NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Conjugative plasmid transfer is the most important mechanism for bacteria to deliver and acquire genetic information to cope with rapidly changing environmental conditions. To transfer genetic information intercellularly mating cell-cell channels between donor and recipient bacteria have to be established. For plasmid transfer in Gram-negative bacteria subassemblies of these mating channels have been discovered, the order in which the transferred DNA contacts the transporter proteins has been determined and crystal structures of key components of the so-called conjugative type IV secretion systems have been solved. In contrast to this, knowledge on conjugative plasmid transfer of sex pheromone-inducible plasmids in Enterococcus faecalis is limited to molecular details on the complex regulation processes whereas for broad-host-range plasmids from Gram-positive bacteria investigations on the structure of the conjugative transfer apparatus and the interplay of the secretion components have recently started. The following chapter has the intention to give an overview of the state of the art on conjugative plasmid transfer in Gram-positive and Gram-negative bacteria.
Introduction
Bacterial conjugation is the most important means of gene delivery enabling adaptation of bacteria to changing environmental conditions including spread of antibiotic resistance genes, thereby generating multiply antibiotic resistant pathogens. Multiply resistant pathogens, such as Pseudomonas aeruginosa, Staphylococcus aureus and Enterococcus faecalis represent a serious threat to antibiotic treatment of hospitalized and immuno-suppressed patients. Therefore, much effort has been and is still made towards elucidating the molecular mechanisms of conjugative plasmid transfer.
Bacterial conjugation systems are specialized types of type IV protein secretion systems (T4SS) dedicated to transport proteins (e.g., virulence factors, toxins) from bacterial pathogens to their mammalian hosts. The conjugative T4SS have evolved to transport DNA substrates in addition to proteins intercellularly.
Relevant progress has been made in deciphering the transport pathway of DNA and protein substrates through the Gram-negative (G-) cell envelope (see refs. 1, 2). Recently the Christie group2 provided evidence for the order of transferred (T)-DNA contact with the T4SS proteins of the prototype T4SS of Agrobacterium tumefaciens during T-DNA export to the plant nucleus. The best characterized T4SS are the Agrobacterium T-DNA transfer system and the conjugative transfer systems of plasmids RP4, F and R388, all originating from G- bacteria. Intense investigations on the transfer mechanisms of plasmids from Gram-positive (G+) bacteria have started only a few years ago. An exception represent the well-studied sex-pheromone responsive plasmids of E. faecalis whose transfer underlies a complex regulatory mechanism exerted by small secreted signal molecules, the so-called pheromones (for recent comprehensive reviews see refs. 3, 4).
The chapter is divided into three parts summarizing the current knowledge of conjugative transfer mechanisms, mating cell-cell channel assembly and structure:
- in G- bacteria;
- of pheromone-responsive conjugative plasmids in E. faecalis; and
- of nonpheromone-responsive plasmids in G+ bacteria.
Conjugative DNA Transfer in Gram-Negative Bacteria
Conjugative DNA transfer systems in G- bacteria, nowadays generally referred to as specialized T4SS, have been extensively studied for more than two decades. T4SS translocate DNA and protein substrates across the bacterial cell envelope. In general, T4SS transport their substrates to recipient cells via direct cell-to-cell contact. But there are also examples of contact-independent protein export and DNA release to and uptake from the extracellular milieu.1,5,6
Considerable progress has been made towards the mechanistic understanding of intercellular DNA transport in the plasmid model systems RP4, R388 and F (for recent reviews see refs. 7-9) and in the T-DNA transport system of A. tumefaciens.1,10 Several models for transenvelope DNA/protein transport have been proposed (for a summary see ref. 11) which match considerably well with the experimental data. Recently the order in which T-DNA contacts T4SS proteins on its way through the A. tumefaciens cell envelope has been determined by Christies´ group.2,12,13 This discovery is a milestone towards the elucidation of the conjugative DNA/protein secretion mechanism.
On account of these very interesting results the state of the art of T4SS in G- bacteria will be presented on basis of the prototype T4SS, the A. tumefaciens T-DNA system (VirB/VirD4 transfer system).
The T4SS Operon Structures
The A. tumefaciens VirB/D4 T4SS is encoded by the virB and virD operons.13 The virB operon codes for 11 genes, virB1 to virB11. The VirB proteins, termed the mating pair formation (mpf) proteins, build a cell envelope-spanning structure required for substrate transfer, and an extracellular filament, the T pilus that mediates attachment to recipient cells.15 The virD operon encodes five genes, virD1 to virD5. virD1 and virD2 encode gene products processing the DNA substrate (T-DNA) for transfer. These are named the DNA transfer and replication (Dtr) proteins.16 virD3 and virD5 encode proteins that are not essential for processing or transfer. virD4 codes for the coupling protein (CP).17,18 The VirD4 CP is not involved in T-DNA processing or formation of the T-pilus but delivers together with the mpf structure substrates across the cell envelope.14,15
T4SS Substrates
The T-Strand-Relaxase Complex
