NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Introduction
The number of sequenced arthropodan lipocalins adds up to over eighty (see Table 1). From our currently fragmented knowledge of arthropodan genomes, the last common ancestor of this phylum is proposed to possess two lipocalins (see Chapter 3). Intra-lineage duplications enlarged the number of lipocalins, with some large family expansions occurring independently in blood-feeding insects and arachnids.
Table 1
Summary of properties of all known arthropodan lipocalins.
Most arthropodan lipocalins share the common signature and structural properties with the rest of the family. They are single modular proteins of around 200 amino acids that fold tightly in a β-barrel with potential for binding small hydrophobic molecules in a central pocket. They usually have two α-helices at the N- and C-terminal sides, packed against the outer surface of the barrel. Nonetheless, we will encounter in this phylum variations around this main theme. While maintaining the lipocalin fold, the arthropodan lipocalins call our attention because of the panoply of interesting structural and functional explorations that have arisen throughout evolution. Because of the recent extensive review on arthropodan lipocalins by Kayser,1 with special emphasis in their structure and function, it is our aim to offer a review focusing on the peculiarities of these lipocalins while framing them in an evolutionary context.
As a first step of analysis, we can simply look at an alignment of amino acid sequences of arthropodan lipocalins. A schematic representation of such an alignment is shown in Figure 1. The location of all important gaps is coincident with the loops (L) of the β-barrel. While L1, L4 and L6 are relatively well conserved, all other loops accommodate significant variations in size. Particularly large extensions in L5 are present in one of the Pallidipin sequences, and in other lipocalins related to Nitrophorins (see below). Other expanded loops (L3 and L7) are also expected in these lipocalins. These three loops (L3, L5 and L7) face the entrance of the binding pocket (‘the open side of lipocalins’, see Chapter 2), and their variations could condition the ligand binding properties of these lipocalins. Other lipocalins such as the Drosophila lipocalins GLaz and Karl show unique extensions in L2. Because this loop faces the closed side of the β-barrel, it is most probably involved in protein-protein interactions. Loop L3 of GLaz is also long, but there is no sequence similarity with the L3 loop of R. prolixus Pallidipin.
Variations are also observed in the length of the N- or C- terminal segments. Most lipocalins can be classified roughly in tree groups, which differ in the length of their C-termini after the last cysteine residue. The longest of these C-terminal extensions have a size similar to the hydrophobic GPI anchoring signal unique to the grasshopper Lazarillo (shown as a zigzag line in Fig. 1). However, the sequence signatures for GPI anchorage are absent in all other cases. In addition, Karl shows a uniquely long N-terminal extension, while some of the insect saliva lipocalins are shorter than average (not shown).
Therefore, we find in arthropods a wide range of variation in particular loops, and in the Nand C-terminal regions of the proteins. These changes are not expected to alter significantly the β-barrel core, and thus provide a substrate of variation for potential functional diversification and specialization.
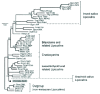
Using the amino acid sequences, we have reconstructed a phylogeny of arthropodan lipocalins (Fig. 2) including the group of chordate lipocalins most related to them (Apolipoprotein D, ApoD), and using lipocalins from other kingdoms (plants, fungi and protoctists lipocalins) as the outgroup. However, the arachnid saliva lipocalins were not included. Because of their highly divergent sequences, the reliability of the alignment and the tree-reconstruction methods decreases due to long-branch attraction artifacts.2 Instead, we have superimposed the branch of arachnid saliva lipocalins (dashed lines) as deduced from the analysis of their intron-exon structure3 (see Chapter 3).
Several groups of arthropodan lipocalins are supported in the tree, and these relationships will guide us through the following sections of the chapter.
At the most basal position stands the group of Lazarillo (Laz) related lipocalins, which, in turn, closely relates to the chordate ApoDs currently found in many species ranging from tunicates to humans. The Lazarillo clade also shows, so far, the widest species representation within insects, with one lipocalin found in Orthoptera and Lepidoptera, and two in several Diptera species (fruitflies and mosquitoes) and hymenoptera (bees). Some of the new sequences retrieved from mosquitoes and bees can be ascribed as orthologous to one of the four lipocalins found in the fruitfly Drosophila melanogaster. However, divergence rates are either unequal or very high, so that the sequence of an older Drosophila species, D. yakuba, is set apart in the phylogeny.
The group of tick saliva lipocalins might have stemmed out of an ancestor similar to these Lazarillo lipocalins, since their gene structure is most similar to the Drosophila Neural Lazarillo (NLaz).3 This group represents a big expansion of the lipocalin family through gene duplication and divergence, probably associated to the adaptation to blood-feeding habits. They are all expressed in the salivary glands. Whether more “conventional” lipocalins are also present in other tissues of ticks, remains to be investigated.
A clade relates the only fully sequenced lipocalins reported in crustaceans: the Crustacyanins (CRCs). They have been well characterized in the lobster Homarus gammarus, but proteins with very similar biochemical properties are now found in more species of lobsters, crayfish and crabs. Two CRCs per species are found at the most (only the fully sequenced CRCs have been included in the tree).
The biliproteins (BPs) and their relatives are monophyletically related. All biliproteins so far established as lipocalins are found in Lepidoptera, a relatively young and very diverse group of insects. BPs have also been isolated from other insect orders but their sequences are not yet known and hence their lipocalin nature is still open (reviewed in ref. 1). A few of the lepidopteran lipocalins from this clade are well characterized by having a bilin bound as a specific ligand resulting in a blue protein. The ligands, if any, of the related members of this clade are not known; we therefore refer to them as biliprotein-related proteins (BPRPs). As it is the case for CRCs, a maximum of two lipocalins have been found in each species sampled.
The insect saliva lipocalins relate to the biliproteins clade. This group represents another independent expansion of lipocalins associated with hematophagy adaptation in insects. Interestingly, among them are the Nitrophorins (NPs) that are heme-containing lipocalins. The branches of the insect saliva lipocalins clade are very long, reflecting their high sequence divergence. Caution should thus be taken, since mistaken relationships due to long branch attraction are not ruled out: sequences that are more dissimilar tend to group together. Finding more lipocalins in the same species of blood-sucking insects, as well as the use of other phylogenetically informative characters, should help to establish their relationship to the rest of the family.
In the next sections we will review each of the five groups of lipocalins. We have found big contrasts in the level of knowledge about arthropodan lipocalins. From very fine details on protein structure, ligand-protein interactions and physiological functions, to the vastly unexplored functional and biochemical properties of new sequenced lipocalins. In any case, arthropods are an excellent arena in which to scrutinize and admire the enormous versatility of such an apparently simple protein fold.
The Lazarillo/Apolipoprotein D Related Lipocalins
According to the consensus pattern of gene structure and protein sequence motifs of the family, Lazarillo is a conventional lipocalin with the three conserved protein motifs of the family (see Chapter 2) and its gene ORF intervened by three introns,4 two of which are conserved in every metazoan lipocalin found so far (see Chapter 3). As shown in Figure 2, phylogenetic analyses support the orthologous relationship of Laz and the vertebrate ApoD lipocalin.4-6 In addition, Laz molecular features, expression pattern and function closely resemble those of ApoD.
Lazarillo in Grasshopper (Schistocerca americana)
The grasshopper Lazarillo (Laz) was identified by two of us thanks to a monoclonal antibody (mAb) raised against embryonic neural proteins. This mAb allowed the study of the expression profile and of a number of biochemical properties, and the protein purification that led to the Laz transcript by standard cloning techniques.7,8 Laz gene is transcribed as a long (3 kb) mRNA, with a fairly long (> 2 Kb) 3'-UTR.
Grasshopper Laz shows some striking biochemical features, since it is, so far, the only lipocalin bound to the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor. However, the functional relevance of this singular type of membrane attachment remains unknown. The protein is also heavily glycosylated: Cleavage with PNGase F demonstrates that around 40% of Laz molecular weight is due to N-linked oligosaccharides. Interestingly, the five Asn residues predicted to be glycosylated appear to be polarized to one side of the protein by homology-modeling of the tertiary structure of Laz.9 The shift in electrophoretic mobility of the protein under reducing vs. nonreducing conditions, and Laz homology-modeling suggest that the four cysteine residues of Laz form two alternating disulfide bonds and contribute to its folding.
Although we are in the process of analyzing the ligand capabilities of the grasshopper Laz and other relatives, the study of the Laz 3D-model and preliminary experiments (Ganfornina, Ortega, Åkerström and Sanchez unpublished observations) suggest that Laz can bind small and rod-shaped hydrophobic ligands, but not the flat-shaped bilins bound by other insect lipocalins (see above).
Due to its GPI tail, Laz is expected to interact homophilically, or with other proteins, in order to transduce signals from the cell surface to the intracellular compartment. We have evidence that various recombinant forms of Laz can oligomerize in solution (Ganfornina et al, unpublished observations), although no information is available for interactions with other proteins such as potential receptors.
In terms of the spatial and temporal pattern of expression, the grasshopper Laz appears on the surface of a subset of neural precursors, the neuroblasts, as well as in many postmitotic neurons along the embryonic central, peripheral, and enteric nervous system.8,10,11 An interesting observation is the transient expression observed in the neuroblasts, and in some neurons that express Laz during the period of axon outgrowth. The restricted expression of Laz within the developing nervous system has made the anti-Laz mAb an excellent tool for the characterization of the NS in economically important species like Locusta migratoria or Schistocerca gregaria.11-13
Outside the nervous system, Laz is expressed by the undifferentiated tips of the excretory Malpighian tubules, by sessile mesodermal cells that originate at the tip of the invaginating proctodeum and then migrate anteriorly to the amnioserosal membrane, and by nephrocytes of the subesophageal body.
