progressively with the increase of telomerase inhibition (Fig. 5, right panel). Therefore, at least in this case, R-TRAP showed excellent reproducibility in those conditions in which drug-induced tumor suppression were investigated.
In order to perform successful R-TRAP assays, with reasonably predictive value for clinical use, several conditions must be fulfilled. One of the most important requirements is that the agent should not be able to interfere directly with telomerase activity of target tumor cells, at least at concentrations in the range of PPC values. Actually, In some experiments, R-TRAP showed higher sensitivity of tumor cells to CDDP than that detectable by the MTT assay (9 and Fig. 4). This could be interpreted as a result of a direct effect of CDDP on telomerase activity with a decline of TA/cell. However, previous studies performed in our laboratory and by other authors showed that telomerase activity is not modulated when CDDP and other common anti-cancer agents are added to cell extracts just before starting the in vitro elongation step of the first “TS” primer13 for the TRAP assay.9,14,31,32 In any case, the possibility that some chemotherapeutic agents could influence the enzyme during cell culture is still open. For example, in human testicular cells, Burger et al found that high concentrations of CDDP could down regulate telomerase activity.31 In contrast, no decrease of telomerase activity following treatment with CDDP was observed in nasopharyngeal carcinoma cells32 and in ovarian cancer cells.33 It must be pointed out, however, that in all these experiments equal amounts of protein extract relative to control and treated cells were tested. Using such experimental design, interpretation of the results appears to be rather difficult. Actually, tumor suppression by CDDP is often the result of programmed cell death. However, the drug can produce also cell senescence-like growth arrest32 and necrosis,34 especially in case of defects in the apoptotic pathway.35 Moreover, studies performed in different laboratories indicate that telomerase itself could play a role in apoptotic cell death.36,37 Protein content of target cells could be sensibly different between control and treated cells. In the case of treated cells, also, the alternative outcomes of exposure to CDDP could affect differently the total amount of proteins present in cells undergoing death processes.
In other studies, the effects on telomerase activity of cytostatic concentrations of CDDP, etoposide, mitomycin C, daunorubicin were investigated in leukemic cells. In this case, the assay was performed adjusting the number of drug-treated cells to that of controls. The results indicate the TA/cell was not affected by treatment with the antitumor agents.14 On the other hand, Lin and coworkers38 evaluated the antineoplastic effects of CDDP, VCR, etoposide, and γ-rays delivered by1,37 Cesium, in three leukemic cell lines. In their work, the results obtained with the TRAP were compared with those obtained in parallel with the MTT assay. The observed telomerase activity was consistent with the results shown by MTT in most but not in all cases. In this study the authors found that in selected circumstances etoposide, VCR and gamma-radiation are able to severely down-regulate the telomerase activity of leukemic cells. However, TRAP assay was performed using a fixed amount of proteins for each cell extract, thus rendering, as already mentioned, these data not easily interpretable. Surprisingly, other studies showed that exposure to etoposide of three pancreatic cancer cell lines, relatively resistant to this agent, induced up-regulation of TA/cell.39 However, in two other cell lines that were sensitive to etoposide-induced apoptosis, no change of TA/cell was detected.39 Thus, the same drug (i.e., etoposide) was found to produce up-regulation, no influence, or possibly down-regulation on TA/cell in different cancer cell populations. These results suggest that drug-mediated modulation of TA/cell could be under the control of cellular factors that are not presently fully elucidated. The most recent view on regulation of telomerase activity depicts a very complex scenario that involves control at the level of gene transcription,40 post-traslational regulation of protein synthesis,41 and protein phosphorylation.42 Moreover, oncogenes and tumor suppressor genes have been found to influence telomerase activity.43–45 In addition, evidence indicates that telomerase is reversibly regulated during cellular proliferation, cell cycle progression, differentiation, apoptosis and cellular senescence.46 Therefore, to avoid misinterpretation of R-TRAP assays, it is required to perform an accurate analysis on the effects that could be produced on AT/cell in a large number of different tumors by the agents to be tested.