The key enzyme of conjugative plasmid transfer is the DNA relaxase, a transesterase which cleaves a specific phosphodiester bond in the origin of transfer (oriT) thereby initiating the conjugative transfer.The relaxase preserves its energy from cleavage of the phosphodiester backbone of theT-strand as a stable phosphotyrosyl intermediate with the T-strand.19 The complex of processing proteins at oriT (relaxase and accessory proteins) is termed the relaxosome. Conjugative DNA transfer proceeds in a 5´-3´ direction16 suggesting that the relaxase, covalently bound to the 5´ end of the T-strand, supplies substrate recognition signals and possibly also exerts a piloting function to direct DNA transport through the secretion channel.21,21
Protein Substrates
Conjugation systems also export proteins independently of DNA.22-28 The A. tumefaciens T-DNA transfer system translocates the VirE2 protein in a chaperone-independent and the VirE3 and VirF protein in a chaperone-assisted way into the recipient cell. Indirect evidence was also obtained for translocation of the relaxase-T-DNA complex to plant cells.20 T4SS substrates contain potential secretion signals at their C-termini. The C-termini of A. tumefaciens VirB/D4 T4SS substrates carry a conserved Arg-X-Arg motif, whereas many T4SS substrates—including relaxases from various conjugation systems—carry many positively charged residues, mostly Arg within the last 30-50 residues.10 Recently it was shown that the C-Terminus mediates an interaction between the VirE2 secretion substrate and the VirD4 CP of the VirB/D4 T4SS.29 It can be suggested that the charged C-termini of T4SS substrates probably contribute to substrate recognition by mediating productive contacts with the CP of the T4SS.10
Initiation of T-Strand Transfer: Substrate Recruitment by the Coupling Protein
It is well established that a givenT4SS, e.g., encoded by plasmids R388 or RP4 or the A. tumefaciens VirB/D4 system, translocates a restricted set of substrates including the cognate plasmid or oncogenic T-DNA, one or a few mobilizable plasmids such as ColE1 or RSF1010, and one or a few proteins.10,16
The selectivity of the T4SS is exerted by the respective CPs.18,30-32 Many experimental data on chimeric T4SS (composed of a CP from one T4SS and a mpf structure from a second T4SS) suggest that the CP links the Dtr processing proteins bound at oriT- the relaxosome- to the T4SS, hence the origin of the term “coupling protein”.10 In vitro and in vivo studies have demonstrated several CP-relaxase interactions.33,34
Upon recruitment of the relaxosome, how does the CP mediate the next step of transfer? Structural studies have begun to shed light on the answer to this question. Topology studies have shown that CPs consist of an N-proximal region that includes two transmembrane helices and a small periplasmic domain, and a large C-terminal region that resides in the cytoplasm.35, 36 The crystal structure has been solved for the soluble domain (TrwBΔN70) of the TrwB CP of the IncW plasmid R388. The TrwBΔN70 crystal consists of six equivalent protomers that form a spherical particle of overall dimensions of 110 Å in diameter and 90 Å in height. This ring-like structure possesses a central channel of 20 Å in diameter, which traverses the structure, possibly connecting cytoplasm with periplasm.37,38 Based upon the collected data on CP interactions with mpf subunits the role of the CP can be summarized as follows:
The CP delivers DNA substrates to the secretion apparatus through contacts with the DNA-processing proteins. Then, through contact with VirB10, a putative structural scaffold protein for assembly of the transenvelope T4SS, the CP coordinates passage of the T-strand through the mpf channel. Due to the known structure of the CP one possibility how this translocation occurs would be that the CP acts as a translocase to transport the substrate across the inner membrane.10
A Transenvelope Secretion Channel: The mpf Structure
All of the VirB proteins except for the VirB1 lytic transglycosylase are required for substrate export.39 Thus far, no supramolecular organelles at cellular junctions of mating cells have been identified by high-resolution electron microscopy. However, several experimental findings support the existence of an envelope-spanning secretion apparatus. For example, the presumptive VirB/ VirD4 mating channel or subcomplexes thereof have been isolated by membrane solubilization with nonionic detergents. At least two large complexes were detected, one composed of the T-pilus associated proteins VirB2, VirB5, and VirB7, and the second consisting of several other VirB proteins and the VirD4 CP.40 Independent studies have also reported the isolation of a VirB2/VirB5/VirB7 complex and subcomplexes of other VirB proteins, such as VirB7/VirB9, VirB6/VirB7/VirB9 and VirB7/VirB9/VirB10 by detergent solubilization and immunoprecipitation or GST-pull-down as-says (summarized in ref. 10).These results were confirmed by two-hybrid screens and further pairwise interactions were detected in vivo.41,42
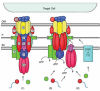
Taken the above mentioned results together with computer-based predictions and topology studies of individual VirB subunits, a general architecture for the VirB/VirD4 T4SS can be presented: The VirD4 CP and the two mpf ATPases, VirB4 and VirB11, are localized predominantly or exclusively at the cytoplasmic face of the inner membrane. VirB6 is a highly hydrophobic protein predicted to span the inner membrane several times. VirB8 and VirB10 are bitopic proteins with short N-terminal cytoplasmic domains, a transmembrane helix and large C-terminal periplasmic domains. VirB2 (the major pilin protein), VirB3 and VirB5 are located in the periplasm,VirB2, VirB5, and VirB7 assemble as the extracellular T pilus. VirB7 is a small lipoprotein, which forms together with VirB9 a covalently cross-linked dimer. This dimer or a higher-order VirB7-VirB9 multimer assemble at the outer membrane. VirB9 has nine possible β-sheet outer membrane-spanning segments according to the Schirmer-Cowan algorithm.43 Therefore, it is the best candidate for forming an oligomeric secretion-like pore mediating secretion of substrates and/or protrusion of the T pilus across the outer membrane.10 A model for the general architecture of the T4SS is shown in Figure 1.