Lazarillo in the Fruitfly (Drosophila melanogaster)
The Drosophila genome sequencing project recovered two regions in the second chromosome of the fruitfly with sequence similarity to the lipocalins. The transcription of the two putative genes was confirmed by RT-PCR from Drosophila embryos,14 and their deduced amino acid sequence showed two conventional lipocalins (Neural Laz, NLaz and Glial Laz, GLaz) with the three lipocalin motifs, and the four cysteines involved in forming alternate disulfide bonds.
Both Drosophila lipocalins show the highest sequence similarity to grasshopper Laz. However, they do not show the GPI tail. They are secreted to the extracellular environment as predicted by their signal peptide and suggested by the difficulty of recognizing the NLaz protein by whole-mount immunohistochemistry unless the secretory machinery is inhibited by the use of monensin.14 No further details of their biochemical properties are known at present.
The Drosophila Laz genes pose a typical problem of orthology ascription. Two character sets closely relate GLaz to grasshopper Laz: amino acid sequence similarity14 and exon-intron arrangement4 (see Chapter 3). However, other protein properties support NLaz as the grasshopper Laz ortholog:
- Sequence alignment (Fig.1, see also Chapter 2) suggests the existence of two unique loops in GLaz that could be involved in specific functional interactions with a ligand or other proteins. These features make GLaz more dissimilar to Laz, while the alignment of NLaz and Laz has only minor gaps (1-2 residues).
- NLaz is more glycosylated (five predicted sites) than GLaz (only one site).
- NLaz is predicted to have an acidic pI (4.5), similar to the grasshopper protein (4.8) and distinct from the strongly basic GLaz (pI 9.0).
- NLaz, but not GLaz, has a long C-terminal segment similar in size to the GPI anchoring signal of Laz (Fig.1).
Neither of the other Drosophila lipocalins is more related to grasshopper Laz. Thus, the Laz ancestor gene probably gave rise to NLaz and GLaz by duplication in the fruitfly, the traces of which have been subsequently blurred by their divergence and acquisition of new functions.
Looking at the spatio-temporal expression pattern of these lipocalins, we have collected more data to ascertain their relationships to the grasshopper Laz. Time wise, the transcripts of both lipocalins are abundant at late stages of embryogenesis, mainly when the nervous system is being formed, in the pupal stage, and in adulthood. In addition, GLaz mRNA is present all along the embryonic period, with a maternal contribution at early stages. The low abundance during larval development and the burst observed after pupariation for both lipocalins is correlated with the profound tissue reorganization that occurs at this critical period of metamorphosis.
In terms of the tissue expression profile, NLaz holds its name because of its expression in a subset of CNS neuroblasts and neurons, while GLaz is expressed by the longitudinal glia of the ventral nerve cord and a specific glial precursor. Besides, a subset of neurons expresses NLaz in larval and adult CNS (Sanchez et al, unpublished observations). The pattern of NLaz transcription is particularly interesting, as it changes drastically when considering different neuromeres and even hemisegments. This indicates a very dynamic transcription for NLaz and suggests that different neurons need this lipocalin for particular time periods. NLaz protein is observed in axons, which suggests that the protein is transported along them and is possibly secreted to the extracellular environment by the growth cone. Outside the nervous system, only the embryonic expression of these lipocalins has been explored. NLaz appears in the amnioserosa, the fat body and the developing gut, while GLaz is mainly expressed in the developing gut and salivary glands.
Lazarillo Function, or Functions?
Laz perturbation by in vivo incubation of grasshopper embryos with the anti-Laz mAb resulted in axonal growth delay and growth cone misrouting.8 Thus, the grasshopper Laz is functionally related to the modulation of growth and guidance of developing axons.
While functional analyses of the Drosophila Laz lipocalins are underway, a hypothesis based on their molecular properties had been proposed.14 They differ mostly in surface regions of the proteins (see above), while an analysis by homology-modeling predict that the binding pocket environment of NLaz and GLaz is very similar. Because they are secreted and expressed in nearby embryonic cells, it is reasonable to propose that their functional specificity is due to protein-protein interactions. The binding of a ligand could serve as a modulator for those interactions.
We are currently analyzing the effect of loss-of-function mutations for the genes NLaz and GLaz (Ganfornina, Sanchez et al, unpublished observations). Surprisingly, no drastic effects are observed after abolishing the function of these two lipocalins during development, thus reducing support for an essential role in axonal pathfinding and growth during normal development. However, during adulthood, the lack of NLaz or GLaz causes a decrease of life span and more sensitivity to the oxidative stress produced by paraquat, a generator of free radicals. Therefore, in terms of neural development, it remains as a possibility that these lipocalins are needed under particular developmental conditions that pose a threat to the proliferation and survival of neurons and/or to their axonal growth. In this sense, ApoD, the chordate ortholog of Laz, is functionally involved in the cell-proliferation/cell-death balance, and in cell migration upon injury (reviewed in Chapter 13). Most data support that Laz/ApoD is a multifunctional protein used in different cellular or physiological contexts.
Other Lipocalins Related to Lazarillo
The lipocalin Karl was found when screening for genes specifically expressed in the blood cells15 (and Bailey's unpublished observations). The first expression is observed in plasmatocytes during late embryonic development. Later, during larval stages, Karl is strongly expressed in the lymph glands (the larval hematopoietic organ) and in circulating hemocytes. Blood cells are important for the immune system of insects, since they act as macrophages and as producers of antimicrobial peptides. A potential role of Karl in the immune system of insects is being investigated.
The EST and genome sequencing projects are helping to identify more Laz related lipocalins. In addition to Karl, GLaz and NLaz, we have found a fourth lipocalin in the genome of D. melanogaster, yielding four as the maximum number of lipocalins in this well studied species. An equal number of lipocalins have been found in the hymenoptera Apis mellifera, the honeybee (although the full sequence is not available yet for all of them). It is worth noting that at least one of them is also expressed in the nervous system.
Several mosquito genome projects are underway and at least two lipocalins in Anopheles gambiae show sequence similarity to Laz. However, other mosquito sequences become associated with the more divergent group of insect saliva lipocalins in our protein phylogeny reconstruction (Fig. 2).
Finally, in the fall webworm moth Hyphantria cunea, we found the only example of a Laz related lipocalin within Lepidoptera. Interestingly, Hyphantrin was initially regarded as a biliprotein due to its (distant) similarity to the BPRPs clade of lipocalins and to its expression in the epidermis of pupae. However, the Hyphantrin sequence is more related to the Laz-like lipocalins (Fig. 2). In contrast, other BPRPs (like Gallerin or Bombyrin) are neuronal lipocalins (see below), reflecting that nervous system expression is not an exclusive character of the Laz-related lipocalins.
Crustacyanins: Blue Lipocalins in Crustaceans
As the name suggests, Crustacyanin (CRC) denotes a blue protein from a crustacean. Proteins with biochemical properties of CRC have been isolated from several species of lobsters, crabs and crayfishes, although the CRCs from Homarus gammarus are by far the best known. By contrast to the insect biliproteins discussed below, the blue color of this protein is not due to a bilin, but to a carotenoid. Hence, CRC represents a blue carotenoprotein. Over the past 50 years, many attempts have been undertaken to uncover the building principle and the physical basis of the blue color of CRC, but only recent work succeeded to solve some key features of its structure. The many facets of this long-standing endeavor has been well reviewed recently.16
CRC represents a protein complex with astaxanthin (3,3'-dihydroxy-β,β-carotene-4,4'-dione), a red carotenoid derived from β-carotene by metabolic oxidation that takes place in many marine arthropods. In the native protein complex, however, the absorption spectrum of the carotenoids is red-shifted by about 150 nm, resulting in an absorption maximum of 632 nm and hence in a blue carotenoprotein. The red color of the free ligand comes back upon protein denaturation, what happens when the lobster is cooked.
The native CRC complex with a molecular mass of ˜320 kDa is called a-Crustacyanin. It is composed of eight β-Crustacyanin subcomplexes that are dimers of 20-kDa monomers, the ultimate protein subunits. As each subunit contains one molecule of astaxanthin, the 16-mer α-Crustacyanin complex contains 16 molecules of astaxanthin. Unusually, the dissociation into the β-Crustacyanin dimers is irreversible. Even more surprising is the shift of the absorption maximum of astaxanthin from 472 nm (in hexane) in the unbound form to about 585 nm in β-Crustacyanin and further to 632 nm in α-Crustacyanin. While the resolved structure of β-Crustacyanin provides a reasonable basis to explain the spectral properties of the carotenoid in the dimer (a 100-nm shift), the further shift of about 50 nm awaits the crystal analysis of the α-complex.16,17 According to a very recent study, the tight packing of the subunits (yet unknown in detail) allowing exciton coupling between excited states of astaxanthin molecules is the likely cause for this impressive bathochromic effect.18
When Crustacyanin protein is separated electrophoretically, it appears as five distinct monomers. However, they are encoded by only two genes (CRC1 and CRC2). Recent results suggest that the nonencoded differences result from nonphysiological alterations during purification and storage of α-crustacyanin.19 The protein sequences of CRC1 and 2 predict a typical lipocalin folding with two conserved cysteine bridges. The structures of four different β-Crustacyanin complexes have been solved,19-22 with two bound astaxanthin per dimer. In fact, the crystal structures demonstrate a β-barrel made up of eight antiparallel strands and the presence two helices. In the dimers, the monomers are oriented in such a way that their cavity openings are facing each other. Even more surprisingly, each of the two astaxanthin ligands is bound to both cavities by bridging both protein subunits thus sharing each cavity. This unusual mode of ligand binding is a consequence of the large extended conformation of the carotenoid that does not fit into a single lipocalin cavity. The specific arrangement of the two carotenoid chromophores positioned fairly in parallel and close together, explains the high stability of the dimeric pigment complex. The structures further reveal a conformational change of the bound carotenoid that, together with protein contacts, is thought to result in the observed shift of the absorption spectrum of the dimeric β-Crustacyanin versus free astaxanthin.
