NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Introduction
Life History: Sexual and Asexual Reproduction
Those who study ciliates have struggled over the years to establish a place in the pantheon of model organisms.1 Eukaryotic cell biologists have turned profitably to yeast models for the powerful genetic tools at their disposal, while developmental biologists have cultivated a gallery of metazoan embryos with contributions from the plant and fungal worlds. Yet ciliates continue to contribute to fundamental aspects of both cell biology and development, often by extreme example, and among the ciliates, Tetrahymena has emerged as one of the stars.
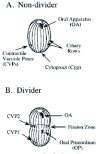
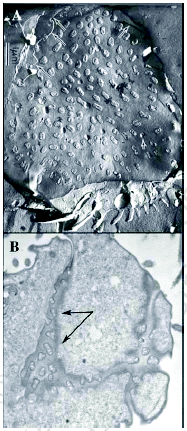
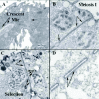
Tetrahymena is a freshwater, unicellular organism, approximately 40 to 60 microns in length. They feed on bacteria which they capture by means of four ciliated “membranelles” that form a set of three combs that brush particles into a curved buccal cavity rimmed by a fourth “undulating membrane” (Fig. 1). Ciliates are characterized by three conspicuous features, ciliature that can be specialized for locomotion or food capture, alveolar membranes that lie just beneath the plasma membrane forming a set of flattened sacks, and “nuclear duality”.2 Ciliates posses both a somatic, transcriptionally active macronucleus, and a germinal, transcriptionally silent micronucleus. It has been attractive to borrow the language of germline and soma from the world of metazoan embryos and recent findings suggest that this may reflect more than creative license. Indeed, ciliates may represent the simplest form of life to generate distinct somatic and germinal nuclear lineages.
During vegetative growth, ciliates reproduce by binary fission. In Tetrahymena, this involves a remarkable reorganization of the cortical cytoskeleton. The equatorial fission zone separates the anterior division product (proter) containing the functional oral apparatus from the posterior fission product (opisthe) containing the functional water-expulsion organelle, (the contractile vacuole system) along with the cell “anus” or cytoproct (Fig.1). Each division product would perish without the coordinate synthesis of a new oral structure (Oral Primordium, see Fig. 2) in the posterior opisthe, and a new Contractile Vacuole Pore system in the proter. Events that attend the cortical patterning associated with vegetative cell division have been studied extensively.3
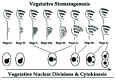
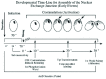
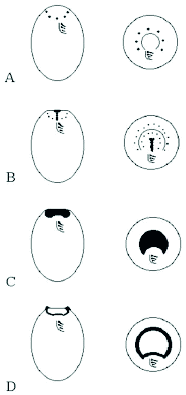
Figure 2
This figure depicts stages in oral development (stomatogenesis) and corresponding stages in nuclear division and cytokinesis (after Lansing et al). Dots represent individual basal bodies.
When Tetrahymena cells are removed from nutrient medium and starved, they have a repertoire of morphogenetic responses that seem adaptive. Starved cells frequently undergo a process of “oral replacement”.4 This involves disassembling the existing oral apparatus and reforming a new one through de novo basal body synthesis. Cells that are maintained under nutritional stress may undergo a second type of transformation to a “dispersal” form, or fast swimmer.5,6 Finally, nutritional challenge can predispose Tetrahymena for sexual reproduction including meiosis, chromosomal recombination and the exchange of meiotic products with another cell. Sex as a strategy for grooming ones' genome has also been richly discussed in ciliates.7
Nuclear exchange in Tetrahymena (and in ciliates in general) is performed in a distinctive way. Metazoan embryos achieve fertilization through whole-cell fusion of sperm and egg. Whole cell fusion also mediates sexual events in haploid, unicellular yeasts (Saccharomyces) and algae such as Chlamydomonas. Tetrahymena, on the other hand, generate a transient region of limited cell fusion. 8,9 Two diploid cells form this fusion junction anterior to their oral apparatus, and emerge from conjugation 12 hours later having exchanged haploid nuclei, while reestablishing cortical integrity.
Developmental Biology
Time Line for Assembly and Disassembly
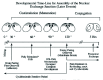
Events leading to assembly of a nuclear exchange junction have a rich laboratory history. Tetrahymena exhibit three requirements for pair formation. First cells must be nutritionally deprived for a minimum of 2 hrs at 30°C. This starvation step has been named “initiation”.10 Second, cells of complementary mating types must be present. Tetrahymena possess seven different mating types,11 determined by the mating-type locus.12 Any two of these can mate with one another, but under normal circumstances none will mate with themselves. There are, however, cases of “self-mating” cultures associated with clonal senescence.13,14 Finally, after starvation-induced initiation, cells of complementary mating types must be allowed to collide with one another in a nonshaking culture. This period of cell-cell contact has been referred to as “costimulation”.10,15 The events of initiation and costimulation are illustrated on a time line in Figures 3 and 4.
Starvation-Induced Initiation
The first step in preparing cells for conjugation is starvation. This has been accomplished in one of two ways. Tetrahymena can be allowed to deplete a monoxenic culture of bacteria such as Klebsiella pneumoniae13 or, with the development of a defined axenic medium,16 one can transfer cells from nutrient medium to a nonnutritive medium by centrifugation and resuspension.17,18 There are obvious advantages to transferring cells from a defined nutrient medium to starvation medium by centrifugation in that one gains control over the synchrony of subsequent developmental events. Initiation can take as little as 70 minutes, (though laboratories routinely starve cells overnight). Initiation can be blocked by exposure to hyperosmotic conditions after 34 minutes of initiation.19,20 Following initiation, a 15-minute exposure to food can erase most of the program of initiation.21 This means that starved, mixed cells will require an additional 45 minutes of starvation plus costimulation, (see below) before they are once again ready to pair. Curiously, no single nutrient can block initiation by itself. Only a nutritional upgrade to complete media will inhibit initiation suggesting it is the total nutritional state of the cell that determines readiness for mating, and no single molecular trigger.22
Costimulation
Contact-Mediated or Soluble Signal Molecule?
After starvation, cells of complementary mating types can be mixed thereby provoking the mating reaction. This involves a series of developmental events that remodel the anterior cell membrane and its associated cytoskeleton, and results in the formation of a remarkable and dynamic system of cell-cell junctions. This period of obligatory cell-cell interaction lasts between one and two hours.15,21,23 There has been some controversy as to whether or not costimulation involves soluble signal molecules, or strictly physical cell-cell contact. In reviewing the literature, both appear to play a legitimate role, though cell-cell contact seems the principle trigger.