Energy Supply-VirB4 and VirB11
VirB11 belongs to a large family of ATPases associated with macromolecule secretion systems.44
Homologues are widely distributed among the G- bacteria, and they are also functional in secretion systems of G+ bacteria and in Archaea.10 VirB11 is highly insoluble and has been difficult to analyze biochemically. But soluble forms of VirB11 homologues have been characterized enzymatically and structurally. The VirB11-homologue of the Helicobacter pylori pathogenicity island, HP0525Cag and several plasmid-encoded VirB11-homologues were shown to assemble as homohexameric rings as demonstrated by electron microscopy.45 HP0525Cag also presented as a homohexamer by X-ray crystallography. It is a double-stacked ring with a central cavity of about 50 Å in diameter.46 The overall HP0525Cag structure appears to be highly conserved, even among distantly related NTPases encoded by other transport or fimbrial biogenesis systems.10 HP0525Cag is also structurally similar to members of the AAA ATPase superfamily.47 Many AAA ATPases act as energy-dependent unfoldases in substrate remodelling. Considering that the T4SS are export systems, the VirB11-like ATPases might act as chaperones to unfold the protein substrates at the channel entrance.10 VirB11-like ATPases associate tightly but peripherally at the inner face af the inner membrane.48,49 Genetic studies showed the coordinated action of VirB11 and the VirD4 CP for substrate transfer.10 Mutagenesis analyses of VirB11 proved that it participates both in pilus biogenesis and assembly or function of the secretion machine.50 It is likely that VirD4 and VirB11 hexamers localize next to each other at the inner membrane as depicted in Figure 1. Llosa et al51 described a model that presents conjugation systems as two separately acting inner membrane translocases: the CP functions as a general recruitment factor for all T4SS substrates which is in agreement with all other prominent T4SS models, as shown in Figure 2. Although it is tempting to assume that the CP delivers the whole substrate to the mpf complex, they propose that the CP translocates the T-strand across the inner membrane while simultaneously transferring the relaxase to VirB11 and the other mpf proteins for secretion. The model of Llosa and coworkers explains very well most experimental data but it still remains difficult to envision how two transport proteins localized next to each other at the inner membrane coordinate their activities to mediate secretion of one substrate, the T-DNA-relaxase complex across the inner membrane.10
VirB4 possesses two putative membrane-spanning domains, one close to the N terminus and another located more centrally near the Walker A nucleoside triphosphate binding motif.48 VirB4 self-interacts as shown by Dang et al52 and Ward et al.42 ATP hydrolysis has not been convincingly shown for VirB4-like proteins, but these proteins require intact Walker A motifs to mediate substrate export.53,54 Bohne et al55 reported that the presence of a subset of VirB proteins including VirB4 in Agrobacterium recipient cells increases the efficiency of plasmid uptake in intraspecies matings significantly. These data led to the proposal that these VirB proteins assemble as a complex that stabilizes mating junctions or perhaps they directly facilitate DNA transport across the recipient cell envelope.56 However, it was also demonstrated that a Walker A mutation does not decrease the capacity of VirB4 to stimulate plasmid DNA acquisition by recipient cells. Dang et al52 argued that VirB4 probably contributes structural information required for substrate transfer in either direction across the cell envelope. But an intact ATP-binding motif is necessary for configuring this T4SS specifically for substrate export. Christie concludes that the VirD4 CP, VirB11, and VirB4 must interact in complex and dynamic ways—probably through ATP-powered conformational changes— to energize substrate transfer to and across the inner membrane (ref. 10 and Fig. 1).
Working Models for T4SS
Three different working models describing the possible T4SS architecture and translocation routes are discussed at present (Fig. 2). The first model proposed by Christie14 suggests that the mpf proteins assemble as a transenvelope channel for substrate export in one step. This model predicts that the T4CP recruits DNA and protein substrates to the translocation apparatus and then coordinates its activity with a VirB11-type ATPase to drive substrate transfer through the mpf channel.11
The second model is a generalized version of the two-step routing pathway described for the export of the Bordetella pertussis toxin.57 In the first step, an inner membrane translocase delivers substrates across the inner membrane. In the second step, the T4SS translocase, composed of mpf proteins, transports substrates across the outer membrane.56 This model predicts that the CP, when present, acts as an inner membrane translocase for both DNA and protein substrates. This activity is exerted completely independent of the mpf proteins.11 An alternative two-step model, termed the “shoot and pump” model also suggests two inner membrane transporters. However, in this model the T4CP acts as a DNA translocase, whereas the mpf complex translocates protein substrates. In the periplasm, both pathways converge for mpf-dependent transport across the outer membrane.51 The “shoot and pump” model does not exclude the proposed function of the T4CP as a general recruitment factor. But it postulates that upon recruitment the T4CP translocates DNA and delivers the protein substrate to the mpf channel. The “shoot and pump” model is especially attractive: first, because it accommodates most experimental findings to date and second, because it nicely explains why T4SS evolved to be so highly flexible.11,51
Pheromone-Responsive Conjugative Plasmids in E. faecalis
Though the enterococcal pheromone-inducible conjugative plasmids such as pCF10, pAD1, and pPD1 represent a unique class of mobile genetic elements spreading virulence traits readily with high transfer efficiency even in aqueous systems, the mechanism of their conjugative DNA secretion system has not been studied intensively. However, the complex regulatory mechanisms underlying specific efficient plasmid exchange has been investigated in great detail. The hereafter presented state of the art will focus on the regulatory machinery that interacts specifically with the pheromone peptides thereby controlling plasmid acquisition of plasmid-free enterococcal recipient cells. The data were derived mainly from a comprehensive review on enterococcal peptide sex pheromones by Chandler and Dunny.4
The pheromone plasmids are induced to transfer from donor cells by mating pheromones that are produced by potential recipient cells. The pheromone plasmids have evolved a fascinating and complex regulatory system to ensure their maintenance and stable existence in a population. Their transfer genes are induced by small (7-8 amino acids (aa)) peptides chromosomally encoded by all known enterococcal strains. Each peptide is highly specific for a cognate plasmid or for a family of closely related plasmids. All pheromones analyzed so far are produced by proteolytic processing of the cleaved signal sequences of secreted lipoproteins. The processed peptides are secreted into the growth medium and are utilized by the plasmid-containing donor cells to sense the presence of a nearby recipient cell. The pheromone-responsive plasmids use an interesting combination of host and plasmid encoded proteins to sense exogenous pheromone in order to activate the expression of transfer genes and to avoid self-induction by pheromone that is encoded on the chromosome of the host cell.4 An overview of the main steps involved in pheromone-induced conjugation in enterococci will be given based upon data obtained from plasmid pCF10, the model plasmid of the Dunny group. pCF10-transfer is induced when the recipient-produced pheromone is detected by the donor ell at the cell surface by the plasmid-encoded lipoprotein PrgZ.58 PrgZ acts together with the chromosomally encoded oligopeptide permease system (Opp) to import the pheromone into the cytoplasm of the recipient cell.59 Import of the pheromone is necessary for pheromone response. The interaction process that likely initiates the induction in donor cells is binding of the imported pheromone to the plasmid-encoded cytoplasmic protein PrgX. PrgX is a negative regulator of expression of conjugative functions. Binding of pheromone to PrgX abolishes its repression so that transfer genes are synthesized. Two additional pCF10-encoded proteins, PrgY and iCF10, are required to keep the transfer system off in pCF10-harbouring cells grown in the absence of exogenous pheromone. PrgY and iCF10 are supposed to block self-induction of donor cells by endogenous pheromone.