It is obvious, that the presence of the blue CRC in the carapace of the lobster significantly contributes to camouflage coloration in its marine habitat that is dominated by blue to blue-green light. This is why other marine arthropods (and also other animals) have developed a similar protective coloration in their exoskeleton or underlying tissues. Representative species are the brine shrimp Artemia, some other shrimps and the water flea Daphnia, from which a blue high-molecular weight glycoprotein, called artemocyanin, has been purified.23,24 In contrast to CRC, however, the blue color of artemocyanin is not due to astaxanthin or any other carotenoid but due to a bilin, reported to be similar to biliverdin IXα. An N-terminal sequence obtained from one of these biliproteins did not reveal any relationship to insect biliproteins that have been identified as lipocalins. So, these crustacean biliproteins seem to have evolved independently as functional equivalent means of protection.
The Bilin-Binding Lipocalins and Their Relatives
The term biliproteins denotes proteins that are specifically associated with a bilin as ligand. They are widespread among insects though mostly known from lepidopteran species. A number of these typically blue proteins have been purified and characterized in more or less detail, but crystal structures have been obtained only for the two proteins discussed below in detail (for an overview, see the review by Kayser in ref. 1). However, other lipocalins, for which the ligand binding properties are not yet known, appear grouped with the biliproteins in a strongly supported clade (Fig. 2). Whether they represent cases of functional divergence without much sequence variations will be discussed below.
Bilin-Binding Protein of Pieris brassicae
The name ‘bilin-binding protein’ (BBP) specifically refers to the biliprotein from Pieris brassicae, the large white cabbage butterfly. BBP is mainly found in the hemolymph of the last larval instar and in the wings of the adult stage, while it is also present in other tissues such as fat body and epidermis. The blue protein in the wings is masked by the large amounts of white pterins in the overlaying scales.
BBP was isolated from whole butterflies by one of us, crystallized and unexpectedly identified as a prototypic lipocalin, the first from an invertebrate source.25,26 This contrasts to the well known cyanobacterial biliproteins, which are of mainly helical structure and function in photosynthetic light harvesting.27,28 BBP is found predominantly as a ˜19.8 kDa monomer composed of 174 amino acids,29,30 is not associated with sugar or lipid, and has one molecule of biliverdin IXγ noncovalently bound.
BBP crystallizes as a tetramer, i.e., as a dimer of dimers, with the monomers showing the typical lipocalin folding of an eight-stranded antiparallel β-barrel with two cysteine bridges at positions known to be conserved in lipocalins.25,26 Besides of two short helical structures before the first β-strand and close to the C-terminus, respectively, there is one long α-helix attached to a side of the barrel (Fig. 3). The bilin ligand of BBP is bound in the cavity of the barrel and displays the cyclic helical, porphyrin-like structure that is normally adopted also by open-chain tetrapyrroles (for a review, see ref. 31). The orientation of the bilin is such that the two carboxyl groups at the outer pyrrole rings point to the opening of the cavity, i.e., to the solvent. Most remarkably, the bilin, whose two enantiomeric cyclic conformations are rapidly interconverted in solution and therefore optically inactive, is fixed by the protein in one enantiomeric form (Fig. 3) resulting in an optically active bound state.32 This specific mode of binding affects also the visible absorption spectrum of the bilin, which shows a red shift of about 25 nm to a plateau around 670 nm in the bound state; the UV-absorption band of the bilin in BBP is at 383 nm.
According to studies employing radiolabeled precursors for the bilin and the apoprotein, respectively,33,34 there are two major developmental periods of BBP synthesis, one in the last larval instar during the feeding period, and another one during development of the adult insect starting at mid-pupal stage. These studies further revealed that the bilin and the protein are made in parallel and that opening of the porphyrin ring must immediately follow its synthesis. This developmental pattern of biosynthesis has been confirmed by Northern analysis of BBP messenger RNA, which also identified the fat body as the major site of BBP synthesis in the larva.29 Interestingly, holo-BBP in the developing butterfly is produced in the wings, as revealed in isolated wings in culture (Sehringer and Kayser, unpublished results).
The fact that BBP biosynthesis is under strict developmental and tissue control and, furthermore, that most of the tetrapyrrole precursor 5-aminolevulinate is used for bilin synthesis34 suggest a vital role of this lipocalin in this insect. However, the functional implications of this program remain to be discovered. While the blue biliproteins of insects are generally said to play a role in camouflage coloration, which is certainly true in many species (see below for Insecticyanin), this is not obvious in P. brassicae, for example, where the coloration of larvae and adults is dominated by other pigments (for details, see ref. 1). A hypothetical role of BBP based on light absorption, as in cyanobacterial biliproteins,28 can be excluded as it lacks the required photochemical properties.35 This is likely true for all insect biliproteins. More realistic, though unproved roles of insect biliproteins related to metabolic regulation are discussed in a recent lipocalin review (see ref. 1), and they include prevention of cellular damage by reactive oxygen and nitrogen species, CO signaling cascades, and regulation of soluble guanylyl cyclase by biliverdin.
Insecticyanin of Manduca sexta
Insecticyanin (INS) is the counterpart of the butterfly BBP in the tobacco hornworm moth, Manduca sexta. This insect biliprotein was the first one to be purified and physicochemically characterized as a major hemolymph constituent.36 INS occurs in larval hemolymph as a tetrameric complex of ˜23-kDa subunits and, like BBP, has noncovalently bound biliverdin IXγ at a stoichiometry of 1:1, and is also free of sugar and lipid. Sequencing confirmed the larger size of the monomer, compared to BBP, as it comprises 189 amino acids residues making up a molecular mass of ˜21.4 kDa.37
In the crystallized state, INS, again like BBP, is obtained as a tetramer with the subunits exhibiting the characteristic lipocalin folding38 that first became known for vertebrate retinol-binding protein (see Chapter 2). While the barrel with the two sheets of orthogonally arranged up-and-down β-strands is almost identical to that of BBP, the moth protein shows additional helical structures, not only before and after the eight strands (for structural details, see ref. 1). The bilin of INS is bound to the cavity of the barrel in a way comparable to that of BBP, i.e., it displays the same ‘frozen’ cyclic helical conformation with the carboxyl groups directed to the solvent. As generally observed in members of the lipocalin family, the high similarity in the crystal structures of INS and BBP contrasts with the low similarity in their amino acid sequences, which are about 40% identical. This discrepancy is even higher when vertebrate lipocalins with the same 3D structure, e.g., retinol-binding protein, are included.
INS occurs in two isoelectric forms, a more acidic form (INS-a) and a more basic form (INS-b). Both molecular variants are present in the larval integument, while INS-b is the only form in the hemolymph.39 The isoforms are encoded by two distinct INS genes that are both expressed mainly in the epidermis and to a lesser degree in the fat body.40,41 The two gene products, which differ in 13 amino acid residues, are differentially exported into the two compartments: cuticle and hemolymph. More detailed developmental studies revealed that the two INS genes are differentially regulated in epidermis and fat body, show different temporal expression patterns and undergo significant cyclic changes in their messenger RNAs during larval development.39 INS is synthesized from the second larval instar up to end of last instar, when its messenger RNA is lost as a consequence of the secretion of 20-hydroxyecdysone, which follows a decline of juvenile hormone marking the transition to the pupal stage.42 In accordance with this observation, expression of the two INS genes is enhanced in the presence of juvenile hormone.43 No expression takes place in the pupal and adult stages. Despite the cease of synthesis at pupation, INS is present in the hemolymph of the pupa and even the adult insect. In the young female moth, the larval biliprotein is sequestered into developing oocytes via a membrane receptor that specifically binds INS with high affinity.44,45
The uptake of INS into the eggs of M. sexta suggests a significant role of the biliprotein in embryogenesis or related processes. However, any role in the egg is open to speculation. During the larval stages, on the other hand, the presence of the blue INS together with the yellow carotenoids in hemolymph and epidermis create a green coloration that conceivably provides a protective camouflage effect (see also ref. 1). This is in contrast to the situation in larvae of P. brassicae, as discussed above.
The Biliverdin-Binding Proteins of Samia cynthia ricini
Two biliproteins have been purified from larval hemolymph of the moth Samia cynthia ricini and described as biliverdin-binding proteins BBP-I and BBP-II.46 In contrast to the two Insecticyanins from M. sexta, the two proteins from S. cynthia share only 43% sequence identity and the length of their branches in the phylogenetic tree (Fig.2) suggests that more divergence has taken place after the underlying gene duplication in this silkworm than in the tobacco hornworm. The subsequent independent evolution resulted in a significant difference in molecular size and in oligomeric state: while BBP-I occurs as a monomer of 20.5 kDa, BBP-II behaves as a dimer of 22.7-kDa subunits. Moreover, these two proteins are immunologically distinct from each other. BBP-II is most likely identical to the biliverdin-binding protein that is present in the molting fluid during larval-pupal ecdysis in this silkmoth. Both BBPs seem to be synthesized in the epidermis and secreted into the hemolymph and molting fluid, respectively. Each monomer of BBP is associated with one molecule of biliverdin tentatively identified as the IX? isomer, which is also bound in the biliproteins of P. brassicae and M. sexta.