The first suggestion that there might be a soluble factor associated with mating in Tetrahymena came from Takahashi.24 More extensive evidence came from the Wolfe laboratory.25,26 These early studies demonstrated that both costimulation and pair formation were profoundly delayed when cells were washed into fresh starvation medium. This delay was eliminated if, after washing, cells were returned to a conditioned starvation medium. The factor that was present in conditioned medium was a macromolecule (retained by dialysis) that was heat-stable (30 minutes in boiling water) and secreted constitutively by cells in either nutrient or starvation medium from sexually mature or immature cells. It had no mating-type specificity, and its synthesis was blocked by cycloheximide treatment suggesting that it was an unusually heat-stable, secreted protein. Wolfe named this compound FAC (factor active in conjugation).27 He went on to demonstrate that FAC acts like neither a hormone (as seen in yeast mating systems and the ciliate Blepharisma) nor an agglutinin (as seen in Chlamydomonas and Paramecium). It does not trigger conspicuous changes in the recipient cell, nor does it cause immediate aggregation of cells. Rather, it seems to somehow facilitate those cell-cell contacts that bring about both costimulation and subsequent cell adhesion. Adair et al25 suggested we think of this factor as a type of extra-cellular matrix molecule, a provocative idea in that it would suggest a role for ECM proteins in cells that do not form tissues, yet need to stabilize cell-cell interactions for this one event in their life histories. Consistent with this notion is the demonstration that both initiation and costimulation require trace amounts of calcium ion.22
Recent studies lend further support to the idea of a soluble mating factor, and raise new questions. Fujishima et al28 characterized a very early morphological response in cells undergoing costimulation. Starved Tetrahymena of complementary mating types exhibit a subtle, yet reproducible and consistent “rounding” of their shape within ten minutes of mixing. Stirring a mixed culture (which can prevent costimulation and subsequent pairing) also prevented cell rounding. When starved cells were washed and mixed into fresh starvation medium, there was a 30-minute delay in both rounding and pairing that was eliminated when cells were washed into conditioned medium. Medium from cells exhibiting a complementary mating type could not bring about rounding in starved cells until both mating types were present. In short, rounding was induced when starved cells of complementary mating types were able to undergo collisions in the presence of some secreted factor (possibly Wolfe's FAC).
Finally, in a very recent study Anafi et al (personal communication) demonstrate that ten minutes after one mixes starved cells of complementary mating types, there is a wave of tyrosine phosphorylation on cytoplasmic proteins as revealed by immuno-fluorescence and western blotting using a monoclonal antibody directed against phospho-tyrosine residues. Furthermore, starved cells of one mating type show elevated tyrosine phosphorylation in response to cell-free, conditioned medium from another mating type. Significantly, conditioned media only stimulates phosphorylation in cells of a different mating type from the cells that were used to condition the medium. This result, if valid, may force us to reopen the question of whether or not there are mating-type specific signal molecules secreted by Tetrahymena in addition to soluble factors that promote costimulation and pair formation.
Activation and Maturation
Costimulation can be divided into two stages: activation (first half hour following mixing) and maturation (30 to 120 minutes following mixing,29 see Fig. 3, Fig. 4). During the initial “activation stage,” costimulation is sensitive to cycloheximide (10 μg/mL), actinomycin D (20 μg/mL), physical agitation and nutritional enhancement of the medium. After this initial period of activation, loose pairs begin to form that can still be disrupted by either feeding, or mechanical agitation. Curiously, there is a brief period in which loose, “homotypic” pairing can occur.30 That is, cells of the same mating type form brief associations. These are intrinsically unstable, however, and give way to heterotypic pairing that is reinforced during the subsequent maturation step.
By one hour, mixed cells become “fully stimulated”.10 This condition is revealed when cells are maintained in physical contact with one another for a full hour while weak-pair associations are disrupted through periodic bouts of mechanical agitation. Such cells will form pairs immediately following cessation of the last bout of agitation, but only if they have accumulated one hour of nonshaken contact with one another.
Ninety minutes after mixing, pairs that are violently disrupted (as opposed to gentle disruption) produce cell isolates that can complete the full 12-hour developmental program involving meiosis, mitosis, self-fertilization and differentiation of the macronuclear anlagen.31,32 This strange scenario suggests that after only one and a half hours of interaction, the conjugation program can be initiated and maintained even in the absence of a mating partner. Two hours after mixing, (1.5 hrs after initial cell pairs are observed), pairs have fully matured. They are now resistant to feeding, cycloheximide, and agitation. This period may be associated with the formation of actual pores connecting the cytoplasm of the mating partners22 (see below).
Tip Transformation
Membrane Events
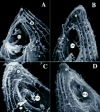
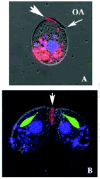
In response to costimulation, Tetrahymena remodel their anterior tips in a process called tip transformation.33 Specifically, physical collisions induce a shape change in the anterior region of the cell that begins approximately 30 minutes after mixing. The anterior pole changes from a pointed cap that forms from the convergence of numerous longitudinal ciliary rows and intervening ciliary ridges (Fig. 5A), to a region of smooth, naked membrane (Fig. 5D, asterisk). In costimulated cells, the intervening ciliary ridges converge in a seam, or chevron just anterior to the Oral Apparatus. This seam appears to be the site of novel membrane synthesis. In nonmating cells, the endo-membrane system is dispersed predominantly in the form of dictyosomes, or simplified membrane lamellae associated with ciliary basal bodies and the “deep face” of cortical mitochondria.34 The only time a well-organized, multi-lamellate golgi apparatus has been reported in Tetrahymena is during mating when it forms adjacent to the developing conjugation junction (Fig. 6).35 As tip transformation proceeds, the ridge-seam broadens, producing a blunt, naked, protruding region of membrane at the anterior tip that extends down the ventral face just anterior to the Oral Apparatus (Fig. 5B-D). When a sufficient number of costimulated cells complete tip transformation, loose adhesions form between mating partners (Fig. 7A). As mentioned earlier, transient adhesions can and do form between homotypic pairs (same mating types), but these fail to stabilize and are replaced by heterotypic pairings.30 As the region of adhesion broadens it forms the “fusion plate”9 (Fig. 7BB). This becomes perforated with hundreds of 0.1-0.2 μm pores that join the cytoplasm of the two mating partners (Fig. 8). Rows of intra-membrane particles form at the perimeter of the fusion plate, as revealed by freeze fracture electron microscopy.9
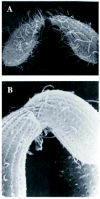
Figure 7
Scanning electron micrographs of mating pairs (reprinted with permission from the publishers from Wolfe and Grimes, and Wolfe et al). A) This figure shows loosely paired cells. B) Mating pairs are shown with a mature conjugation junction.
Membrane Proteins and Cell Adhesion: Concanavalin a Receptors
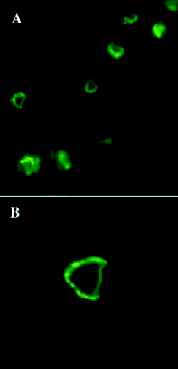
In 1976, Ofer et al36 demonstrated that the plant lectin concanavalin A (25 micrograms per mL) could inhibit pairing in costimulated Tetrahymena, and cause loosely bound pairs to dissociate. ConA is known to bind to mannose-containing glycoproteins. When conA was conjugated to Fluorescein (FITC-conA) and delivered to a mixed, mating culture, it was found to bind strongly to the developing conjugation junction in Tetrahymena.37 The pattern of conA receptors on Tetrahymena during costimulation and pairing has been carefully documented using both FITC-conjugated conA with fluorescence microscopy,37-41 and ferritin-conjugated conA with transmission electron microscopy.37,42 FITC-conA decorates the oral region in nonpairing cells, and is also incorporated into food vacuoles. As cells undergo costimulation, labeling of food vacuoles diminishes (ceasing by 20 minutes),37 suggesting that mating cells cease to feed. Within 15 minutes of mixing, conA-receptors appear in “small clusters near the anterior end, but posterior to the actual tip” (Fig. 9A). Subsequently, punctate conA labeling appears within the region of membrane remodeling located at the anterior tip of the cell (the future conjugation junction), and consolidates into a vertical line (Fig. 9B). Morphologically, this region appears as a region of smooth membrane devoid of the cilia, or membrane ridges that characterize the anterior surface of costimulated, nonmating cells.9,33 y one hour after mixing, conA labeling is dense and uniform within the, now broadened and transformed tip (Fig. 9C). Pairing follows shortly, and fluorescence becomes restricted to a heart-shaped ring around the perimeter of the conjugation junction39 (Fig. 9D, Fig. 10, Fig. 11).