Exposure of pCF10-containing cells to exogenous pheromone, cCF10, is phenotypically visible by aggregation of the culture resulting from upregulation of the expression of aggregation substance PrgB from the pCF10-encoded prgB gene.60 The aggregation results in close contact between donor- and recipient cells probably enabling effective plasmid transfer, even in liquid medium. Approximately 15 additional genes are encoded 3´ from prgB. Data from the Dunny group suggest that many of these genes are upregulated by pheromone.4
The PrgZ pheromone binding protein is critical in the first step of pheromone induction: recognition of pheromone and import into the cytoplasm. All pheromone-responsive conjugative plasmids encode a PrgZ-like protein. They are homologues of the peptide-binding OppA proteins found in a wide range of bacterial species.61 The PrgZ-type proteins are cell surface proteins anchored to a lipid moiety on the outer surface of the cytoplasmic membrane.58 The PrgZ family of pheromone-binding proteins have been shown to increase the sensitivity of each plasmid system to its cognate pheromone.58,62
The pheromones themselves are encoded within the chromosome of E. faecalis, processed by host proteins and secreted inlextremely small amounts into the culture medium. In the case of pCF10, cCF10 is released at ˜10-11 M and can induce a donor cell at concentrations of 2 × 10-12 M corresponding to less than five molecules per cell under the conditions tested.63 Despite multiple pheromones with different spectra of activities secreted by a single cell, each plasmid responds specifically to its cognate pheromone with surprising sensitivity.64,65 The nucleotide sequence of the E. faecalis strain V58366 showed that the pheromone precursors lie within the N-terminal signal sequences of predicted surface lipoproteins.67 Proteolytic processing of the pheromone precursor is required prior to release of the mature pheromone into the culture medium. Signal peptidase II cleaves at a specific cysteine residue liberating the signal peptide from the lipoprotein.68
How does the plasmid prevent a response to its own host´s endogenously produced pheromone? The pheromone plasmids have evolved two independent mechanisms of avoiding this self-induction such that the transfer response is only induced if a nearby recipient is detected. One mechanism is exerted by the synthesis of a plasmid encoded inhibitor peptide, iCF10 in the case of pCF10, which neutralizes endogenously produced pheromone in the culture medium.4 The inhibitor peptides are proposed to compete with the pheromone for binding to the surface binding protein PrgZ.69,70 The level of iCF10 in donor cultures has been found to be 10-100-fold above the pheromone level. This molar ratio is just sufficient to neutralize the cCF10 activity released by the same cells. The inhibitor peptides are very similar to each other and to the inducing pheromones. They are 7-8 aa hydrophobic peptides likely being processed from 22 to 23 aa precursors. Despite the apparent homology between the inhibitors and their peptides, their functions are very specific. Data on the pPD1 and the pCF10 plasmid imply that the function of the inhibitor may include more than just competitive inhibition of PrgZ and may instead involve specificity at some other level, since the response of the systems to their cognate pheromones and inhibitors is highly specific.64,65 This specificity determinant/mechanism remains to be unravelled.
PrgY is the other pCF10-encoded element involved in control of endogenous pheromone. While the inhibitors control endogenous pheromone in the culture medium, the membrane protein PrgY controls endogenous pheromone activity that remains associated with the cell.4 Buttaro et al71 showed that a significant amount of pheromone in cCF10 producing cells remains associated to the cell wall. The average plasmid-free recipient cell appears to have twice as much cCF10 in its cell wall than the amount that is secreted into the supernatant. When pCF10 is acquired, the concentration of cCF10 in the supernatant is not affected70 whereas the cell wall-associated cCF10 is decreased
8-fold from that of plasmid-free recipient cells.71 PrgY was shown to be involved in this reduction of envelope-associated cCF10 after acquisition of the plasmid, but its mechanism of action is not clear. Other PrgY-like proteins have been identified in recent genome sequencing projects. Curiously, the bacterial species that have been found to encode a PrgY-type protein, do not have characterized peptide-signal systems and are quite distantly related to E. faecalis. This finding further deepens the mystery surrounding the role of this protein in the regulation of the pheromone induction process.4
So far, no data are available on the DNA transport mechanism of pCF10 and the other pheromone-inducible enterococcal plasmids. However, it seems reasonable to argue that the DNA secretion process of these plasmids might proceed via a T4S-like mechanism as proved for all known G-systems and currently being studied for the broad-host-range conjugative plasmids from G+ bacteria (see below): The following findings are in favour of a T4S-like mechanism: On two pheromone plasmids, namely pAD1 and pAM373, oriTs have been found. pAD1 has two oriTs, oriT1 and oriT2 and encodes a relaxase, TraX, which has been demonstrated to specifically nick in oriT2. oriTpAM373 has been shown to be similar to oriT2pAD1. Both plasmids are able to mobilize the non conjugative plasmid pAMα1, which encodes two relaxases that are involved in transfer.72 ORF53 encoded by pAD1 is a protein essential for conjugation, which exhibits structural similarities to TraG-like CPs.73 Recently nucleotide sequencing of the 67,673-bp pheromone plasmid pCF10 has been completed. pCF10 contains 57 orfs, orf35 encodes a relaxase, pcfG74 with highest homology to LtrB, the relaxase of the Lactococcus lactis conjugative plasmid pRS01.75
Nonpheromone-Responsive Plasmids in G+ Bacteria
Enterococci harbour also a pheromone-independent conjugative plasmid, namely the 65.1-kb pMG1, that transfers efficiently in broth matings. Interestingly, Southern hybridization of pMG1 DNA showed no homology to pheromone-responsive plasmids and the broad-host-range conjugative plasmids pAMß1 and pIP501.76
Aggregation-mediated plasmid transfer in Bacillus thuringiensis and in lactic acid bacteria has been summarized in Grohmann et al77 and will not be discussed further here. Recently Belhocine et al78 demonstrated that conjugation is one of the mechanisms by which group II introns, originally discovered on a L. lactis conjugative plasmid (pRS01) and within a chromosomally located sex factor in L. lactis 712, are broadly disseminated between widely diverged G+ organisms.