Biliproteins of about 20 kDa, the size of typical lipocalins, have also been described from at least four other lepidopteran species, as reviewed in.1 As their N-terminal sequences only are known to date, their definite identification as lipocalins is still open.
Biliprotein-Related Lipocalins with Unknown Ligand
Some insect proteins, whose amino acid sequence identifies them as lipocalins, are monophyletically related to the known biliproteins described above (see Fig. 2). The one first described was Gallerin, which is mainly expressed in the nervous system of Galleria mellonella, the wax moth,47 particularly in glial cells of the nerve cord. The sequence of the Gallerin precursor, comprising 203 amino acid residues, shows a close relationship to BBP and INS (see Fig. 2).
The clade of biliprotein-related proteins (BPRPs) also includes Bombyrin, a lipocalin from the silkworm Bombyx mori.48 As Gallerin, it is also expressed in the nervous system (pupae brain). Whether Gallerin or Bombyrin have the potential to bind a bilin or if they occur as true biliproteins in the insect is not known. Their nervous system expression also links them, at least functionally, with the Laz related lipocalins, expressed in glial and neuronal cells.
Quite recently, lipocalin sequences from the moth Lonomia oblique became known (Acc. No. AY829833,49 AY829809,49 and AY90898650).Two of them have been included in our phylogenetic tree (Lip1 and Lip4, see Fig. 2) and the third one shows only two amino acid substitutions with respect to the second. Their closest relatives within the BPRPs clade are the lipocalins from S. cynthia ricina. Lip1 and Lip4 were identified as transcripts in the larval spicule and integument, respectively, being Lip1 postulated to play a role in the hemorrhagic syndrome. The third sequence has been reported as a prothrombin activator. Their function as well as their possible association with a bilin awaits further characterization, but it would be interesting to see if a set of lipocalins with a functional relationship with the hemostatic system of vertebrates is also present in Lepidoptera as is the case of the saliva lipocalins from hematophagous arthropods (see below).
Expansions of the Lipocalin Family in Blood-Feeding Arthropods
We have described so far many interesting functional explorations that lipocalins have undergone through arthropod evolution. Now, two independent expansions of the family have been found when exploring proteins or mRNA expressed in the salivary gland of two distantly related arthropods, insects and arachnids, that share a common way of life: feeding on vertebrate's blood.
Their assignment to the lipocalin family has been a difficult task, since they share little or almost undetectable protein sequence similarity with other lipocalins. However, evidence of their common ancestry keeps accumulating, and these data are helping to frame the following evolutionary hypothesis: A history of intra-lineage duplications accompanied by high divergence, while both groups of lipocalins (from insects and arachnids) converge to control the hemostatic system of their host to make the most out of a blood meal.
Both, blood-feeding insects and ticks, produce in their saliva a cocktail of proteins specialized in the control of their host hemostatic machinery (reviewed by refs. 51,52). In Table 2, we relate each antihemostatic activity with the protein that plays that role in the cocktail. In Hemiptera and Acari, most of these functions are carried out by an independently evolved set of lipocalins. In Diptera (mosquitoes) these roles have been taken by completely unrelated proteins. 53,54 This is certainly a striking case of convergent evolution, in which each parasite organism has adopted a different solution to fulfill the same task, probably evolving in parallel to the evolution of hematophagy (dating around 90 My55). It also represents a case of coevolution with the host hemostasis and inflammation systems, which have evolved very complex and redundant reactions.
Table 2
Different strategies for the control of the host's hemostatic machinery by blood feeding arthropods.
The Saliva Lipocalins in Hematophagous Insects
At least 28 proteins that share the lipocalin fold are expressed in the salivary glands of hematophagous insects. They comprise the Nitrophorins (NPs), the Biogenic Amine Binding Protein (BABP), the Rhodnius Platelet Aggregation Inhibitors (RPAIs), all of them found up to date only in one species (Rhodnius prolixus); and the Triabins, Pallidipins, and Procalins (found in three species of blood-feeders).
Their amino acid sequences have low similarity with the lipocalins. However, their eight-stranded β-barrel structure shows a striking similarity and constitutes the first hint to family relationship.56-59 When secondary structure constraints are imposed into the alignment, a few conserved sites are then evident, and the SCR1 is recognized, although with some variations. An attempt to place them within the family phylogeny is shown in Figure 2, and proposes a biliprotein-like lipocalin as the common ancestor of the blood feeding-related lipocalins. However, the tree position of this clade must await confirmation from other sources (such as intron-exon structure3,4).
The Nitrophorins: Carriers of NO
Four Nitrophorins (NPs) were purified out of the salivary glands of R. prolixus60-64 that constitute ˜50% of the total protein content in the insect saliva. They were characterized as NO-transporters, histamine binding and anti-coagulation factors (see Table 2). Recent transcriptome and proteome analyses of the salivary glands have revealed up to 11 NPs (or NP-related proteins65).
The expression of NPs is developmentally regulated66 through molting. New NPs are added in each molting step, been the fifth instar nymph the stage with a higher number of NPs. Some NPs, like NP3, are specific of this stage. Other NPs are more abundant in the first instar nymph and decrease through molting (like NP5 and NP6), been undetectable in adulthood. NP2, on the contrary, is the only one described to be present in all stages. As we will see below, NP2 has a functional specialization that would make it indispensable.
The crystal structure of several NPs have been resolved.56-59,67 They show the typical lipocalin barrel fold with three short alpha-helices and two disulfide bonds (at conserved positions with respect to other insect lipocalins).
Functional Interactions through the Binding Pocket of Nitrophorins
NPs have a ferric heme that binds NO or histamine62 (reviewed by ref. 51). The heme is deeply buried in their hydrophobic pocket, and one histidine residue constitutes the fifth iron ligand.51 Binding and stabilization of NO is analogous to oxygen-binding myoglobins from vertebrate. However, NPs use the lipocalin fold, different from the α helical globin fold.
The complex NP-NO is stable at low pH (around 5), being the NO moiety buried by hydrophobic side chains and protected during storage at the entrance of the NP pocket.67 However, when the NPs encounter a pH of ˜7.5 in the host blood, a conformational change debilitates the NO binding.60,62,68 NO is released and exchanged for histamine, that binds in the same location with a 100-fold higher affinity.67 These properties make NPs excellent bifunctional shuttles bringing in NO and removing histamine from the wound, fulfilling two objectives at the same time: vasodilation and avoidance of the inflammatory response of the host.
The shuttle function of NPs depends on the properties of the hydrophobic pocket, but variations in the NPs binding pocket have been described.56,69,70 Thus, NPs are not completely redundant transporters, and a potential for functional specialization exist.
In addition, the binding pocket of NPs could carry out other functions, like enzymatic activities71,72 where NPs oxidize cysteine with H2O2 as reaction product. During this thiol-oxidase reaction NPs can also destroy norepinephrine, conferring additional vasodilation potential. NPs have also been shown to have a heme-peroxidase activity in vitro. However, the physiological significance of these enzymatic activities remains to be confirmed.
Molecular Interactions of Nitrophorins not Involving the Hydrophobic Pocket
NP2 is able to interfere in a heme-independent manner with the blood coagulation cascade at the factor X maturation step:61 NP2 inhibits the conversion of factor X to factor Xa. Even though the NP2 mechanism of action has to be elucidated, its proteolysis regulation role is carried out through protein-protein interactions.
NP7 binds anionic phospholipids with high affinity in activated platelets.73 Modeling of the NP7 three-dimensional structure shows a positively charged area on the surface of the protein that could bind negatively charged membrane surfaces containing phosphatidylserine. This binding competitively blocks the prothrombin activation, since the prothrombinase complex needs to bind to the same membrane surfaces.
These roles, not related to the binding pocket of lipocalins, seem to have evolved independently many times along the evolution of the family, showing once again the versatility of the lipocalin fold for the acquisition of novel functions.74
In summary, the cocktail of NPs released in blood-feeding insects saliva shows a high degree of molecular and functional diversification, from minor changes in the binding pocket leading to differences in affinity for their ligands, to completely novel interactions through their external protein surface.
The Biogenic Amine Binding Protein of Rhodnius Prolixus
The Biogenic Amine Binding Protein (BABP) was identified in the salivary glands of fifth instar nymphs.65,75 Its amino acid sequence and biochemical properties set this protein aside of the NPs within the set of insect saliva lipocalins. As NPs, BABP has four cysteines probably forming two disulfide bonds.
BABP does not contain a heme moiety in the hydrophobic pocket, and is unable to bind ADP (a ligand for other insect saliva lipocalins, see below). Instead, it is able to bind norepinephrine (Kd = 24 nM), serotonin (Kd = 102 nM), and epinephrine (Kd = 354 nM). The molecular modeling of BABP shows a narrower binding pocket that easily accommodates serotonin, but that would explain its inability to bind heme.
By sequestering serotonin and catecholamines from the wound site, BABP acts as a vasodilator and platelet aggregation inhibitor. Its function has been tested in tissue and cell culture assays. Addition of BABP inhibits the serotonin-mediated contraction of the rat uterine horn, the norepinephrine-mediated contraction of the rabbit aorta, and it eliminates the potentiation by serotonin and epinephrine of ADP- or collagen-induced platelet aggregation.75 Therefore, BABP adds a different activity to the cocktail of saliva lipocalins controlling the host hemostatic system.