It has been pointed out that conA molecules exhibit cross-linking activity, and this could drive the observed “tipping” phenomenon in which conA receptors form aggregates. This has been ruled out by first fixing cells with a variety of agents, and then labeling with conA.37-39 The study by Watanabe's group is of particular interest in that they demonstrated that when conA labeling is performed prior to observation or fixation, surface labeling is underestimated. Only the most intensely labeled regions appear. When cells are fixed (3 % glutaraldehyde) and then labeled with FITC-conA, a more extensive labeling appears in the anterior end of the cell, though the details of conA tipping and ring formation remain unchanged.
Several studies set out to search for newly synthesized conA-binding glycoproteins as cells underwent costimulation and pairing. Van Bell43 performed extensive SDS-PAGE analysis on mating and costimulating cells and probed gels with 125I-conA. Although a number of conspicuous conA-binding proteins were detected, none showed changes in their level of expression during prepairing or conjugation. Wolfe and Feng,41 isolated cytoskeletal frameworks (triton-extracted cortical residues) and analyzed conA-binding proteins associated with these by Western Blots using conA as a probe. Again, there were no changes associated with pair formation. Other studies identified a number of conA labeling surface proteins (Dentler, 1992). The most recent study by Driscoll and Hufnagel44 discovered a conA-labeling 28kD protein associated with ciliary membranes that appears to be down regulated in response to starvation, but again, no conA receptors seemed to be synthesized during the period of costimulation. These studies suggest that costimulation induced conA receptor tipping is brought about not by novel synthesis of the conA receptors themselves, but by reorganization of existing surface glycoproteins resulting in recruitment and aggregation of these membrane proteins at the anterior, conjugation junction. Curiously, general membrane protein synthesis is essential for conA receptor tipping to occur. Pair formation can be blocked by general inhibitors of protein synthesis such as cycloheximide.22 More specifically, tunicamycin, a reagent that blocks protein glycosylation, blocks pair formation as well.45 These studies suggest that though conA receptors may already exist diffusely distributed on the surface membrane prior to costimulation, their redistribution and aggregation at the future conjugation junction may depend upon synthesis of other surface membrane glycoproteins. In 1987, Pagliaro and Wolfe46 demonstrated that binding of conA to membrane receptors causes certain conA binding proteins to become tightly associated with the underlying cytoskeletal frameworks. Specifically a 23 kD and a 25 kD protein become triton-insoluble when cells are first exposed to conA prior to extraction. This becomes intriguing when coupled with the later discovery that two calcium-binding proteins (TCBP23 and TCBP25) sharing the same molecular weights can be isolated from cortical fractions and appear to decorate the conjugation junction (see below).
One can envision a model in which costimulation induces rapid synthesis of a spatially localized “anchor protein” in the newly formed membrane of the conjugation junction. ConA receptors that were diffusing freely in the plasma membrane would then aggregate at this site and help to mediate cell adhesion while triggering cytoskeletal association of cytoplasmic proteins. In short, conA receptor tipping is one of the first biochemical changes associated with pairing, and it appears to be essential for mediating early cell adhesion events that precede construction of the conjugation junction.
Ultrastructure of the Early Junction and Pore Formation
The finest studies documenting the ultrastructure of the early conjugation junction come from two laboratories, that of Jason Wolfe9,47 and Judy and Eduardo Orias.48,49 Normal cortical architecture in Tetrahymena consists of three membranes: a plasma membrane and alveolar sacs which appear as an outer and inner alveolar membrane in cross section.50 Beneath the inner-most membrane (the inner alveolar membrane) is a proteinaceous layer referred to as the epiplasm.51 This layer can be isolated as an intact, cell-ghost structure, and has undergone a great deal of biochemical analysis. 52-58 During tip transformation, the novel membrane that forms the conjugation junction is devoid of cilia, and has none of the typical cortical organelles: basal bodies, mitochondria, mucocysts, and alveolar sacks. The epiplasm layer that lines the cell cortex appears continuous with a denser, tighter layer that lines the membrane of the conjugation junction. This has been referred to as the sub-membrane scaffold (ss),47 and it too can be isolated as an intact structure (Cole, unpublished,Fig. 11).
As cells become firmly attached, their plasma membranes are separated by a 50 nm gap with strands of “matrix material” extending between them47 (Fig. 12). On the cytoplasmic side the sub-membrane scaffold appears as a 50 nm thick layer of electron dense material.47 Transmission electron microscope profiles of the developing conjugation junction reveal that, two hours after mixing, numerous independent membrane fusion events are initiated all across the fusion plate. These result in the formation of hundreds of well-spaced 0.1-0.2 μm diameter pores. Wolfe explains that the scaffolding material becomes thin over regions of future pore formation, the apposed membranes indent towards one another and the space between the membranes fills with some electron dense material.47 In certain sections, it appears that membrane indentation originates in one cell partner, and somehow evokes response in the apposed membrane.
As pore formation proceeds, the 50nm gap collapses bringing the two apposed membranes into intimate proximity just prior to fusion. Details of the actual membrane fusion event are lacking. After membrane fusion results in pore formation, these appear to be occluded by a “wispy filamentous material”.47 In TEM, these occlusions appear as wheels of electron dense material filling the pore and extending out into the cytoplasm (Fig. 8). In freeze fracture, SEM preparations these appear as plugs of material (Fig. 8A). This material, as well as the “extracellular matrix” material and the sub-membrane scaffolding survive detergent extraction suggesting linkage between sub-membrane scaffolding, integral membrane proteins, and extracellular matrix materials within the conjugation junction. One problem that remains to be explained is how a “fusion plate” perforated by hundreds of 0.1-0.2 micron pores permits transfer of a migrating pronucleus that is over 10 times the diameter of any given pore.
Transformation of the Fusion Plate into the Nuclear Exchange Junction
Membrane Events
At the time of pore formation, the junction becomes stabilized and begins to permit cytoplasmic exchange.59 The initial pores show fairly consistent dimensions (0.1-0.2 μm in diameter). From casual examination of a number of TEM images, it appears that more pores form over regions of membrane that are adjacent to a nucleus. Over the next 2 hours, pores broaden (possibly fusing with neighboring pores) creating ever larger apertures separated by ever thinner regions of double membrane reticulum. As the transfer pronucleus associates with the membrane of the conjugation junction, that is where we see the most pronounced broadening and fusing of adjacent membrane pores. The largest pores documented by Wolfe,9 had diameters of nearly 0.45 μm, though more typically 0.32 μm (ten times the area of the smaller, earlier pores). This transformation (depicted in Fig. 16) reaches an extreme when the membrane reticulum separating the apertures is reduced to a network of branching inter-connected tubules with circular cross section of uniform, 90 μm diameter (Fig. 13). This transformation has been beautifully captured by Judy Orias employing both conventional, and high voltage TEM.48,49 As junction membranes are transformed into this reticulum of branching spaghetti, the electron dense scaffold proteins disappear, and the membranes appear naked (Fig. 13B,D). It is likely that the curtain-organization of the membrane at this point represents an energetically favored state, and does not require protein scaffolding to maintain it (E. Orias, personal communication).