Another parameter relative to the R-TRAP assay, concerns optimal drug concentrations that should be used in vitro. The importance of drug concentration emerges from the analysis of chemosensitivity profiles of individual primary tumor samples exposed to different antitumor agents.10 It was shown that R-TRAP allowed detection of different chemosensitivity patterns in 9 ovarian carcinomas and 8 non small cell lung cancers. However, increasing concentrations of CDDP tended to reduce the differences in chemosensitivity among different tumors of the same histological type.10 This observation suggests that the concentrations of CDDP to be used for R-TRAP should be lower or equivalent to PPC (i.e., 2,5–3,8 mg/ml, refs. 21,47). In contrast, chemosensitivity patterns relative to 5-fluorouracil and vinorelbine were found to be essentially concentration-independent. In this case the assay can be performed with drug concentrations equivalent or even higher respect to the PPC (20 mg/ml and 1,5 mg/ml respectively, refs. 21,48). The mechanism underlying the different patterns of concentration/effect that have been noted between CDDP and 5-fluorouracil or vinorelbine, is presently unknown. However, it is reasonable to start from the idea that the profound differences in the mode of action of these three drugs should play a role. Actually, CDDP, 5-fluorouracil and vinorelbine belong to alkylating agents, to fluoropyrimidine antimetabolites and to agents acting on microtubules, respectively. It follows that prospective clinical studies based on R-TRAP assays should be preceded by preliminary tests on a series of human cancers exposed to graded concentrations of the antitumor agents under investigation. This would allow to select optimal concentration ranges to be used for the in vitro chemosensitivity assays that will be performed on cancer cells of each single patient.
A widely applicable chemosensitivity assay is characterized by a high feasibility rate. In the case of R-TRAP, the assay can be considered feasible when freshly isolated tumor cells, not subjected to drug treatment, survive in tissue culture and provide a reasonably good TRAP signal for at least 24–48 h. This is the shortest time possibly required for sensitivity/resistance determination with most of antitumor agents. A total of 52 human tumor samples has been tested for R-TRAP feasibility.10 Marked differences have been found among tumors originated from different organs. In particular ovarian cancers, melanomas and gastrointestinal tumors provide a good percentage of feasibility (100, 67 and 75% respectively). Consistent with this finding is that ovarian tumors and melanomas are known to survive rather easily in tissue culture, generating long-term cultured tumor lines with a relatively high frequency. On the other hand, sarcomas and breast cancer showed low feasibility rate. This finding seems also to be in line with the observation that it is notoriously difficult to maintain these two types of tumor in tissue culture.
In any case, feasibility of R-TRAP does not imply tumor cell proliferation, as required by assays based on the incorporation of radioactive precursors into DNA or by tumor cloning assays, but just tumor survival accompanied by adequate TRAP signal. However, like for other chemosensitivity assays, a substantial technical progress to improve tissue processing and cell culture conditions are needed in order to extend reliable R-TRAP tests to a large number of different tumor types.
Although other questions remain to be addressed, the most crucial matter relative to the R-TRAP approach concerns the possibility that the results of the in vitro test could be predictive of the in vivo responses. No data are presently available on this subject, and the uncertainty, common to other chemosensitivity assays, still remains.15,16 Actually, some authors state that uncontrollable factors, such as tumor vascularization, altered host's metabolism or other host-dependent resistance mechanisms, would limit the in vivo predictability of in vitro assays.
In order to solve these controversies, randomized clinical trials should demonstrate that drug treatments selected on the basis of chemosensitivity testing provide significantly higher response rates and survival with respect to those attained with standard protocols. Previous prospective clinical studies on this matter did not furnish encouraging results.16 It must be pointed out that chemosensitivity assays, utilized in these trials on primary tumor specimens, appear to be technically questionable, as discussed in the first part of the present Chapter. Therefore, the novel approach with the use of R-TRAP assays, although entirely speculative at this time, could open up new therapeutic strategies. However, large scale randomized clinical studies are warranted to establish the potential value of information provided by R-TRAP-based technology for “tailored” drug treatment of cancer patients.
The in Vitro R-TRAP Assay for Antitumor Cell-Mediated or Antibody-Mediated Cytotoxicity
As previously pointed out in this Chapter, tumor specimens of various clinical sources (e.g., biopsies or fragments of tumor tissues obtained from surgical removal of the neoplasia) contain a variety of cells of all types, and debris. In these conditions it is hard to test immune-mediated cytotoxic effects directed against the tumor. Actually, selective labeling of neoplastic cells with radioactive isotopes (e.g., 51Cr) with the intent to run isotope-release assays, is difficult. If release of cytokines (that are usually produced by effector cells following interaction with target cells, e.g., interferon-γ) is used as a cytotoxicity assay, possible interference of bystander cells in tumor specimens could alter release modalities and mechanisms.