The Rhodnius Platelet Aggregation Inhibitors
With at least eleven NPs and one BABP, three important mechanisms that the host implements to avoid blood loss are counterattacked: vasoconstriction, immune response, and blood coagulation (including platelet aggregation). However, another set of lipocalins is put forward by Rhodnius prolixus in the feeding site: the Rhodnius Platelet Aggregation Inhibitors (RPAIs). A total of 7 RPAIs have been identified,65,76,77 that show a set of 6 conserved cysteines and share sequence similarity with other saliva lipocalins (Triabin and Pallidipin, see below). No crystal structure is available for RPAIs, but biochemical analyses suggest they can bind two ADP molecules.
RPAIs inhibit platelet aggregation by strongly binding ADP (in the nanomolar range76). ADP, which is released from activated platelets, injured endothelial cells, and erythrocytes, is able to decrease the threshold for the collagen-induced platelet aggregation even at very low concentrations. Apyrase, an enzyme also present in the insect saliva (not a lipocalin, see Table 2), would degrade ADP to AMP and Pi. However, Apyrase is not an efficient scavenger of ADP due to its high Km (>20 μM). When the concentration of ADP is too low for the Apyrase to work, RPAIs do their job, sequestering ADP and increasing the threshold for collagen-induced platelet aggregation.
Triabins, Pallidipins and Procalins
Triabin, first described in the saliva of Triatoma pallidipennis78 has also close relatives in Rhodnius prolixus65 and in Triatoma brasiliensis.79 Its protein sequence is most similar to RPAIs and Pallidipins, with six conserved Cys residues that form three disulfide bonds.
The crystal structure of Triabin has been solved,80 and it shows an important peculiarity: the β-barrel has strands B and C exchanged, altering the antiparallel organization common to all other lipocalin structures solved to date.
The T. pallidipennis Triabin functions as an anti-coagulation factor by directly binding to Thrombin through a protein-protein interaction. The details of this interaction are well known thanks to the crystallization of the complex Triabin-Thrombin.80 Triabin represents another example (like NP2 and NP7) of the use of lipocalins for functional interactions not related to the hydrophobic binding pocket.
Pallidipin is another inhibitor of the collagen-induced platelet aggregation,81,82 found in T. pallidipennis, R. prolixus,65 and T. brasiliensis.79 It has six conserved cysteines and a few other conserved amino acid residues and are also related to RPAIs and Triabins. The mechanism of action of Pallidipin is not known.
Finally, Procalin, has been described as an allergen found in the saliva of T. pallidipennis,83 and related proteins have been found in R. prolixus65 and T. brasiliensis.79 The role of this lipocalin during blood feeding is not known. However, because of their known allergenicity, they are playing a role in the host response to the blood-feeder bite. In this case, we are looking at the coevolution process from the point of view of the host, since the immune response to salivary proteins is detrimental for the blood-feeder. Turning these parasite-host interactions into something useful in the control of diseases transmitted by blood-feeders arthropods is an important area of biomedical research.
The Saliva Lipocalins in Hematophagous Arachnids (the tick saliva lipocalins)
Lipocalin-related proteins have also been found in the saliva of a different group of hematophagous arthropods, the ticks (Acari). They are the only lipocalins described so far in the class Arachnida. The saliva of ticks has been of interest both, for the study of parasite-host interactions in terms of hemostasis control,84 and for the toxicity of some ticks, provoking important livestock losses in certain areas.85
A total of 20 tick lipocalins have been described to date, being the hard tick Histamine Binding Proteins (HBPs) the ones whose crystal structure established the first hint for a possible relationship with lipocalins.86,87 Other lipocalins found in the saliva of hard ticks (family Ixodidae) are the Serotonin and Histamine Binding Protein (SHBP), and 11 new proteins from Ixodes scapularis. Finally, Moubatin and a set of four Tick Salivary Gland Proteins (TSGPs) have been found in soft ticks (family Argasidae).
The tick saliva lipocalins share some sequence similarity among them, but when comparing their sequences with more conventional lipocalins, or even with the salivary proteins of blood-feeding insects, any traces of common ancestry seem to have been erased. However, one can use characters derived from the intron-exon arrangement of the genes to perform phylogenetic inferences.4 This method has been useful to suggest a common origin for tick saliva proteins and lipocalins.3 Therefore, we currently view this group of tick saliva proteins as another highly divergent extension of the lipocalin family, that originated during adaptation to blood feeding. In Figure.2, we show the branching point of tick saliva lipocalins (resulting from a gene structure based phylogeny3) superimposed to the amino acid sequence based phylogeny.
The Hard Tick Histamine Binding Proteins and Their Relatives
Three closely related Histamine Binding Proteins (HBPs) have been isolated from the salivary glands of Rhipicephalus appendiculatus.86 They sequester histamine released by the host in response to tissue damage. Therefore, they fulfill one of the roles described for NPs in hematophagous insects: reducing the immune and inflammatory host responses.
However, the histamine binding of HBPs uses a different mechanism.86,87 No heme moiety is present and, most unusually within lipocalins, HBPs have two internal and separate binding sites for histamine: the H (high-affinity) and L (low-affinity) sites. These two sites are lined with acidic residues, quite useful for binding a basic ligand. The hydrophilicity of these pockets represents another striking difference with the binding pockets of most lipocalins, suited for housing hydrophobic ligands. The H site occupies the position expected for other lipocalins, but the entrance of histamine to this site is anomalous when comparing it with the open side of other lipocalin pockets (reviewed by ref. 87). On the other hand, the L site occupies the closed end of the barrel in other lipocalins, a region that bears the structurally conserved regions (SCRs), three amino acid sequence motifs clustered together in the barrel closed end region (see Chapter 2). This region is completely remodeled in HBPs, since it accommodates the L site and its entrance.
As it was the case for NPs (see above) the expression of HBPs is stage and sex specific: HBP1 and 2 are secreted only by adult females, and HBP3 by larvae, nymphs and adult males. They also differ in their glycosylation and macromolecular complexes: HBP1 and 2 are nonglycosylated monomers, while HBP3 forms a disulfide-linked dimer. The functional significance of their temporal and sex-dependent regulation or of their other molecular attributes are not yet understood.
An HBP related protein, the Serotonin and Histamine Binding Protein (SHBP) has been described in the hard tick Dermacentor reticularis.88 SHBP also has two internal binding sites. Binding of histamine to the H site has been tested, while the ligand for the L-like site is unknown. However, the analysis of its structure makes serotonin the most likely candidate ligand.
Up to 11 new sequences with similarity to previously known tick saliva lipocalins have been retrieved from database searches85 in another hard tick, Ixodes scapularis. The analysis of their primary sequences suggests that they are secreted proteins. However, more information needs to be collected on their structure and biochemical properties in order to understand their role in blood-feeding and their potential for functional specialization.
Moubatin and the Tick Salivary Gland Proteins Expressed in the Saliva of Soft Ticks
Moubatin was found in the saliva of the soft tick Ornithodoros moubata because of its platelet aggregation inhibitory activity.89,90 Its amino acid sequence shows some similarity with HBPs, placing Moubatin as a member of this expansion of the lipocalin family. Moubatin fulfills a role that blood-sucking insects perform with a different set of lipocalins: inhibition of the collagen-stimulated aggregation of platelets.
In the closely related species Ornithodoros savignyi, four highly abundant Tick Salivary Gland Proteins (TSGPs) have been proposed to have a role in salivary gland granule biogenesis.91 All proteins destined to be secreted have to be tightly packed into the secretory vesicles (or granules). Mans and Neitz92 have shown that TSGPs take up most of the accessible volume in secretory granules, and propose macromolecular compaction of TSGPs as an important part of the packaging mechanism under the slight acidity and high calcium concentration that prevail in the granules. Since the cocktail of material secreted upon feeding contains not only proteins destined to control the hemostatic system of the host, but can also contain pathogens and toxins, a close understanding of the mechanisms that sort which proteins are secreted, could help to control the pathogens transmitted by ticks.
The role of TSGPs once they are secreted is unknown. They do not bind histamine nor any of the other mediators involved in the control of the host response to the tick bite. They have been tested for effects on the blood coagulation cascade and on ADP- and collagen-induced platelet aggregation assays85 with negative results. As summarized in Table 2, these activities are carried out in this tick by nonlipocalin proteins like Savignygrin and Savignin.
However, two of the TSGPs (TSGP2 and TSGP4) are toxins that affect the cardiovascular system of the host93 and are therefore involved in the pathogenesis of toxicosis caused by O. savignyi bites. Since the closely related TSGPs and Moubatin are not toxic, the properties that render a lipocalin toxic seem to be a recent acquisition in the evolution of ticks. The toxicity of these lipocalins might be considered as detrimental for the feeding parasite, as was the case for the allergenic Procalin, especially for ticks that have to spend longer periods of time on the host to complete a meal.
While the three dimensional structures of Moubatin or TSGPs have not been solved, TSGPs models have been constructed using the known structure of HBP2 with a reasonable fit. It is therefore expected that their tertiary structure will show commonalities with lipocalins. When the phylogenetic relationship among tick saliva lipocalins is assayed,85 the major conclusion is that they are the result of recent gene duplications since they become grouped into genus-specific clades, reflecting gene duplication after tick speciation.