Nuclear-Cytoskeletal Events
As events remodel the membranes of the conjugation junction, the micronucleus of each mating partner undergoes a series of three divisions, meiosis I, meiosis II and a third gametogenic nuclear division that is mitotic in nature.60-65 During meiosis, there is some form of nuclear assessment going on.66 When a nucleus is found “defective” by some, as yet ill-understood criterion (aneuploidy, loss of chromosomes, broken chromosomes), meiosis terminates, and all four meiotic products are destroyed. When a nucleus “passes” assessment, one of the four haploid products becomes tethered to the conjugation junction via microtubules67 and/or 14 nm filaments,68,69 where it is shielded from signals that trigger destruction of the other three meiotic products. The surviving, nucleus initiates a third, gametogenic nuclear division. This tethering has been named “nuclear selection”.67 The third nuclear division produces what may be accurately thought of as two gametic pronuclei. The resident pronucleus remains in the cytoplasm of its parent cell, while the transfer pronucleus becomes pressed against the conjugation junction. At this point, both resident and transfer pronuclei of a single mating partner show strands of microtubules between them67 and they (and the conjugation junction) are decorated with the proteins fenestrin, and TCBP-25 (see below). The transfer pronucleus bears a striking cytological asymmetry in the electron microscope. On the cytoplasmic side, the pronucleus has acquired a thatched cap of short microtubules cris-crossing to form a type of net (Fig. 13C). Some microtubules even appear to form “T”-junctions with one another (Fig. 14). On the junction side, the pronucleus is nearly devoid of microtubules, and closely pressed against the, now-perforated plasma membrane. It is widely assumed that some form of microtubule-based motility is responsible for the impending nuclear exchange event. Vinblastine and nocodazole (drugs that destabilize microtubules) are effective at preventing nuclear exchange.70-73 Both fluorescence microscopy using antisera raised against tubulin,67 and electron microscopy49 reveal microtubule involvement. Some type of mechanical pressure appears to be generated in that (A) the transfer pronuclei lose their spherical shape and become flattened or lens-shaped as they press up against the conjugation junction, and (B) the junction itself bows out, deforming as the microtubule driven pronuclei press into it from opposing sides (Fig. 13A). The transfer pronuclei pass, side by side at the same anterior-posterior level within the cell74 migrate towards the partner's resident pronuclei, and undergo karyogamy (pronuclear fusion). After pronuclear transfer, the conjugation junction is severely disrupted, and the cap of short microtubules and fenestrin that had previously decorated the nucleus remains behind as a “patch” in the gap within the conjugation junction (Fig.15). How the two plasma membranes are reestablished prior to pair separation has never been studied, though membrane pores persist long after nuclear exchange. A model depicting membrane changes during nuclear exchange appears in Figure 16.

Figure 14
This TEM image (courtesy of Judy Orias) reveals a remarkable series of microtubule “T”-junctions associated with the pronuclear basket.
The Conjusome: A p-Granule Nuage Assemblage?
In 1999, Janetopolous et al,75 discovered a novel, nonmembrane bound, electron-dense structure located adjacent to the nuclear exchange junction. It appeared late in conjugal development at the time that the developing macronucleus was being remodeled. Antibodies to the chromodomain protein pdd1 (implicated in chromatin remodeling during development)76-78 labeled this structure.75 This structure, the conjusome, begs further inquiry. It resembles p-granules seen in the embryos of Caenorhabditis and Drosophila, or the so-called nuage of higher metazoan embryos. These structures have been implicated in distinguishing germ cells from somatic cell lineages early in the development of numerous metazoan embryos. The molecular machinery that is involved in germ-cell determination of Drosophila has been found in Tetrahymena, where it is involved in the developmental remodeling of the somatic nucleus.79-81 It is tantalizing to contemplate that the conjusome may represent a ciliate, unicellular version of the nuage assemblage that drives this fundamental developmental decision that distinguishes nuclear fates. It is also interesting to note that the conjusome assembles in the anterior cytoplasm that is associated with the conjugation junction, very near where the golgi assembles early in development.
Biochemical Studies
PAGE Analysis
A number of extensive studies have characterized novel protein synthesis in association with initiation, costimulation, and pair formation.43,82-85,86 Despite these efforts, little progress has been made to assign functions to even the most likely candidates. During starvation –induced initiation, it would seem that there are no new proteins synthesized. Three proteins show a significant reduction in abundance, however: a 43kD, 47kD and 56kD protein.85 This has lead to speculation that mating readiness may involve removal of inhibitory proteins rather than the acquisition of pair-stimulating proteins. This view is consistent with data from Paramecium, suggesting that “immaturin” is expressed in cells that are not yet competent to pair, and vanishes from pairs that are fully mature.87,88
During costimulation, numerous authors have seen up-regulation of a protein (or proteins) with the approximate molecular weight of 80kD.43,82,83 Garfinkel and Wolfe noticed that their 80kD protein was present in isolated nuclear preparations, and speculated on its role as a transcription factor. Van Bell used 2-D PAGE to identify two sets of induced proteins; one set that showed induction early in development (15-60 min after mixing), and a second set that was induced later in development (peaking at 4-6 hrs). In both sets the most prominent protein shares a molecular weight of 80kD, and the author speculates that these may represent post-translational modifications of the same molecule. Although this protein has become a familiar marker of mating readiness, no substantive information has been forth-coming regarding its function or identity.
Immunofluorescence
A different quest for proteins of the conjugation junction has involved generation of monoclonal antibodies or polyclonal antisera to novel Tetrahymena proteins. Three potentially interesting proteins have been identified.
Fenestrin
This 64kD protein was first identified from a screening of monoclonal antibodies generated against cortical residues of Tetrahymena pyriformis.74 The antibody 3A7 cross-reacted with proteins from both T. pyriformis and T. thermophila. In vegetative cells, the 3A7 antibody binds to all the “windows” in the cell: the oral apparatus, contractile vacuole pores, the cytoproct and the fission ring. In mating cells this antibody decorates the conjugation junction on the cytoplasmic face of the membrane in much the same pattern as that seen by FITC-conA on the extracellular face of the membrane, though it remains in the central disk and is not excluded to the perimeter as seen with conA labeling. 3A7 also decorates both the migratory and stationary pronuclei, though not before the third gametogenic nuclear division. Fenestrin appears to adhere to the cytoplasmic face of the sub-membrane scaffolding (ss) first recognized by Wolfe.9 It also appears to decorate filaments that extend out from the conjugation junction and appear to anchor the pronuclei. Recently, a group claims to have sequenced the fenestrin gene by mass spectrometry of the protein fragments.89
Calmodulin Homologs
Takemasa et al90,91 cloned the genes for two Tetrahymena calmodulin homologs: TCBP-25 and TCBP-23 respectively. These genes encode EF – hand, Ca2+ binding proteins. Polyclonal antisera were generated directly against these proteins that had been expressed in E. coli.92,93 Both proteins appear to associate with the epiplasm. Furthermore, the antisera to TCBP-25 labels migratory and stationary pronuclei in mating cells, and faintly decorates the exchange junction as well.94 This pattern is quite similar to the fenestrin labeling of the conjugation junction, though other “windows” into the cell were not labeled. A similar protein was found in Paramecium, (PCBP 25α)where it was shown to be a target for phosphorylation.95 These findings are significant in that they may implicate calcium ions in the dynamic processes associated with nuclear exchange during conjugation.