Conjugative Transfer of Broad-Host-Range Plasmids
Transfer of broad-host-range G+ plasmids occurs at a variable frequency (generally in the range of 10-3 to 10-6) depending on the plasmids and the mating-pair genotype, and mating requires cocultivation of donor and recipient cells on a solid surface.77 Most conjugative plasmids identified so far in streptococci and enterococci actually show a broad host range (and hence are referred to as broad-host-range plasmids,79,80 while those found in staphylococci seem to be limited to the genus Staphylococcus.
The complete nucleotide sequences of the staphylococcal plasmid pSK41,81,82 the lactococcal plasmid pMRC01,83 the enterococcal plasmid pRE25, the streptococcal plasmid pIP50184,85 and the complete tra region of the staphylococcal plasmid pGO186 have been determined. Sequence comparisons revealed interesting similarities of the tra regions of these self-transmissible plasmids.77
All of the tra regions show a highly modular organization so that the arrangement of the first seven genes is well conserved among the compared tra regions, with the exception of an insertion of two genes of unknown function between the putative relaxase gene traA and gene traB in pMRC01. The pMRC01 tra region is the most distantly related and contains seven unique genes. Interestingly, a traG gene homologue coding for a putative lytic transglycosylase is present in all plasmids except for pMRC01, while traK homologues coding for a putative CP are present in all five plasmids (Fig. 3).
Information about the regulatory processes involved in gene transfer of nonpheromone-responsive plasmids in G+ bacteria is scarce: TrsN, a 7.2-kDa protein encoded by pGO1, was shown to repress the synthesis of essential tra genes by binding to promoter-like sequences upstream of trsA, the first gene of the conjugative gene cluster trs.87
Tanimoto and Ike88 detected a gene, traA, in the unrelated plasmid pMG1, which is upregulated during conjugation. They found that the traA gene product is associated with the formation or stabilization of mating aggregates during broth mating.
Regulation of the pIP501 tra Region
The operon organization of the pIP501 tra region was elucidated recently by reverse transcription PCR of mRNA isolated from E. faecalis harbouring pIP501. All 15 pIP501 tra genes, orf1-orf15, are transcribed as a single operon of 15.1 kb (Kurenbach and Grohmann, unpublished data). The compact organization of the pIP501 tra region makes autoregulation of the tra operon by the TraA protein likely. The -10 region of the operon promoter Ptra overlaps an inverted repeat structure, proposed to represent the binding site for the TraA relaxase.89 TraA-binding to Ptra has been roved by gel retardation and DNase I footprinting assays. Ptra-lacZ fusions showed strongly reduced promoter activity when TraA was supplied in trans (Kurenbach and Grohmann, unpublished data). Environmental stimuli for pIP501 tra gene expression have not been detected so far. The tra genes are expressed throughout the life cycle of E. faecalis and the expression level is independent of the growth phase (Kurenbach and Grohmann, unpublished data). Thus, we conclude that the pIP501 tra operon is negatively autoregulated at the transcriptional level by the conjugative DNA relaxase TraA.
Homologies to T4SS Components
Macromolecular transfer systems ancestrally related to the conjugative mpf complexes are called T4SS, as originally proposed by Salmond.90 T4SS include conjugative transfer apparatus, protein secretion systems of G-pathogens, and natural transformation systems. T4SS are widely distributed among the G- bacterial world (for recent reviews see refs. 1, 10, 91). We have also found T4 homologues on conjugative elements of G+ bacteria.77 Exemplarily, the pIP501-encoded T4 homologues will be discussed here. pIP501 encodes one T4 homologue of each of the protein families involved in T-DNA transfer and in G- bacterial plasmid transfer.92
Energy Supply
ORF5 (pIP501) encodes a putative VirB4-like ATPase, which could deliver energy for DNA/ protein transport by hydrolysis of ATP. ORF5 shows a score of 71.2 and E value of 3 × 10-13 as a member of the VirB4 family of intracellular trafficking and secretion proteins (COG3451). ORF5 (653 aa) is the largest protein encoded by the pIP501 tra region, consistent with the fact that VirB4-homologues of G- bacteria also represent the largest gene product (e.g., VirB4 of the Agrobacterium Ti plasmid: 789 aa) of the respective tra region. VirB4-type proteins are ubiquitous among the T4SS and are sometimes present in two or more copies. Experimental evidence for a structural contribution of VirB4 to mpf channel formation that is independent of the VirB4 ATPase activity has been provided (for a review see ref. 10).