Therefore, in spite of the structural and biochemical differences of tick saliva lipocalins, the remaining resemblance to the β-barrel of standard lipocalins and the data on their gene structure3 have shown more similarities than previously thought, and therefore had been assigned as lipocalins. Quoting Paesen et al,87 ‘tick lipocalins are very eccentric members of the lipocalin family’, which highlights the versatility of the lipocalin fold to carry out many functions. Lipocalins seem to be perfect ‘moonlightlers’94,95 and the phylum Arthropoda has certainly been a great arena for lipocalin functional explorations.
More Expansions of the Lipocalin Family? The Case of Cockroach Milk
Recently, Williford et al96 have described a new set of lipocalins that are secreted by the brood sack epithelium of the viviparous cockroach Diploptera punctata as part of a nutritive secretion or “milk” for the embryos. In species of ovoviviparous cockroaches, embryos develop within an infolding of the intersegmental membrane at the posterior end of the abdomen and are born as first instar larvae. This poach provides protection against desiccation and parasitism. The appearance of viviparity is linked to the production of “milk”, now known to be full of lipocalins. Although only one of them has been included in our phylogenetic tree (Dpun.Milk in Fig.2), a total of 22 distinct lipocalin protein sequences have been identified and the intron-exon structure of one of them has been described.96
The presence of four introns intervening the coding sequence of this cockroach milk lipocalin would relate their gene structure to the tick saliva lipocalins3 and to NLaz4 (see Fig.4 in Chapter 3). However, their protein sequence becomes grouped with the insect saliva lipocalins and other highly divergent lipocalin sequences like Drosophila yakuba lipocalin, or the cockroach lipocalins of Blatella germanica (with allergenic activity97) and of Leucophaea maderae. Curiously, the later is part of an aphrodisiac secretion from the male tergal gland,98 another infolding of the intersegmental membrane that could be considered as a morphological and physiological equivalent to the brood sac of D. punctata, but serving a different function at the organismal level.
Multiple gene copies of the milk lipocalins are detected in the genome of D. punctata, therefore representing one more case of gene duplication associated to a recent cooption event:74 the acquisition of the new nutritive function which requires the production of these proteins in large amounts, and that embryos develop the ability to drink. The analysis of this new expansion of the lipocalin family should help to understand the transition of ovoviviparity to viviparity in this group of arthropods, in which morphological, physiological and molecular changes are linked and result in a completely new reproductive strategy.
Concluding Remarks
The main conclusion of this work is that lipocalins show and extraordinary ability to acquire novel functions, even though they are apparently simple, single modular proteins. The pattern of duplication and divergence of this protein family in the phylum Arthropoda is characterized by an initial low number of lipocalins maintained in most species sampled, in contrast with large intra-lineage multiplication of genes clearly linked to the appearance of new functions (e.g., feeding or reproductive strategies). This pattern is the result of an evolutionary pathway independent and quite distinct from the one followed by lipocalins in the other best sampled phylum, the chordates (see Chapter 3). However, the properties of lipocalins that grant evolutionary access to such an amazing panoply of functions are the same: their robust folding combined with a flexibility for change which allows lipocalins to interact with different ligands in their binding pocket and make contact with different molecules through their protein surface.
References
- 1.
- Kayser H. Lipocalins and structurally related ligand-binding proteins. In: Gilbert LI, Iatrou K, Gill S, eds. Comprehensive Molecular Insect Science. Oxford: Elsevier. 2005;4:267–306.
- 2.
- Philippe H, Laurent J. How good are deep phylogenetic trees? Curr Opin Genet Dev. 1998;8:616–623. [PubMed: 9914208]
- 3.
- Mans BJ, Neitz AWH. Exon-intron structure of outlier tick lipocalins indicate a monophyletic origin within the larger lipocalin family. Insect Biochem Mol Biol. 2004;34(6):585–594. [PubMed: 15147759]
- 4.
- Sanchez D, Ganfornina MD, Gutierrez G. et al. Exon-intron structure and evolution of the Lipocalin gene family. Mol Biol Evol. 2003;20(5):775–783. [PubMed: 12679526]
- 5.
- Ganfornina MD, Gutierrez G, Bastiani M. et al. A phylogenetic analysis of the lipocalin protein family. Mol Biol Evol. 2000;17(1):114–126. [PubMed: 10666711]
- 6.
- Gutierrez G, Ganfornina MD, Sanchez D. Evolution of the lipocalin family as inferred from a protein sequence phylogeny. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 2000;1482(1-2):35–45. [PubMed: 11058745]
- 7.
- Ganfornina MD, Sanchez D, Bastiani MJ. Lazarillo, a new GPI-linked surface lipocalin, is restricted to a subset of neurons in the grasshopper embryo. Development. 1995;121:123–134. [PubMed: 7867494]
- 8.
- Sanchez D, Ganfornina MD, Bastiani MJ. Developmental expression of the lipocalin Lazarillo and its role in axonal pathfinding in the grasshopper embryo. Development. 1995;121:135–147. [PubMed: 7867495]
- 9.
- Sanchez D, Ganfornina MD, Bastiani MJ. Lazarillo, a neuronal lipocalin in grasshoppers with a role in axon guidance. Biochimica Et Biophysica Acta. 2000;1482(1-2 SU-):102–109. [PubMed: 11058752]
- 10.
- Ganfornina MD, Sanchez D, Bastiani MJ. Embryonic development of the enteric nervous system of the grasshopper Schistocerca americana. J Comp Neurol. 1996;372:581–596. [PubMed: 8876455]
- 11.
- Graf S, Ludwig P, Boyan G. Lazarillo expression reveals a subset of neurons contributing to the primary axon scaffold of the embryonic brain of the grasshopper Schistocerca gregaria. J Comp Neurol. 2000;419(3):394–405. [PubMed: 10723013]
- 12.
- Boyan G, Reichert H, Hirth F. Commissure formation in the embryonic insect brain. Arthropod Structure & Development. 2003;32(1):61–77. [PubMed: 18088996]
- 13.
- Boyan GS, Braunig P, Posser S. et al. Embryonic development of the sensory innervation of the clypeo-labral complex: Further support for serially homologous appendages in the locust. Arthropod Structure & Development. 2003;32(4 SU-):289–302. [PubMed: 18089013]
- 14.
- Sanchez D, Ganfornina MD, Torres-Schumann S. et al. Characterization of two novel lipocalins expressed in the Drosophila embryonic nervous system. Int J Dev Biol. 2000;44(4):349–359. [PubMed: 10949044]
- 15.
- Bailey UM. The drosophila lipocalin, Karl, is specifically expressed in the blood cell compartment. Paper presented at: Benzon symposium No. 50: The lipocalin protein superfamily. Copenhagen, Denmark. 2003
- 16.
- Chayen NE, Cianci M, Grossmann JG. et al. Unravelling the structural chemistry of the colouration mechanism in lobster shell. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 12):2072–2082 (Epub 2003 Nov 2027). [PubMed: 14646064]
- 17.
- Dellisanti CD, Spinelli S, Cambillau C. et al. Quaternary structure of alpha-crustacyanin from lobster as seen by small-angle X-ray scattering. FEBS Letters. 2003;544(1-3):189–193. [PubMed: 12782314]
- 18.
- van WijkAA, Spaans A, Uzunbajakava N. et al. Spectroscopy and quantum chemical modeling reveal a predominant contribution of excitonic interactions to the bathochromic shift in alpha-crustacyanin, the blue carotenoprotein in the carapace of the lobster homarus gammarus. J Am Chem Soc. 2005;127(5):1438–1445. [PubMed: 15686376]
- 19.
- Habash J, Helliwell JR, Raftery J. et al. The structure and refinement of apocrustacyanin C2 to 1.3 Å resolution and the search for differences between this protein and the homologous apoproteins A1 and C1. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 3):493–498 (Epub 2004 Feb 2025). [PubMed: 14993674]
- 20.
- Gordon EJ, Leonard GA, McSweeney S. et al. The C1 subunit of alpha-crustacyanin: The de novo phasing of the crystal structure of a 40 kDa homodimeric protein using the anomalous scattering from S atoms combined with direct methods. Acta Crystallogr D Biol Crystallogr. 2001;57(Part 9):1230–1237. [PubMed: 11526314]
- 21.
- Cianci M, Rizkallah PJ, Olczak A. et al. The molecular basis of the coloration mechanism in lobster shell: Beta-crustacyanin at 3.2-Å resolution. Proc Natl Acad Sci USA. 2002;99(15):9795–9800. [PMC free article: PMC125020] [PubMed: 12119396]
- 22.
- Cianci M, Rizkallah PJ, Olczak A. et al. Structure of lobster apocrustacyanin A1 using softer X-rays. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 9):1219–1229. [PubMed: 11526313]
- 23.
- Krissansen GW, Trotman CNA, Tate WP. Identification of the blue-green chromophore of an abundant biliprotein from the haemolymph of Artemia. Comp Biochem Physiol. 1984;77B:249–252.
- 24.
- Peeters K, Brendonck L, Moens L. The occurrence of artemocyanin in Branchiopoda (Crustacea). Comp Biochem Physiol A Physiol. 1994;109(3):773–779. [PubMed: 8529016]
- 25.
- Huber R, Schneider M, Epp O. et al. Crystallization, crystal structure analysis and preliminary molecular model of the bilin binding protein from the insect Pieris brassicae. J Mol Biol. 1987;195(2):423–434. [PubMed: 3656419]
- 26.
- Huber R, Schneider M, Mayr I. et al. Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 Å resolution. J Mol Biol. 1987;198(3):499–513. [PubMed: 3430616]
- 27.