49 kD Filament-Forming Protein
This curious protein was isolated from Tetrahymena thermophila by in vitro assembly/disassembly.96 It appears to form 14 nm filaments in the presence of calcium and ATP. A polyclonal antiserum was generated against it97,98 and its gene was later cloned.99 The sequence revealed it to have homology to citrate synthase and it has been touted as a dual-function protein.100 Immuno-fluorescence revealed that this protein forms fine cables that radiate from the conjugation junction at the time of nuclear selection, and appear to associate with the gametic pronuclei.68,69 This protein is of interest for two reasons. First, this protein may be involved in capturing one of the meiotic nuclei, shielding it from programmed nuclear elimination, and tethering it to the conjugation junction for a subsequent gametogenic mitosis. Second, if its assembly is mediated by calcium, then we may see one reason for calmodulin family proteins to also be assembled at the conjugation junction.
Genetic Studies
Two types of mutant analyses have been conducted in Tetrahymena. First, clonal senescence produces cell lines (referred to as “star” cells) which show varying degrees of micronuclear aneuploidy, and are infertile. Star-cells reveal interesting defects during conjugation. Second, chemical mutagenesis using nitrosoguanidine, has produced a growing gallery of conjugation mutants in a series of mutant screens.64,65
Aneuploid Cells and Their Developmental Phenotype
When an aneuploid, “star” cell partner is mated to a diploid partner, conjugation is aberrant. Pairing occurs, and micronuclei in both partners undergo meiosis. In the diploid partner, one of the meiotic products is “selected”, becomes tethered to the conjugation junction, and undergoes the third gametogenic division. The other three haploid nuclei are destroyed. In the aneuploid, “star” partner, none of the meiotic products is selected, and all four are destroyed. The diploid partner transfers its gametic pronucleus across to the star partner, but receives nothing in exchange. After unilateral nuclear transfer, development arrests, and pairs separate prematurely (9 hours rather than 12 hours at 30 C). The exconjugants are capable of remating immediately, and the second round of pairing proceeds normally establishing whole genome homozygotes. This alternative developmental program has been called “genomic exclusion”.101
Two points are worth emphasizing about genomic exclusion pairs. First, such pairs are mechanically weakened, and easily disrupted.102 One could predict, that unilateral nuclear association would result in the formation of fewer nuclear pores on the unoccupied site, and that this would result in a less robust conjugation junction. This could be tested by performing TEM sectioning on Star X Star matings. Second, it has been shown that star-matings produce a transferable factor that inhibits the developmental program in diploid X diploid matings.66 Furthermore, it has been argued that some form of micronuclear assessment occurs during first meiosis (Cole et al, in preparation). When mating involves an aneuploid “star” cell, micronuclear assessment results in the generation of a dominant, conjugal arrest activity that prevents the pair from entering the postzygtotic developmental program.
bcd Mutants Affect Nuclear Selection and Fusion
Nitrosoguanidine mutagenesis produced bcd, (broadend cortical domains), in a 1987 screen for pattern mutants. These cells exhibit supernumerary cortical organelles: multiple oral apparatuses, extra contractile vacuoles and extra cytoprocts.103 Subsequent studies revealed that they were also sterile, and exhibited an interesting, pleiotropic conjugal phenotype.104 First, in matings between bcd homozygotes, nuclear selection frequently captured two meiotic nuclei rather than the typical one. These each divided and one saw pairs with two pronuclei on each side of the conjugation junction. Second, such pairs arrested shortly after nuclear exchange, and multiple pronuclei aggregated at the conjugation junction without undergoing nuclear fusion. One interpretation, is that the bcd mutant results in a broadening of the nuclear-selection field as well as all the other cortical domains. It has not escaped our attention that every cortical domain affected by the bcd mutation is also a target for fenestrin involvement. It is attractive to consider that the bcd mutation may affect fenestrin distribution, either directly or indirectly.
Pair Separation Failures
A second pattern mutant that exhibits a conjugation phenotype is janus A. This mutant produces a mirror image duplication of its ventral cortical pattern.105 Mating is substantially normal up to 12 hours, when pair separation normally occurs. At this point, pairs remain attached at the conjugation junction.106 This proves fatal, in that pairs are unable to rebuild their oral apparatus and enter the vegetative feeding pathway. Other mutants have been shown to produce the same phenotype: cnj6 and mra.64,107 Actinomycin D or cycloheximide treatments delivered after macronuclear anlagen formation also result in a pair-separation failure.108 This phenotype is difficult to understand without more facts. The janA connection to this (a cortical phenotype derangement) remains mysterious, though potentially, janA pairs produce an expanded region of pair fusion, resulting in two-cell parabiosis.
cnj10 Pleiotropy
This mutant exhibits a conjugal phenotype that is quite intriguing.64f cnj10 X cnj10 matings show three instances where there is a failure of nuclear-cortical interaction. First, nuclear selection frequently fails. Second, even in pairs that complete nuclear selection and generate gametic pronuclei, nuclear transfer fails. Nuclear fusion also fails in most partners. Finally, even though postzygotic development proceeds relatively normally in some pairs, none of the post-zygotic nuclei become anchored at the posterior cortex (the normal fate of two postzygotic nuclear products destined to become micronuclei). Hence, all the nuclei wander freely in the cytoplasm and differentiate into a plethora of macronuclear anlagen. It is compelling to suggest that the gene identified by the cnj10 mutation is necessary for nuclear-cortical interactions and nuclear-nuclear interactions during fertilization.
Epilogue
The Tetrahymena conjugation junction presents a remarkable biological system promising insight into a number of fundamental problems of cell biology and development. It represents a rare case in which one sees specific, developmental induction of a golgi complex. Moreover, it also represents a unique event that somehow coordinates hundreds of independent membrane fusion events within a confined membrane domain. It reveals a potentially novel microtubule-based motility system in the appearance of “T-junctions” within the pronuclear transfer basket. It may participate in the developmental assembly of a p-granule/ nuage-like organelle that directs differentiation of a somatic nucleus from a germ-line nucleus, and finally, it represents a rich, dynamic dialog between nucleus and cell cortex. This dynamic structure will bear watching in the years to come.
Acknowledgements
I wish to thank Joseph Frankel, for leading me into the spectacular world of ciliates, Jason Wolfe for his encouragement in writing this chapter on a subject near and dear to him, and a special thanks to Judy and Ed Orias, whose keen observations and boundless generosity fill these pages. This work was supported by an NSF grant # MCB 0444700 “The Gene Stream”.
References
- 1.
- Sapp J. Beyond the Gene: Cytoplasmic inheritance and the Struggle for Authority in Genetics. New York: Oxford University Press. 1987
- 2.
- Lee JJ, Leedale GF, Bradbury PC. An Illustrated Guide to the Protozoa: Organisms Traditionally Referred to as Protozoa, or Newly Discovered Groups. 2nd ed. Lawrence: Society of Protozoologists. 2000
- 3.
- Frankel J. Pattern Formation: Ciliate Studies and Models. New York: Oxford University Press. 1989
- 4.
- Frankel J. Participation of the undulating membrane in the formation of oral replacement primordia in Tetrahymena pyriformis. J Protozool. 1969;16:26–35. [PubMed: 5806199]
- 5.
- Nelsen EM. Transformation in Tetrahymena pyriformis: Development of an inducible phenotype. Dev Biol. 1978;66:17–31. [PubMed: 109331]
- 6.
- Nelsen EM, DeBault LE. Transformation in Tetrahymena pyriformis—Description of an inducible phenotype. J Protozool. 1978;25:113–119. [PubMed: 96253]
- 7.
- Bell G. Sex and death in protozoa: The history of an obsession. Cambridge: Cambridge University Press. 1988
- 8.
- Wolfe J. Cytoskeletal reorganization and plasma membrane fusion in conjugation Tetrahymena. J Cell Sci. 1985;73:69–85. [PubMed: 3894390]
- 9.