It is logical, therefore, to propose an R-TRAP protocol (hereafter called R-TRAPCTX) based on the same principles adopted for chemosensitivity assays, to evaluate cell-mediated cytotoxicity against telomerase-positive target tumor cells. The theoretical advantages of this approach over other techniques can be listed as follows: (a) no limits of time are required for effector/target cell interaction to run R-TRAPCTX. In the majority of radioactive assays, spontaneous release of the isotope taken up by target cells, increases with the incubation time. Therefore, the interpretation of the results could be very difficult when effector/target cell cocultures are carried out for excessively long times; (b) R-TRAPCTX provides direct evidence of target tumor cell killing; (c) in a number of cases, R-TRAPCTX does not require long-term survival of tumor cells in tissue culture. Actually, if effector cells are appreciably cytotoxic, few hours of effector/target coculture performed shortly after tumor cell collection, are sufficient to detect effector cell activity; (d) immune-mediated killing of few target tumor cells in the context of large amounts of telomerase-negative non-neoplastic cells, can be monitored by R-TRAPCTX .
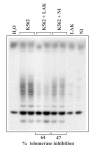
Preliminary studies on R-TRAPCTX have been performed in our laboratory, Different tumor cell lines were exposed to NK, LAK or cytotoxic allosensitized T lymphocytes. Figure 5 shows a representative experiment in which K562 target cells were co-cultured with non-activated (i.e., “NK”) or interleukin-2-activated (i.e., LAK) effector lymphocytes for 4 h. Parallel 51Cr-release assays and R-TRAPCTX showed a good correlation between target cell cytolysis and decline of telomerase activity following effector/target cell interaction. In other experiments, similar results were obtained with cytotoxic allosensitized lymphocytes co-cultured with the relevant tumor targets (data not shown). It should pointed out, however, that interpretation of the results appears to be substantially difficult if effector cells express high telomerase activity. In this case, subtraction of “background” telomerase signal due to immune lymphocytes does not always provide a satisfactory answer to this problem (Faraoni I. et al, in preparation).
In conclusion, R-TRAPCTX could represent a novel and reliable test for measuring natural or antigen-dependent immunity against target tumor cells. However, additional studies are required to validate this technique in different clinical situations.
References
- 1.
- Kim NW, Piatyszek MA, Prowse KR. et al. Specific association of human telomerase activity with immortal cell and cancer. Science. 1994;266:2011–2015. [PubMed: 7605428]
- 2.
- Krupp G, Klapper W, Parwaresch R. Cell proliferation, carcinogenesis and diverse mechanisms of telomerase regulation. Cell Mol Life Sci. 2000;57:464–486. [PubMed: 10823247]
- 3.
- Faraoni I, Bonmassar E, Graziani G. Clinical applications of telomerase in cancer treatment. Drug Resist Updat. 2000;3:161–170. [PubMed: 11498381]
- 4.
- Poremba C, Willenbring H, Hero B. et al. Telomerase activity distinguishes between neuroblastomas with good and poor prognosis. Ann Oncol. 1999;10:715–721. [PubMed: 10442195]
- 5.
- Clark GM, Osborne CK, Levitt D. et al. Telomerase activity and survival of patients with node-positive breast cancer. J Natl Cancer Inst. 1997;89:1874–1881. [PubMed: 9414175]
- 6.
- Meyerson M. Role of telomerase in normal and cancer cells. J Clin Oncol. 2000;18:2626–2634. [PubMed: 10893296]
- 7.
- White LK, Wright WE, Shay JW. Telomerase inhibitors. Trends Biotechn. 2001;19:114–120. [PubMed: 11179805]
- 8.
- Faraoni I, Turriziani M, Bonmassar E. et al. Development of a novel in vitro chemosensitivity assay: telomerase as a possible marker of tumor cell survival. J Chemother. 1996;8:394–398. [PubMed: 8957721]
- 9.
- Faraoni I, Turriziani M, Masci G. et al. Decline in telomerase activity as a measure of tumor cell killing by antineoplastic agents in vitro. Clin Cancer Res. 1997;3:579–585. [PubMed: 9815723]
- 10.
- Faraoni I, Graziani G, Turriziani M. et al. Suppression of telomerase activity as an indicator of drug-induced cytotoxicity against cancer cells: in vitro studies with fresh human tumor samples. Lab Invest. 1999;79:993–1005. [PubMed: 10462037]
- 11.