- Schirmer T, Bode W, Huber R. Refined three-dimensional structures of two cyanobacterial C-phycocyanins at 2.1 and 2.5 Å resolution. A common principle of phycobilin-protein interaction. J Mol Biol. 1987;196(3):677–695. [PubMed: 3119857]
- 28.
- MacColl R. Cyanobacterial phycobilisomes. J Struct Biol. 1998;124(2-3):311–334. [PubMed: 10049814]
- 29.
- Schmidt FS, Skerra A. The bilin-binding protein of Pieris brassicae. cDNA sequence and regulation of expression reveal distinct features of this insect pigment protein. Eur J Biochem. 1994;219(3):855–863. [PubMed: 8112337]
- 30.
- Suter F, Kayser H, Zuber H. The complete amino-acid sequence of the bilin-binding protein from Pieris brassicae and its similarity to a family of serum transport proteins like the retinol-binding proteins. Biol Chem Hoppe Seyler. 1988;369(6):497–505. [PubMed: 3202956]
- 31.
- Kayser H. Pigmens. In: Kerkut GA, Gilbert LI, eds. Comprehensive insect biochemistry, physiology and pharmacology. Pergamon Press. 1985;10:367–415.
- 32.
- Scheer H, Kayser H. Conformational studies of biliproteins from the insects Pieris brassicae and Cerura vinula. Z Naturforsch. 1988;43c:84–90.
- 33.
- Kayser H. De novo synthesis and levels of cytochrome c and a biliprotein during pupal-adult development in a butterfly, Pieris brassicae. Z Naturforsch. 1984;39c:938–947.
- 34.
- Kayser H, Krull-Savage U. Development-specific incorporation of [14C]5-aminolevulinate and [3H]leucine into cytochrome c and biliprotein in the butterfly, Pieris brassicae. Correlation with the ecdysteroid titer in the pupa. Z Naturf. 1984;39c(948-957)
- 35.
- Schneider S, Baumann F, Geiselhart P. et al. Biliproteins from the butterfly Pieris brassicae studied by time-resolved fluorescence and coherent anti-stokes Raman spectroscopy. Photochem Photobiol. 1988;48:239–242.
- 36.
- Cherbas P. Biochemical studies of insecticyanin. Ph.D. thesis, Harvard University, Cambridge, Massachusets. 1973
- 37.
- Riley CT, Barbeau BK, Keim PS. et al. The covalent protein structure of insecticyanin, a blue biliprotein from the hemolymph of the tobacco hornworm, Manduca sexta L. J Biol Chem. 1984;259(21):13159–13165. [PubMed: 6386809]
- 38.
- Holden HM, Rypniewski WR, Law JH. et al. The molecular structure of insecticyanin from the tobacco hornworm Manduca sexta L. at 2.6 Å resolution. EMBO J 1987. 6(6):1565–1570. [PMC free article: PMC553525] [PubMed: 3608987]
- 39.
- Riddiford LM, Palli SR, Hiruma K. et al. Developmental expression, synthesis, and secretion of insecticyanin by the epidermis of the tobacco hornworm, Manduca sexta. Arch Insect Biochem Physiol. 1990;14(3):171–190. [PubMed: 2134175]
- 40.
- Li W, Riddiford LM. Two distinct genes encode two major isoelectric forms of insecticyanin in the tobacco hornworm, Manduca sexta. Eur J Biochem. 1992;205(2):491–499. [PubMed: 1572353]
- 41.
- Li WC, Riddiford LM. The two duplicated insecticyanin genes, ins-a and ins-b are differentially expressed in the tobacco hornworm, Manduca sexta. Nucleic Acids Res. 1994;22(15):2945–2950. [PMC free article: PMC310259] [PubMed: 8065906]
- 42.
- Kiely ML, Riddiford LM. Temporal programming of epidermal cell protein synthesis during the larval-pupal transformation of Manduca sexta. Roux's Arch Devel Biol. 1985;194:325–335.
- 43.
- Li W, Riddiford LM. Differential expression of the two duplicated insecticyanin genes, ins-a and ins-b, in the black mutant of Manduca sexta. Arch Biochem Biophys. 1996;330:65–70. [PubMed: 8651705]
- 44.
- Kang Y, Kulakosky PC, Van Antwerpen R. et al. Sequestration of insecticyanin, a blue hemolymph protein, into the egg of the hawkmoth Manduca sexta. Evidence for receptor-mediated endocytosis. Insect Biochem Mol Biol. 1995;25(4):503–510. [PubMed: 7742835]
- 45.
- Kang Y, Ziegler R, van Antwerpen R. et al. Characterization of the solubilized oocyte membrane receptor for insecticyanin, a biliprotein of the hawkmoth, Manduca sexta. Biochim Biophys Acta. 1997;1324(2):285–295. [PubMed: 9092715]
- 46.
- Saito H. Purification and characterization of two insecticyanin-type proteins from the larval hemolymph of the Eri-silkworm, Samia cynthia ricini. Biochim Biophys Acta. 1998;1380:141–150. [PubMed: 9545563]
- 47.
- Filippov VA, Filippova MA, Kodrík D. et al. Two lipocalin-like peptides of insect brain. In: Suzuki A, Kataoka H, Matsumoto S, eds. Molecular mechanisms of insect metamorphosis and diapause. Tokyo: Industrial Publishing and Consulting, Inc. 1995:35–43.
- 48.
- Sakai M, Wu C, Suzuki K. Nucleotide and deduced amino acid sequences of a cDNA encoding a lipocalin protein in the central nervous system of Bombyx mori. Nippon Sanshi-gaku Zasshi. 2001;70:105–111.
- 49.
- Veiga ABG, Francischetti IMB, Guimaraes JA. A catalog of Lonomia obliqua transcripts and proteins probably involved in the hemorrhagic syndrome [GenBank database]
- 50.
- Reis CV, Ho PL, Ramos CR. r-LOPAP: A member of lipocalin family which shows serine-protease activity [GenBank database]
- 51.
- Montfort WR, Weichsel A, Andersen JF. Nitrophorins and related antihemostatic lipocalins from Rhodnius prolixus and other blood-sucking arthropods. Biochimica Et Biophysica Acta. 2000;1482(1-2 SU-):110–118. [PubMed: 11058753]
- 52.
- Urata J, Shojo H, Kaneko Y. Inhibition mechanisms of hematophagous invertebrate compounds acting on the host blood coagulation and platelet aggregation pathways. Biochimie. 2003;85(5):493–500. [PubMed: 12763308]
- 53.
- Ribeiro JMC, Charlab R, Pham VM. et al. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34(6):543–563. [PubMed: 15147756]
- 54.
- Valenzuela JG, Pham VM, Garfield MK. et al. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem Mol Biol. 2002;32(9):1101–1122. [PubMed: 12213246]
- 55.
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19(5):748–761. [PubMed: 11961108]
- 56.
- Andersen JF, Champagne DE, Weichsel A. et al. Nitric oxide binding and crystallization of recombinant nitrophorin I, a nitric oxide transport protein from the blood-sucking bug Rhodnius prolixus. Biochemistry. 1997;36(15):4423–4428. [PubMed: 9109649]
- 57.
- Andersen JF, Weichsel A, Balfour CA. et al. The crystal structure of nitrophorin 4 at 1.5 Å resolution: Transport of nitric oxide by a lipocalin-based heme protein. Structure. 1998;6(10):1315–1327. [PubMed: 9782054]
- 58.
- Andersen JF, Montfort WR. The crystal structure of nitrophorin 2. A trifunctional antihemostatic protein from the saliva of Rhodnius prolixus. J Biol Chem. 2000;275(39):30496–30503. [PubMed: 10884386]
- 59.
- Weichsel A, Andersen JF, Champagne DE. et al. Crystal structures of a nitric oxide transport protein from a blood-sucking insect. Nat Struct Biol. 1998;5(4):304–309. [PubMed: 9546222]
- 60.
- Ribeiro JM, Hazzard JM, Nussenzveig RH. et al. Reversible binding of nitric oxide by a salivary heme protein from a bloodsucking insect. Science. 1993;260(5107):539–541. [PubMed: 8386393]
- 61.
- Ribeiro JM, Schneider M, Guimaraes JA. Purification and characterization of prolixin S (nitrophorin 2), the salivary anticoagulant of the blood-sucking bug Rhodnius prolixus. Biochem J. 1995;308(Pt 1):243–249. [PMC free article: PMC1136869] [PubMed: 7755571]
- 62.
- Ribeiro JM, Walker FA. High affinity histamine-binding and antihistaminic activity of the salivary nitric oxide-carrying heme protein (nitrophorin) of Rhodnius prolixus. J Exp Med. 1994;180(6):2251–2257. [PMC free article: PMC2191789] [PubMed: 7964498]
- 63.
- Champagne DE, Nussenzveig RH, Ribeiro JM. Purification, partial characterization, and cloning of nitric oxide-carrying heme proteins (nitrophorins) from salivary glands of the blood-sucking insect Rhodnius prolixus. J Biol Chem. 1995;270(15):8691–8695. [PubMed: 7721773]
- 64.
- Sun J, Yamaguchi M, Yuda M. et al. Purification, characterization and cDNA cloning of a novel anticoagulant of the intrinsic pathway, (prolixin-S) from salivary glands of the blood sucking bug, Rhodnius prolixus. Thromb Haemost. 1996;75(4):573–577. [PubMed: 8743181]
- 65.
- Ribeiro JMC, Andersen J, Silva-Neto MAC. et al. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem Mol Biol. 2004;34(1):61–79. [PubMed: 14976983]
- 66.