Mating-Channel Proteins
ORF7 (pIP501) is weakly similar to the family of lytic transglycosylases (pfam01464, score, 36.1; E value 0.007) encoded by bacteriophages and T3 and T4SS. ORF7 contains the soluble bacterial lytic transglycosylase (SLT) domain at its N-terminus. This domain is present in SLTs and in “goose-type” lysozymes (GEWL). It catalyzes the cleavage of the β-1,4-glycosidic bond between acetylmuramic acid and N-acetylglucosamine. At the C-terminus ORF7 possesses high similarity with the COG3942 family of surface antigens (score, 108; E value, 1e–24) and with the pfam05257 family (score 96.6; E value, 4e–21) consisting of amidases involved in the cell wall metabolism of bacteria. The pfam05257 family is also known as CHAP (cystein, histidin-dependent amidase/peptidase) domain family.93,94 The CHAP domain is often found in association with further protein domains involved in cleavage of peptidoglycan.93 Proteins containing a N-terminal lytic transglycosylase domain and a C-terminal CHAP domain or vice versa have been characterized. ORF7 also appears to exhibit such a modular structure: a SLT domain at the amino-terminus, between aa 60 and 165, and a CHAP domain between aa 255 and 369. A transmembrane helix has been predicted for the N-terminal moiety of ORF7, approximately between aa 19-35 by several computer programs (HMMTOP, PHDhtm, PROSITE, PSORT etc.). The putative membrane localization s consistent with its proposed role in locally opening the peptidoglycan to facilitate conjugative DNA/protein transport. Interestingly, ORF7 also contains a possible processing site similar to that found in the lytic transglycosylase VirB1. It could be processed to a ORF7* protein (consisting of 210 aa of total 369 aa).
Coupling Proteins
ORF10 (pIP501) belongs to the pfam02534 TraG/TraD family of CPs (score, 291; E value, 1 e-79). These proteins contain a P-loop and a Walker B site for nucleotide binding. Putative homologues of CPs have been detected on the chromosomes of many sequenced G+ bacteria (e.g., E. faecalisV583) as well as on transposons harboured by them.
Functional Characterization and Protein-Protein Interactions of pIP501-Encoded T4-Homologues
The pIP501-encoded T4 homologues have been expressed with N-terminal tags in E. coli and partially purified. ORF7 protein showed hydrolysis activity on peptidoglycan isolated from E. faecalis by zymogram analysis (Arends and Grohmann, unpublished data). ATP-binding and ATP-hydrolysis assays for the ORF5 and ORF10 protein are in progress.
The following interactions between putative T4 components have been detected by the yeast two-hybrid assay so far (Abajy and Grohmann, unpublished data). The putative ATPase ORF5 interacts with itself which is consistent with dimerization/oligomerization of the VirB4 protein. ORF5 also binds to ORF7 in the in vivo yeast system. This finding is in agreement with detected VirB4-VirB1 interaction.42 ORF7 interacts with itself, with ORF5 and with the putative CP ORF10. ORF7 self-association is consistent with VirB1-dimerization shown by yeast two-hybrid assay.42
Interaction of ORF7 with ORF5 and ORF10 might possibly help incorporate these components in the T4SS structure. ORF10 has been shown to self-associate in vitro (Chmielinska and Grohmann, unpublished data). Further interactions have been detected between T4 homologues and other proteins encoded by the pIP501 tra region (Abajy and Grohmann, unpublished data).
Working Model of a T4S-Like Conjugative Mechanism in G+ Bacteria
Based upon our protein-protein interaction data, the predicted localization and function of the (T4SS-like) proteins and preliminary functional characterization of ORF7 we developed a working model for a simplified T4S-like mechanism in G+ bacteria: We suggest that the putative lytic transglycosylase ORF7 should be more important than the VirB1-homologues in G- bacteria, because of the multilayered peptidoglycan in G+ bacteria in contrast to the thinner monolayer of peptidoglycan in G- bacteria: We propose that ORF7 locally opens the peptidoglycan thereby facilitating the establishment of the transenvelope secretion channel. By interaction with ORF5 and ORF10 ORF7 might recruit these proteins and enable their incorporation in the transport apparatus. The putative ATPase ORF5 with high homology to the COG3451 family of intracellular trafficking and secretion proteins could deliver energy for the DNA/protein secretion process by hydrolysis of NTP. The putative CP ORF10 which interacts with the pIP501-encoded relaxase TraA could link the relaxosome consisting of TraA bound at oriTpIP501, with the transport apparatus. The transport channel presumably consists of ORF5 and non T4SS-like proteins with proposed transmembrane helices such as ORF8, ORF12 and ORF15. Which of these proteins actually act(s) as transporter is currently under investigation.
References
- 1.
- Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;2:137–149. [PMC free article: PMC3873781] [PubMed: 15035043]
- 2.
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. [PMC free article: PMC3882297] [PubMed: 15155952]
- 3.
- Clewell DB, Dunny GM. Conjugation and genetic exchange in enterococci. In: Gilmore MS, ed. The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. 1st ed. Washington: ASM press. 2002:265–300.
- 4.
- Chandler JR, Dunny GM. Enterococcal peptide sex pheromones: Synthesis and control of biological activity. Peptides. 2004;25:1377–1388. [PubMed: 15374642]
- 5.
- Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–277. [PubMed: 11454218]
- 6.
- Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. [PubMed: 11489125]
- 7.
- Schroeder G, Savvides SN, Waksman G. et al. Type IV secretion machinery. In: Waksman G, Caparon M, Hultgren S, eds. Structural Biology of Bacterial Pathogenesis. 1st ed. Washington: ASM press. 2005:179–221.
- 8.
- Llosa M, O'Callaghan D. Euroconference on the biology of type IV secretion processes: Bacterial gates into the outer world. Mol Microbiol. 2004;53:1–8. [PubMed: 15225298]
- 9.
- Lawley TD, Klimke WA, Gubbins MJ. et al. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. [PubMed: 12855161]
- 10.
- Christie PJ. Type IV secretion: The Agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta. 2004;1694:219–234. [PMC free article: PMC4845649] [PubMed: 15546668]
- 11.
- Ding Z, Atmakuri K, Christie PJ. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 2003;11:527–535. [PMC free article: PMC4844353] [PubMed: 14607070]
- 12.
- Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–1211. [PMC free article: PMC3869561] [PubMed: 15554962]
- 13.
- Jakubowski SJ, Krishnamoorthy V, Cascales E. et al. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol. 2004;341:961–977. [PMC free article: PMC3918220] [PubMed: 15328612]
- 14.
- Christie PJ. The Agrobacterium T-complex transport apparatus: A paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. [PMC free article: PMC179082] [PubMed: 9150199]
- 15.
- Lai EM, Chesnokova O, Banta LM. et al. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J Bacteriol. 2000;182:3705–3716. [PMC free article: PMC94541] [PubMed: 10850985]
- 16.
- Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996:197–251. [PubMed: 8768076]
- 17.
- Zhu J, Oger PM, Schrammeijer B. et al. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. [PMC free article: PMC94570] [PubMed: 10869063]
- 18.
- Hamilton CM, Lee P, Li PL. et al. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol. 2000;182:1541–1548. [PMC free article: PMC94450] [PubMed: 10692358]
- 19.
- Byrd DR, Matson MW. Nicking by transesterification: The reaction catalysed by a relaxase. Mol Microbiol. 1997;25:1011–1022. [PubMed: 9350859]
- 20.
- Howard EA, Zupan JR, Citovsky V. et al. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: Implications for nuclear uptake of DNA in plant cells. Cell. 1992;68:109–118. [PubMed: 1732061]
- 21.
- Bravo-Angel AM, Gloeckler V, Hohn B. et al. Bacterial conjugation protein MobA mediates integration of complex DNA structures into plant cells. J Bacteriol. 1999;181:5758–5765. [PMC free article: PMC94097] [PubMed: 10482518]
- 22.
- Rees CED, Wilkins BM. Protein transfer into the recipient cell during bacterial conjugation: Stud ies with F and RP4. Mol Microbiol. 1990;4:1199–1205. [PubMed: 2172695]
- 23.
- Vergunst AC, Schrammeijer B, den Dulk-Ras A. et al. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. [PubMed: 11062129]
- 24.
- Wilkins BM, Thomas AT. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol Microbiol. 2000;38:650–657. [PubMed: 11069687]
- 25.
- Schrammeijer B, den Dulk-Ras A, Vergunst AC. et al. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: Evidence for transport of a novel effector protein VirE3. Nucl Acids Res. 2003;31:860–868. [PMC free article: PMC149200] [PubMed: 12560481]
- 26.
- Vergunst AC, Van Lier MC, den Dulk-Ras et al. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 2003;133:978–988. [PMC free article: PMC281595] [PubMed: 14551327]
- 27.
- Simone M, McCullen CA, Stahl LE. et al. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol Microbiol. 2001;41:1283–1293. [PubMed: 11580834]
- 28.
- Luo Z-Q, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–846. [PMC free article: PMC321768] [PubMed: 14715899]
- 29.
- Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at the cell poles of Agrobacterium tumefaciens. Mol Microbiol. 2003;49:1699–1733. [PMC free article: PMC3882298] [PubMed: 12950931]
- 30.
- Cabezon E, Lanka E, de la Cruz F. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J Bacteriol. 1994;176:4455–4558. [PMC free article: PMC205661] [PubMed: 8021231]
- 31.
- Cabezon E, Sastre JI, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. [PubMed: 9180693]
- 32.
- Sastre JI, Cabezon E, de la Cruz F. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J Bacteriol. 1998;180:6039–6042. [PMC free article: PMC107681] [PubMed: 9811665]
- 33.
- Schroeder G, Krause S, Zechner EL. et al. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: Inner membrane gate for exported substrates? J Bacteriol. 2002;184:2767–2779. [PMC free article: PMC135038] [PubMed: 11976307]
- 34.
- Szpirer CY, Faelen M, Couturier M. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol Microbiol. 2000;37:1283–1292. [PubMed: 10998162]
- 35.
- Das A, Xie YH. Construction of transposon Tn3PhoA: Its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol Microbiol. 1998;27:405–414. [PubMed: 9484895]
- 36.
- Lee MH, Kosuk N, Bailey J. et al. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J Bacteriol. 1999;181:6108–6113. [PMC free article: PMC103640] [PubMed: 10498725]
- 37.
- Gomis-Ruth FX, Moncalian G, Perez-Luque R. et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. [PubMed: 11214325]
- 38.
- Gomis-Ruth FX, Moncalian G, de la Cruz F. et al. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J Biol Chem. 2002;277:7556–7566. [PubMed: 11748238]
- 39.
- Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: VirB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. [PMC free article: PMC205554] [PubMed: 8206843]
- 40.
- Krall L, Wiedemann U, Unsin G. et al. Detergent extraction identifies different VirB protein sub-assemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2002;99:11405–11410. [PMC free article: PMC123269] [PubMed: 12177443]
- 41.
- Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol. 2000;182:758–763. [PMC free article: PMC94340] [PubMed: 10633111]
- 42.
- Ward DV, Draper O, Zupan JR. et al. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci USA. 2002;99:11493–11500. [PMC free article: PMC123284] [PubMed: 12177441]
- 43.
- Cao TB, Saier MH. Jr. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: Organismal distribution, structural constraints and evolutionary conclusions. Microbiol. 2001;147:3201–3214. [PubMed: 11739753]
- 44.
- Planet PJ, Kachlany SC, DeSalle R. et al. Phylogeny of genes for secretion NTPases: Identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci USA. 2001;98:2503–2508. [PMC free article: PMC30167] [PubMed: 11226268]
- 45.
- Krause S, Barcena M, Pansegrau W. et al. Sequence related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc Natl Acad Sci USA. 2000;97:3067–3072. [PMC free article: PMC16193] [PubMed: 10716714]
- 46.
- Yeo HJ, Savvides SN, Herr AB. et al. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV system. Mol Cell. 2000;6:1461–1472. [PubMed: 11163218]
- 47.