- Park KH, Rha SY, Kim CH. et al. Telomerase activity and telomere lengths in various cell lines: changes of telomerase activity can be another method for chemosensitivity evaluation. Int J Oncol. 1998;13:489–495. [PubMed: 9683783]
- 12.
- Mese H, Ueyama Y, Suzuki A. et al. Inhibition of telomerase activity as a measure of tumor cell Killing by cisplatin in squamous cell carcinoma cell line. Chemotherapy. 2001;47:136–142. [PubMed: 11173816]
- 13.
- Piatyszek MA, Kim NW, Weinrich SL. et al. Detection of telomerase activity in human cells and tumors by a telomeric repeat amplification protocol (TRAP). Methods Cell Sci. 1995;17:1–15.
- 14.
- Akiyama M, Horiguchi-Yamada J, Saito S. et al. Cytostatic concentrations of anticancer agents do not affect telomerase activity of leukaemic cells in vitro. Eur J Cancer. 1999;35:309–315. [PubMed: 10448276]
- 15.
- Bellamy WT. Prediction of response to drug therapy of cancer. A review of in vitro assays. Drugs. 1992;44:690–708. [PubMed: 1280562]
- 16.
- Brown E, Markman M. Tumor chemosensitivity and chemoresistance assay. Cancer. 1996;77:1020–1025. [PubMed: 8635118]
- 17.
- Bosanquet AG. Correlations between therapeutic response of leukaemias and in-vitro drug sensitivity assay. Lancet. 1991;337:711–714. [PubMed: 1672185]
- 18.
- Skehan P, Storeng R, Scudiero D. et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. [PubMed: 2359136]
- 19.
- Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. [PubMed: 2470825]
- 20.
- Zwaan CM, Kaspers GL, Pieters R. et al. Cellular drug resistance profiles in childhood acute myeloid leukemia: Differences between FAB types and comparison with acute lymphoblastic leukemia. Blood. 2000;96:2879–2886. [PubMed: 11023525]
- 21.
- Andreotti PE, Cree IA, Kurbacher CM. et al. Chemosensitivity testing of human tumors using a microplate adenosine thiphosphate luminescence assay: clinical correlation for cisplatin resistance of ovarian carcinoma. Cancer Res. 1995;55:5276–5282. [PubMed: 7585588]
- 22.
- Cree IA. Luminescence-based cell viability testing. Methods Mol Biol. 1998;102:169–177. [PubMed: 9680619]
- 23.
- Hamburger A, Salmon SE. Primary bioassay of human tumor stem cells. Science. 1977;197:461–465. [PubMed: 560061]
- 24.
- Von Hoff DD. He's not going to talk about in vitro predictive assays again, is he? J Natl Cancer Inst. 1990;82:96–101. [PubMed: 2403594]
- 25.
- Von Hoff DD. Send this patient's tumor for culture and sensitivity. N Engl J Med. 1983;308:154–155. [PubMed: 6336821]
- 26.
- Von Hoff DD, Clark G, Stogill B. et al. Prospective clinical trial of a human tumor cloning system. Cancer Res. 1983;43:1926–1931. [PubMed: 6339044]
- 27.
- Faraoni I, Turriziani M, Graziani G. et al. Telomerase as a marker of tumor cell viability: a new approach for in vitro chemosensitivity assay? J Exp Clin Cancer Res. 1996;15:311–312.
- 28.
- D'Atri S, Tentori L, Lacal PM. et al. Involvement of the mismatch repair system in temozolomide-induced apoptosis. Mol Pharmacol. 1998;54:334–341. [PubMed: 9687575]
- 29.
- Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. [PubMed: 10767620]
- 30.
- Kaspers G J L, Pieters R, Van Zantwijk CH. et al. In vitro drug sensitivity of normal peripheral blood lymphocytes and childhood leukaemic cells from bone marrow and peripheral blood. Br J Cancer. 1991;64:469–474. [PMC free article: PMC1977664] [PubMed: 1911186]
- 31.
- Burger AM, Double JA, Newell DR. Inhibition of telomerase activity by cisplatin in human testicular cancer cell. Eur J Cancer. 1997;33:638–644. [PubMed: 9274448]
- 32.
- Wang X, Wong S C H, Pan J. et al. Evidence of cisplatin-induced senescent-like growth arrest in nasopharyngeal carcinoma cells. Cancer Res. 1998;58:5019–5022. [PubMed: 9823301]
- 33.
- Akeshima R, Kigawa J, Takahashi M. et al. Telomerase activity and p53-dependent apoptosis in ovarian cancer cells. Br J Cancer. 2001;84:1551–1555. [PMC free article: PMC2363666] [PubMed: 11384107]
- 34.