- Wolfe J. The conjugation junction of Tetrahymena: Its structure and development. J Morphol. 1982;172:159–178. [PubMed: 30096979]
- 10.
- Bruns PJ, Brussard TB. Pair formation in Tetrahymena pyriformis, an inducible developmental system. J Exp Zool. 1974;188:337–344. [PubMed: 4209195]
- 11.
- Nanney DL, Caughey PA. Mating type determination in Tetrahymena pyriformis. Proc Natl Acad Sci USA. 1953;39:1057–1053. [PMC free article: PMC1063905] [PubMed: 16589373]
- 12.
- Lynch TJ, Brickner J, Nakano KJ. et al. Genetic map of randomly amplified DNA polymorphisms closely linked to the mating type locus of Tetrahymena thermophila. Genetics. 1995;141:1315–1325. [PMC free article: PMC1206869] [PubMed: 8601476]
- 13.
- Elliot AM, Nanney DL. Conjugation in Tetrahymena. Science. 1952;116:33–34.
- 14.
- Nanney DL, Caughey PA. An unstable nuclear condition in Tetrahymena pyriformis. Genetics. 1955;40:388–398. [PMC free article: PMC1209728] [PubMed: 17247559]
- 15.
- Bruns PJ, Palestine RF. Costimulation in Tetrahymena pyriformis: A developmental interaction between specially prepared cells. Dev Biol. 1975;42:75–83. [PubMed: 803462]
- 16.
- Thompson GA. Studies of membrane formation in Tetrahymena pyriformis. I. Rates of phospholipid biosynthesis. Biochemistry. 1967;6:2015–2022. [PubMed: 6049442]
- 17.
- Dryl S. Antigenic transformation in Paramecium aurelia after homologous antiserum treatment during autogamy and conjugation. J Protozool. 1959;6(Suppl):25.
- 18.
- Bruns PJ, Brussard TB. Positive selection for mating with functional heterokaryons in Tetrahymena pyriformis. Genetics. 1974;81:831–841. [PMC free article: PMC1224557] [PubMed: 4217747]
- 19.
- Bruns PJ, Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Meth Cell Biol. 2000;62:501–512. [PubMed: 10503214]
- 20.
- Wellnitz WR, Bruns PJ. The prepairing events in Tetrahymena thermophila. Analysis of blocks imposed by high concentrations of Tris-HCl. Exp Cell Res. 1979;119:175–180. [PubMed: 761602]
- 21.
- Wellnitz WR, Bruns PJ. The prepairing events in Tetrahymena. II. Partial loss of developmental information upon refeeding starved cells. Exp Cell Res. 1982;137:317–328. [PubMed: 7056293]
- 22.
- Allewell JM, Oles J, Wolfe J. A physicochemical analysis of conjugation in Tetrahymena pyriformis. Exp Cell Res. 1976;97:394–405. [PubMed: 814008]
- 23.
- Finley MJ, Bruns PJ. Costimulation in Tetrahymena. II. A nonspecific response to heterotypic cell-cell interactions. Dev Biol. 1980;79:81–94. [PubMed: 6773840]
- 24.
- Takahashi M. Does fluid have any function for mating in Tetrahymena? Scient Rep Tohoku Univ. 1973;36:223–229.
- 25.
- Adair WS, Barker R, Turner Jr RS. et al. Demonstration of a cell-free factor involved in cell interactions during mating in Tetrahymena. Nature. 1978;274:54–55. [PubMed: 661994]
- 26.
- Wolfe J, Turner R, Barker R. et al. The need for an extracellular component for cell pairing in Tetrahymena. Exp Cell Res. 1979;121:27–30. [PubMed: 446529]
- 27.
- Wolfe J, Meal KJ, Soiffer R. Limiting conditions for conjugation in Tetrahymena: Cellular development and factor active in conjugation. J Exp Zool. 1980;212:37–46.
- 28.
- Fujushima S, Tsuda M, Mikami Y. et al. Costimulation-induced rounding in Tetrahymena thermophila: Early cell-shape transformation induced by sexual cell-to-cell collisions between complementary mating types. Dev Biol. 1993;155:198–205. [PubMed: 8416833]
- 29.
- Allewell NM, Wolfe J. A kinetic analysis of the memory of a developmental interaction: Mating interactions in Tetrahymena pyriformis. Exp Cell Res. 1977;109:15–24. [PubMed: 409609]
- 30.
- Kitamura A, Sugai T, Kitamura Y. Homotypic pair formation during conjugation in Tetrahymena thermophila. J Cell Sci. 1986;82:223–234. [PubMed: 3793779]
- 31.
- Virtue MA, Cole ES. A cytogenetic study of devolopment in mechanically disrupted pairs of Tetrahymena thermophila. J Euk Microbiol. 1999;46:597–605. [PubMed: 10568032]
- 32.
- Kiersnowska M, Kaczanowski A, Morga J. Macronuclear development in conjugants of Tetrahymena thermophila, which were artificially separated at meiotic prophase. J Euk Microbiol. 2000;47:139–147. [PubMed: 10750841]
- 33.
- Wolfe J, Grimes GW. Tip transformation in Tetrahymena: A morphogenetic response to interactions between mating types. J Protozool. 1979;26:82–89.
- 34.
- Kurz S, Tiedtke A. The Golgi apparatus of Tetrahymena thermophila. J Euk Microbiol. 1993;40:10–13. [PubMed: 8457796]
- 35.
- Elliott AM. Biology of Tetrahymena. Dowden Hutchinson and Ross Inc. 1973
- 36.
- Ofer L, Levkovits H, Loyter A. Conjugation in Tetrahymena pyriformis. The effect of polylysine, Con A, bivalent metals on the conjugation process. J Cell Biol. 1976;7:287–293. [PMC free article: PMC2109833] [PubMed: 820698]
- 37.
- Frisch A, Loyter A. Inhibition of conjugation in Tetrahymena pyriformis by Con A. Localization of Con A binding sites. Exp Cell Res. 1977;110:337–346. [PubMed: 412685]
- 38.
- Watanabe S, Toyohara A, Suzaki T. et al. The relation of concanavalin A receptor distribution to the conjugation process in Tetrahymena thermophila. J Protozool. 1981;28:171–175.
- 39.
- Wolfe J, Pagliaro L, Fortune H. Coordination of concanavalin-A-receptor distribution and surface differentiation in Tetrahymena. Differentiation. 1986;31:1–9.
- 40.
- Pagliaro L, Wolfe J. Concanavalin A binding induces association of possible mating-type receptors with the cytoskeleton in Tetrahymena. Exp Cell Res. 1987;168:138–152. [PubMed: 3096751]
- 41.
- Wolfe J, Feng S. Concanavalin A receptor “tipping” in Tetrahymena and its relationship to cell adhesion during conjugation. Development. 1988;102:699–708.
- 42.
- Suganuma Y, Yamamoto H. Conjugation in Tetrahymena: Its relation to concanavalin A receptor distribution on the cell surface. Zool Sci. 1988;5:323–330.
- 43.
- van BellCT. An analysis of protein synthesis, membrane proteins, and concanavalin A-binding proteins during conjugation in Tetrahymena thermophila. Dev Biol. 1983;98:173–181. [PubMed: 6862103]
- 44.
- Driscoll C, Hufnagel LA. Affinity-purification of concanavalin A-binding ciliary glycoconjugates of starved and feeding Tetrahymena thermophila. J Euk Microbiol. 1999;46:142–146. [PubMed: 10361735]
- 45.