- Moreira MF, Coelho HSL, Zingali RB. et al. Changes in salivary nitrophorin profile during the life cycle of the blood-sucking bug Rhodnius prolixus. Insect Biochem Mol Biol. 2003;33(1):23–28. [PubMed: 12459197]
- 67.
- Weichsel A, Andersen JF, Roberts SA. et al. Nitric oxide binding to nitrophorin 4 induces complete distal pocket burial. Nat Struct Biol. 2000;7(7):551–554. [PubMed: 10876239]
- 68.
- Gerczei T, Fazekas A, Keseru GM. Theoretical study on the nitric oxide binding to nitrophorin 1, an NO-carrier protein from a blood-sucking insect. J Mol Struct: THEOCHEM. 2000;503(1-2):51–58.
- 69.
- Andersen JF, Ding XD, Balfour C. et al. Kinetics and equilibria in ligand binding by nitrophorins 1-4: Evidence for stabilization of a nitric oxide-ferriheme complex through a ligand-induced conformational trap. Biochemistry. 2000;39(33):10118–10131. [PubMed: 10956000]
- 70.
- Kaneko Y, Yuda M, Iio T. et al. Kinetic analysis on nitric oxide binding of recombinant Prolixin-S, a nitric oxide transport protein from the bloodsucking bug, Rhodnius prolixus. Biochim Biophys Acta. 1999;1431(2):492–499. [PubMed: 10350624]
- 71.
- Ribeiro JC. Salivary thiol oxidase activity of Rhodnius prolixus. Insect Biochem Mol Biol. 1996;26(8-9):899–905. [PubMed: 9014335]
- 72.
- Ribeiro JMC. Rhodnius prolixus salivary nitrophorins display heme-peroxidase activity. Insect Biochem Mol Biol. 1998;28(12):1051–1057.
- 73.
- Andersen JF, Gudderra NP, Francischetti IMB. et al. Recognition of anionic phospholipid membranes by an antihemostatic protein from a blood-feeding insect. Biochemistry. 2004;43(22):6987–6994. [PMC free article: PMC2915585] [PubMed: 15170336]
- 74.
- Ganfornina MD, Sánchez D. Generation of evolutionary novelties by functional shift. Bioessays. 1999;21:432–439. [PubMed: 10376014]
- 75.
- Andersen JF, Francischetti IMB, Valenzuela JG. et al. Inhibition of hemostasis by a high affinity biogenic amine-binding protein from the saliva of a blood-feeding insect. J Biol Chem. 2003;278(7):4611–4617. [PubMed: 12464610]
- 76.
- Francischetti IM, Ribeiro JM, Champagne D. et al. Purification, cloning, expression, and mechanism of action of a novel platelet aggregation inhibitor from the salivary gland of the blood-sucking bug, Rhodnius prolixus. J Biol Chem. 2000;275(17):12639–12650. [PubMed: 10777556]
- 77.
- Francischetti IM, Valenzuela JG, Pham VM. et al. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol. 2002;205(Pt 16):2429–2451. [PubMed: 12124367]
- 78.
- Noeske-Jungblut C, Haendler B, Donner P. et al. Triabin, a highly potent exosite inhibitor of thrombin. J Biol Chem. 1995;270(48):28629–28634. [PubMed: 7499380]
- 79.
- Sant AnnaMR, Araujo JG, Pereira MH. et al. Molecular cloning and sequencing of salivary gland-specific cDNAs of the blood-sucking bug Triatoma brasiliensis (Hemiptera: Reduviidae). Insect Mol Biol. 2002;11(6):585–593. [PubMed: 12421416]
- 80.
- Fuentes-Prior P, Noeske-Jungblut C, Donner P. et al. Structure of the thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug. Proc Natl Acad Sci USA. 1997;94(22):11845–11850. [PMC free article: PMC23629] [PubMed: 9342325]
- 81.
- Noeske-Jungblut C, Kratzschmar J, Haendler B. et al. An inhibitor of collagen-induced platelet aggregation from the saliva of Triatoma pallidipennis. J Biol Chem. 1994;269(7):5050–5053. [PubMed: 8106481]
- 82.
- Haendler B, Becker A, Noeske-Jungblut C. et al. Expression, purification and characterisation of recombinant pallidipin, a novel platelet aggregation inhibitor from the haematophageous triatomine bug Triatoma pallidipennis. Blood Coagul Fibrinolysis. 1996;7(2):183–186. [PubMed: 8735814]
- 83.
- Paddock CD, McKerrow JH, Hansell E. et al. Identification, cloning, and recombinant expression of procalin, a major triatomine allergen. Journal Of Immunology (Baltimore, MD: 1950). 2001;167(5):2694–2699. [PubMed: 11509613]
- 84.
- Mans BJ, Neitz AWH. Adaptation of ticks to a blood-feeding environment: Evolution from a functional perspective. Insect Biochem Mol Biol. 2004;34(1):1–17. [PubMed: 14723893]
- 85.
- Mans BJ, Louw AI, Neitz AWH. The major tick salivary gland proteins and toxins from the soft tick, Ornithodoros savignyi, are part of the tick Lipocalin family: Implications for the origins of tick toxicoses. Mol Biol Evol. 2003;20(7):1158–1167. [PubMed: 12777525]
- 86.
- Paesen GC, Adams PL, Harlos K. et al. Tick histamine-binding proteins: Isolation, cloning, and three-dimensional structure. Mol Cell. 1999;3(5 SU-):661–671. [PubMed: 10360182]
- 87.
- Paesen GC, Adams PL, Nuttall PA. et al. Tick histamine-binding proteins: Lipocalins with a second binding cavity. Biochimica Et Biophysica Acta. 2000;1482(1-2 SU-):92–101. [PubMed: 11058751]
- 88.
- Sangamnatdej S, Paesen GC, Slovak M. et al. A high affinity serotonin- and histamine-binding lipocalin from tick saliva. Insect Mol Biol. 2002;11(1):79–86. [PubMed: 11841505]
- 89.
- Keller PM, Waxman L, Arnold BA. et al. Cloning of the cDNA and expression of moubatin, an inhibitor of platelet aggregation. J Biol Chem. 1993;268(8):5450–5456. [PubMed: 8449907]
- 90.
- Waxman L, Connolly TM. Isolation of an inhibitor selective for collagen-stimulated platelet aggregation from the soft tick Ornithodoros moubata. J Biol Chem. 1993;268(8):5445–5449. [PubMed: 8449906]
- 91.
- Mans BJ, Venter JD, Vrey PJ. et al. Identification of putative proteins involved in granule biogenesis of tick salivary glands. Electrophoresis. 2001;22(9):1739–1746. [PubMed: 11425229]
- 92.
- Mans BJ, Neitz AW. Molecular crowding as a mechanism for tick secretory granule biogenesis. Insect Biochem Mol Biol. 2004;34(11):1187–1193. [PubMed: 15522614]
- 93.
- Mans BJ, Steinmann CM, Venter JD. et al. Pathogenic mechanisms of sand tampan toxicoses induced by the tick, Ornithodoros savignyi. Toxicon. 2002;40(7):1007–1016. [PubMed: 12076655]
- 94.
- Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999;24:8–11. [PubMed: 10087914]
- 95.
- Jeffery CJ. Moonlighting proteins: Old proteins learning new tricks. Trends Genet. 2003;19(8):415–417. [PubMed: 12902157]
- 96.
- Williford A, Stay B, Bhattacharya D. Evolution of a novel function: Nutritive milk in the viviparous cockroach, Diploptera punctata. Evol Dev. 2004;6(2):67–77. [PubMed: 15009119]
- 97.
- Arruda LK, Vailes LD, Hayden ML. et al. Cloning of cockroach allergen, Bla g 4, identifies ligand binding proteins (or calycins) as a cause of IgE antibody responses. J Biol Chem. 1995;270(52):31196–31201. [PubMed: 8537384]
- 98.
- Korchi A, Brossut R, Bouhin H. et al. cDNA cloning of an adult male putative lipocalin specific to tergal gland aphrodisiac secretion in an insect (Leucophaea maderae). FEBS Letters. 1999;449(2-3 SU-):125–128. [PubMed: 10338117]
- 99.
- Thompson JD, Gibson TJ, Plewniak F. et al. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 1997;24:4876–4882. [PMC free article: PMC147148] [PubMed: 9396791]
- Introduction
- The Lazarillo/Apolipoprotein D Related Lipocalins
- Crustacyanins: Blue Lipocalins in Crustaceans
- The Bilin-Binding Lipocalins and Their Relatives
- Expansions of the Lipocalin Family in Blood-Feeding Arthropods
- More Expansions of the Lipocalin Family? The Case of Cockroach Milk
- Concluding Remarks
- References
- Lipocalins in Arthropoda: Diversification and Functional Explorations - Madame C...Lipocalins in Arthropoda: Diversification and Functional Explorations - Madame Curie Bioscience Database
- Role of Superoxide in Post-Ischemic Liver Injury - Madame Curie Bioscience Datab...Role of Superoxide in Post-Ischemic Liver Injury - Madame Curie Bioscience Database
- Structure and Replication of Hepatitis Delta Virus RNA - Madame Curie Bioscience...Structure and Replication of Hepatitis Delta Virus RNA - Madame Curie Bioscience Database
- Neural Crest and the Development of the Enteric Nervous System - Madame Curie Bi...Neural Crest and the Development of the Enteric Nervous System - Madame Curie Bioscience Database
- Calcium Signaling During Phagocytosis - Madame Curie Bioscience DatabaseCalcium Signaling During Phagocytosis - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...