- Pestell KE, Hobbs SM, Titley JC. et al. Effect of p53 status on sensitivity to platinum complexes in a human ovarian cancer cell line. Mol Pharmacol. 2000;57:503–511. [PubMed: 10692490]
- 35.
- Gonzales VM, Fuertes MA, Alonso C, Perez JM. Is cisplatin-induced cell death always produced by apoptosis? Mol Pharmacol. 2001;59:657–663. [PubMed: 11259608]
- 36.
- Kondo Y, Kondo S, Tanaka Y. et al. Inhibition of telomerase increases the susceptibility of human malignant glioblastoma cells to cisplatin-induced apoptosis. Oncogene. 1998;16:2243–2248. [PubMed: 9619833]
- 37.
- Fu W, Begley G, Killen MW, Mattson MP. Anti-apoptotic role of telomerase in pheochromocytoma cells. J Biol Chem. 1999;274:7264–7271. [PubMed: 10066788]
- 38.
- Lin Z, Lim S, Viani MA. et al. Down-regulation of telomerase activity in malignant lymphomas by radiation and chemotherapeutic agents. Am J Pathol. 2001;159:711–719. [PMC free article: PMC1850549] [PubMed: 11485929]
- 39.
- Sato N, Mizumoto K, Kusumoto M. et al. Up regulation of telomerase activity in human pancreatic cancer cells after exposure to etoposide. Br J Cancer. 2000;82:1819–1826. [PMC free article: PMC2363240] [PubMed: 10839297]
- 40.
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. [PubMed: 9553037]
- 41.
- Ulaner GA, Hu JF, Vu TH. et al. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int J Cancer. 2001;91:644–649. [PubMed: 11267974]
- 42.
- Liu K, Hodes RJ, Weng N. Telomerase activation in human T lymphocytes does not require increase in telomerase reverse transciptase (hTERT) protein but is associated with hTERT phosphorylation and nuclear translocation. J Immunol. 2001;166:4826–4830. [PubMed: 11290757]
- 43.
- Wang J, Xie LY, Allan S. et al. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. [PMC free article: PMC316904] [PubMed: 9637678]
- 44.
- Li H, Cao Y, Berndt MC. et al. Molecular interactions between telomerase and the tumor suppressor protein p53 in vitro. Oncogene. 1999;18:6785–6794. [PubMed: 10597287]
- 45.
- Crowe DL, Nguyen DC. Rb and E2F-1 regulate telomerase activity in human cancer cells. Biochim Biophys Acta. 2001;1518:1–6. [PubMed: 11267653]
- 46.
- Elenitoba-Johnson K S J. Complex regulation of telomerase activity. Am J Pathol. 2001;159:405–410. [PMC free article: PMC1850547] [PubMed: 11485897]
- 47.
- Untch M, Sevin BU, Perras JP. et al. Evaluation of paclitaxel (taxol), cisplatin and the combination paclitaxel-cisplatin in ovarian cancer in vitro with the ATP cell viability assay. Gynecol Oncol. 1994;53:44–49. [PubMed: 7909786]
- 48.
- Jehl F, Quoix E, Leveque D. et al. Pharmacokinetics and preliminary metabolic fate of navelbine in humans as determined by high performance liquid chromatography. Cancer Res. 1991;51:2073–2076. [PubMed: 1849042]
- Telomerase Activity as a Marker of Tumor Cell Survival to Evaluate Sensitivity o...Telomerase Activity as a Marker of Tumor Cell Survival to Evaluate Sensitivity of Neoplastic Cells to Cancer Treatment - Madame Curie Bioscience Database
- Nuclear Envelope Dynamics in Drosophila Pronuclear Formation and in Embryos - Ma...Nuclear Envelope Dynamics in Drosophila Pronuclear Formation and in Embryos - Madame Curie Bioscience Database
- Cytokines in the Pathogenesis of Rheumatoid Arthritis and Collagen-Induced Arthr...Cytokines in the Pathogenesis of Rheumatoid Arthritis and Collagen-Induced Arthritis - Madame Curie Bioscience Database
- Evolutionary De Novo Design - Madame Curie Bioscience DatabaseEvolutionary De Novo Design - Madame Curie Bioscience Database
- The Evolution of CDK-Activating Kinases - Madame Curie Bioscience DatabaseThe Evolution of CDK-Activating Kinases - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...