- Frisch A, Levkowitz H, Loyter A. Inhibition of conjugation and cell division in Tetrahymena pyriformis by tunicamycin: A possible requirement of glycoprotein synthesis for induction of conjugation. Biochem Biophys Res Commun. 1976;72:138–145. [PubMed: 825115]
- 46.
- Pagliaro L, Wolfe J. Concanavalin-A binding induces association of possible mating type receptors with the cytoskeleton of Tetrahymena. Exp Cell Res. 1987;168:138–152. [PubMed: 3096751]
- 47.
- Wolfe J. Cytoskeletal reorganization and plasma membrane fusion in conjugating Tetrahymena. J Cell Sci. 1985;73:69–85. [PubMed: 3894390]
- 48.
- Orias E. Ciliate conjugation. In: Gall LG, ed. The Molecular Biology of Ciliated Protozoa. Orlando: Academic Press. 1986:45–94.
- 49.
- Orias JD, Hamilton EP, Orias E. A microtubular meshwork associated with gametic pronuclear transfer across a cell-cell junction. Science. 1983;222:181–184. [PubMed: 6623070]
- 50.
- Allen RD. Fine structure of membranous and microfibrillar systems in the cortex of Paramecium caudatum. J Cell Biol. 1971;49:1–20. [PMC free article: PMC2108203] [PubMed: 4102004]
- 51.
- Sibley JT, Hanson ED. Identity and function of a subcortical cytoskeleton in Paramecium. Arch Protistenk. 1974;116:221–235.
- 52.
- Vaudaux P. Isolation and identification of specific cortical proteins in Tetrahymena pyriformis GL. J Protozool. 1976;23:458–464. [PubMed: 823330]
- 53.
- Williams NE, Honts JE, Jaeckel-Williams RF. Regional differentiation of the membrane skeleton in Tetrahymena. J Cell Sci. 1987;87:457–463. [PubMed: 3429495]
- 54.
- Williams NE, Honts JE. The assembly and positioning of cytoskeletal elements in Tetrahymena. Development. 1987;100:23–30. [PubMed: 3308398]
- 55.
- Williams NE, Honts JE. Isolation and fractionation of the Tetrahymena cytoskeleton and oral apparatus. Meth Cell Biol. 1995;47:301–306. [PubMed: 7476503]
- 56.
- Williams NE, Honts JE, Dress VM. et al. Monoclonal antibodies reveal complex structure in the membrane skeleton of Tetrahymena. J Eukar Microbiol. 1995;42:422–427. [PubMed: 7620468]
- 57.
- Honts JE, Williams NE. novel cytoskeletal proteins in the cortex of Tetrahymena. J Eukar Microbiol. 2003;50:9–14. [PubMed: 12674474]
- 58.
- Williams NE. The epiplasm gene EPC1 influences cell shape and cortical pattern in Tetrahymena thermophila. J Eukar Microbiol. 2004;51:201–206. [PubMed: 15134256]
- 59.
- McDonald BB. The exchange of RNA and protein during conjugation in Tetrahymena. J Protozool. 1966;13:277–285. [PubMed: 5953847]
- 60.
- Nanney DL. Nucleocytoplasmic interactions during conjugation in Tetrahymena. Biol Bull. 1953;105:133–148.
- 61.
- Elliot AM, Hayes RE. Mating types in Tetrahymena. Biol Bull. 1953;105:269–284.
- 62.
- Ray CJ. Meiosis and nuclear behavior in Tetrahymena pyriformis. J Protozool. 1956;3:604–610.
- 63.
- Martindale DW, Allis CD, Bruns PJ. Conjugation in Tetrahymena thermophila: A temporal analysis of cytological stages. Exp Cell Res. 1982;140:227–236. [PubMed: 7106201]
- 64.
- Cole ES, Soelter TA. A mutational analysis of conjugation in Tetrahymena themophila 2. Phenotypes affecting middle and late development: Third prezygotic division, pronuclear exchange, pronuclear fusion and postzygotic development. Dev Biol. 1997;189:233–245. [PubMed: 9299116]
- 65.
- Cole ES, Cassidy-Hanley D, Hemish J. et al. A mutational analysis of conjugation in Tetrahymena themophila 1. Phenotypes affecting early development: Meiosis to nuclear selection. Dev Biol. 1997;189:215–232. [PubMed: 9299115]
- 66.
- Cole ES, Virtue MA, Stuart KR. Development in electrofused conjugants of Tetrahymena thermophila. J Eukar Microb. 2001;48:266–279. [PubMed: 11411835]
- 67.
- Gaertig J, Fleury A. Spatiotemporal reorganization of intracytoplasmic microtubules is associated with nuclear selection and differentiation during developmental process in the ciliate Tetrahymena thermophila. Protoplasma. 1992;167:74–87.
- 68.
- Numata O, Sugai T, Watanabe Y. Control of germ cell nuclear behavior at fertilization by Tetrahymena intermediate filament protein. Nature. 1985;314:192–193. [PubMed: 3883198]
- 69.
- Takagi I, Numata O, Watanabe Y. Involvement of 14-nm filament-forming protein and tubulin in gametic pronuclear behavior during conjugation in Tetrahymena. J Eukar Microbiol. 1991;38:345–351.
- 70.
- Hamilton EP. Dissection of fertilization and development in Tetrahymena using anti-microtubule drugs [Ph.D. dissertation] Santa Barbara: Division of Biological Sciences, University of California Santa Barbara. 1984
- 71.
- Hamilton EP, Suhr-Jessen PB. Autoradiographic evidence for self-fertilization in Tetrahymena thermophila. Exp Cell Res. 1980;126:391–396. [PubMed: 7363953]
- 72.
- Hamilton EP, Suhr-Jessen PB, Orias E. Pronuclear fusion failure: An alternate conjugational pathway in Tetrahymena thermophila induced by vinblastine. Genetics. 1988;118:627–636. [PMC free article: PMC1203318] [PubMed: 3366365]
- 73.
- Kaczanowski A, Gaertig J, Kubiak J. Effect of the antitubulin drug nocodazole on meiosis and postmeiotic development in Tetrahymena thermophila. Induction of achiasmatic meiosis. Exp Cell Res. 1985;158:244–256. [PubMed: 3996478]
- 74.
- Nelsen EM, Williams NE, Yi H. et al. “Fenestrin” and conjugation in Tetrahymena thermophila. J Euk Microbiol. 1994;41:483–495. [PubMed: 7804251]
- 75.
- Janetopoulos C, Cole E, Smothers JF. et al. The conjusome: A novel structure in Tetrahymena found only during sexual reorganization. J Cell Sci. 1999;112:1003–1111. [PubMed: 10198282]
- 76.
- Smothers JF, Mizzen CA, Tubbert MM. et al. Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development. 1997;124:4537–4545. [PubMed: 9409671]
- 77.
- Smothers JF, Madireddi MT, Warner FD. et al. Programmed DNA degradation and nucleolar biogenesis occur in distinct organelles during macronuclear development in Tetrahymena. J Euk Microbiol. 1997;44:79–88. [PubMed: 9109258]
- 78.
- Madireddi MT, Coyne RC, Smothers JF. et al. Pdd1p, A novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. [PubMed: 8858150]
- 79.
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. [PMC free article: PMC540227] [PubMed: 15598983]
- 80.
- Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004;18:2068–2073. [PMC free article: PMC515285] [PubMed: 15314029]
- 81.
- Mochizuki K, Fine NA, Fujisawa T. et al. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. [PubMed: 12297043]
- 82.
- Ron A, Suhr-Jessen PB. Protein synthesis patterns in conjugating Tetrahymena thermophila. Exp Cell Res. 1981;133:325–330. [PubMed: 7238604]
- 83.
- Garfinkel MD, Wolfe J. Alterations in gene expression induced by a specific cell interaction during mating in Tetrahymena thermophila. Exp Cell Res. 1981;133:317–324. [PubMed: 7238603]
- 84.
- Suhr-Jessen PB. Stage-specific changes in protein synthesis during conjugation in Tetrahymena thermophila. Exp Cell Res. 1984;151:374–383. [PubMed: 6705833]
- 85.
- van BellCT, Williams NE. Membrane protein differences correlated with the development of mating competence in Tetrahymena thermophila. J Euk Microbiol. 1984;31:112–116. [PubMed: 6737314]
- 86.
- Suhr-Jessen P, Salling L, Larsen HC. Polypeptides during early conjugation in Tetrahymena thermophila. Exp Cell Res. 1986;163:549–557. [PubMed: 3956587]
- 87.
- Haga N. Transformation of sexual maturity to immaturity by microinjecting immature genomic DNA in Paramecium. Zool Sci. 1991;8:11–24.
- 88.
- Miwa I. Destruction of immaturin activity in early mature mutants of Paramecium caudatum. J Cell Sci. 1984;72:111–120. [PubMed: 6533147]
- 89.
- Joachimiak E, Sikora J, Kaczanowska J. et al. Characterization of the fenestrin, a cytoskeletal protein involved in Tetrahymena cell polarity and of its coding gene. J Euk Microbiol. 2005;52:7S.
- 90.
- Takemasa T, Ohnishi K, Kobayashi T. et al. Cloning and sequencing of the gene for Tetrahymena calcium-binding 25-kDa protein (TCBP-25). J Biol Chem. 1989;264:19293–19301. [PubMed: 2808424]
- 91.
- Takemasa T, Takagi T, Kobayashi T. et al. The third calmodulin family protein in Tetrahymena. Cloning of the cDNA for Tetrahymena calcium-binding protein of 23 kDa (TCBP-23). J Biol Chem. 1990;265:2514–2517. [PubMed: 2303413]
- 92.
- Hanyu K, Takemasa T, Numata O. et al. Immunofluorescence localization of a 25-kDa Tetrahymena EF-hand Ca2+-binding protein, TCBP-25, in the cell cortex and possible involvement in conjugation. Exp Cell Res. 1995;219:487–493. [PubMed: 7641801]
- 93.
- Hanyu K, Numata O, Takahashi M. et al. Immunofluorescence localization of a 23-kDa Tetrahymena calcium-binding protein, TCBP-23, in the cell cortex. J Biochem (Tokyo). 1996;119:914–919. [PubMed: 8797091]
- 94.
- Numata O, Hanyu K, Takeda T. et al. Tetrahymena calcium-binding proteins, TCBP-25 and TCBP-23. Meth Cell Biol. 2000;62:455–465. [PubMed: 10503211]
- 95.
- Kim K, Son M, Peterson JB. et al. Ca2+-binding proteins of cilia and infraciliary lattice of Paramecium tetraurelia: Their phosphorylation by purified endogenous Ca(2+)-dependent protein kinases. J Cell Sci. 2002;115:1973–1984. [PubMed: 11956328]
- 96.
- Numata O, Watanabe Y. In vitro assembly and disassembly of 14-nm filament from Tetrahymena pyriformis. The protein component of 14-nm filament is a 49,000-Dalton protein. J Biochem (Tokyo). 1982;91:1563–1573. [PubMed: 6807972]
- 97.
- Numata O, Hirono M, Watanabe Y. Involvement of Tetrahymena intermediate filament protein, a 49K protein, in the oral morphogenesis. Exp Cell Res. 1983;148:207–220. [PubMed: 6414830]
- 98.
- Numata O, Tomiyoshi T, Kurasawa Y. et al. Antibodies against Tetrahymena 14-nm filament-forming protein recognize the replication band in Euplotes. Exp Cell Res. 1991;193:183–189. [PubMed: 1899830]
- 99.
- Numata O, Takemasa T, Takagi I. et al. Tetrahymena 14-NM filament-forming protein has citrate synthase activity. Biochem Biophys Res Commun. 1991;174:1023–1034. [PubMed: 1993043]
- 100.
- Numata O. Multifunctional proteins in Tetrahymena: 14nm filament protein/citrate synthase and translation elongation factor-1 alpha. Int Rev Cytol. 1996;164:1–35. [PubMed: 8575889]
- 101.
- Allen SL, File SK, Koch SL. Genomic exclusion in Tetrahymena. Genetics. 1967;55:823–837. [PMC free article: PMC1211480] [PubMed: 17248381]
- 102.
- Gaertig J, Kaczanowski A. Correlation between the shortened period of cell pairing during genomic exclusion and the block in post-transfer nuclear development in Tetrahymena thermophila. Dev Growth Diff. 1987;29:553–562.
- 103.
- Cole ES, Frankel J, Jenkins LM. bcd: A mutation affecting the width of organelle domains in the cortex of Tetrahymena thermophila. Wilhelm Roux's Arch Dev Biol. 1987;196:421–433. [PubMed: 28305390]
- 104.
- Cole ES. Conjugal blocks in Tetrahymena pattern mutants and their cytoplasmic rescue. I. Broadened cortical domains (bcd). Dev Biol. 1991;148:403–419. [PubMed: 1743392]
- 105.
- Frankel J, Nelsen EM. Positional reorganization in compound janus cells of Tetrahymena thermophila. Development. 1987. pp. 51–68. [PubMed: 3652989]
- 106.
- Cole ES, Frankel J. Conjugal blocks in Tetrahymena pattern mutants and their cytoplasmic rescue. II. Janus A. Dev Biol. 1991;148:420–428. [PubMed: 1743393]
- 107.
- Kaczanowski A. Mutation affecting cell separation and macronuclear resorption during conjugation in Tetrahymena thermophila: Early expression of the zygotic genotype. Dev Genet. 1992;13:58–65. [PubMed: 1395143]
- 108.
- Ward JG, Herrick G. Effects of the transcription inhibitor actinomycin D on postzygotic development of Tetrahymena thermophila conjugants. Dev Biol. 1994;173:174–184. [PubMed: 8575619]
- 109.
- Lansing TJ, Frankel J, Jenkins LM. Oral ultrastructure and oral development in the misaligned undulating membrane mutant of Tetrahymena thermophila. J Protozool. 1985;32:126–139. [PubMed: 3989745]
- The Tetrahymena Conjugation Junction - Madame Curie Bioscience DatabaseThe Tetrahymena Conjugation Junction - Madame Curie Bioscience Database
- Cell-Free Synthesis of Defined Protein Conjugates by Site-directed Cotranslation...Cell-Free Synthesis of Defined Protein Conjugates by Site-directed Cotranslational Labeling - Madame Curie Bioscience Database
- Regulation of Cytoskeletal Dynamics and Cell Morphogenesis by Abl Family Kinases...Regulation of Cytoskeletal Dynamics and Cell Morphogenesis by Abl Family Kinases - Madame Curie Bioscience Database
- Dynamic Interaction between PARP-1, PCNA and p21waf1/cip1 - Madame Curie Bioscie...Dynamic Interaction between PARP-1, PCNA and p21waf1/cip1 - Madame Curie Bioscience Database
- DNA: Alternative Conformations and Biology - Madame Curie Bioscience DatabaseDNA: Alternative Conformations and Biology - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...