- Lupas AN, Martin J. AAA proteins. Curr Opin Struct Biol. 2002;12:746–753. [PubMed: 12504679]
- 48.
- Dang TAT, Christie PJ. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J Bacteriol. 1997;179:453–462. [PMC free article: PMC178716] [PubMed: 8990298]
- 49.
- Rashkova S, Spudich GM, Christie PJ. Mutational analysis of the Agrobacterium tumefaciens VirB11 ATPase: Identification of functional domains and evidence for multimerization. J Bacteriol. 1997;179:583–589. [PMC free article: PMC178735] [PubMed: 9006008]
- 50.
- Sagulenko Y, Sagulenko V, Chen J. et al. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol. 2001;183:5813–5825. [PMC free article: PMC99657] [PubMed: 11566978]
- 51.
- Llosa M, Gomis-Ruth FX, Coll M. et al. Bacterial conjugation: A two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. [PubMed: 12100543]
- 52.
- Dang TAT, Zhou X-R, Graf B. et al. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on assembly and function of the T-DNA transporter. Mol Microbiol. 1999;32:1239–1253. [PMC free article: PMC3918219] [PubMed: 10383764]
- 53.
- Rabel C, Grahn AM, Lurz R. et al. The VirB4 family of proposed traffic nucleoside triphosphatases: Common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J Bacteriol. 2003;185:1045–1058. [PMC free article: PMC142825] [PubMed: 12533481]
- 54.
- Berger BR, Christie PJ. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993;175:1723–1734. [PMC free article: PMC203967] [PubMed: 8449880]
- 55.
- Bohne J, Yim A, Binns AN. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc Natl Acad Sci USA. 1998;95:7057–7062. [PMC free article: PMC22737] [PubMed: 9618538]
- 56.
- Liu Z, Binns AN. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J Bacteriol. 2003;185:3259–3269. [PMC free article: PMC155385] [PubMed: 12754223]
- 57.
- Burns DL. Type IV transporters of pathogenic bacteria. Curr Opin Microbiol. 2003;6:29–34. [PubMed: 12615216]
- 58.
- Ruhfel RE, Manias DA, Dunny GM. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid pCF10, encoding a sex pheromone-binding function. J Bacteriol. 1993;175:5253–5259. [PMC free article: PMC204993] [PubMed: 8349565]
- 59.
- Leonard BA, Podbielski A, Hedberg PJ. et al. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci USA. 1996;93:260–264. [PMC free article: PMC40218] [PubMed: 8552617]
- 60.
- Kao SM, Olmsted SB, Viksnins AS. et al. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J Bacteriol. 1991;173:7650–7664. [PMC free article: PMC212534] [PubMed: 1938961]
- 61.
- Tame JR, Murshudov GN, Dodson EJ. et al. The structural basis of sequence-independent peptide binding by OppA protein. Science. 1994;264:1578–1581. [PubMed: 8202710]
- 62.
- Nakayama J, Yoshida K, Kobayashi H. et al. Cloning and characterization of a region of Enterococcus faecalis plasmid pPD1 encoding pheromone inhibitor (ipd), pheromone sensitivity (traC), and pheromone shutdown (traB) genes. J Bacteriol. 1995;177:5567–5573. [PMC free article: PMC177366] [PubMed: 7559344]
- 63.
- Mori M, Sakagami Y, Ishii Y. et al. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J Biol Chem. 1988;263:14574–14578. [PubMed: 3139658]
- 64.
- Dunny GM, Antiporta MH, Hirt H. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: Probing the genetic and molecular basis for specificity of the pheromone response. Peptides. 2001;22:1529–1539. [PubMed: 11587782]
- 65.
- Ehrenfeld EE, Kessler RE, Clewell DB. Identification of pheromone-induced surface proteins in Streptococcus faecalis and evidence of a role for lipoteichoic acid in formation of mating aggregates. J Bacteriol. 1986;168:6–12. [PMC free article: PMC213413] [PubMed: 3093466]
- 66.
- Paulsen IT, Banerjei L, Myers GS. et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. [PubMed: 12663927]
- 67.
- Clewell DB, An FY, Flannagan SE. et al. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol Microbiol. 2000;35:246–247. [PubMed: 10632894]
- 68.
- Dev IK, Ray PH. Signal peptidases and signal peptide hydrolases. J Bioenerg Biomembr. 1990;22:271–290. [PubMed: 2202720]
- 69.
- Nakayama J, Ono Y, Suzuki A. Isolation and structure of the sex pheromone inhibitor, iAM373, of Enterococcus faecalis. Biosci Biotechnol Biochem. 1995;59:1358–1359. [PubMed: 7670199]
- 70.
- Nakayama J, Ruhfel RE, Dunny GM. et al. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol. 1994;176:7405–7408. [PMC free article: PMC197135] [PubMed: 7545961]
- 71.
- Buttaro BA, Antiporta MH, Dunny GM. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J Bacteriol. 2000;182:4926–4933. [PMC free article: PMC111373] [PubMed: 10940037]
- 72.
- Clewell DB, Francia MV, Flannagan SE. et al. Enterococcal plasmid transfer: Sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid. 2002;48:193–201. [PubMed: 12460535]
- 73.
- Francia MV, Clewell DB. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: Identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol Microbiol. 2002;45:375–395. [PubMed: 12123451]
- 74.
- Hirt H, Manias DA, Bryan EM. et al. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: Complete sequence and comparative analysis of the transcriptional and phenotypic response of pCF10-containing cells to pheromone induction. J Bacteriol. 2005;187:1044–1054. [PMC free article: PMC545727] [PubMed: 15659682]
- 75.
- Mills DA, McKay LL, Dunny GM. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J Bacteriol. 1996;178:3531–3538. [PMC free article: PMC178122] [PubMed: 8655550]
- 76.
- Ike Y, Tanimoto K, Tomita H. et al. Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth matings. J Bacteriol. 1998;180:4886–4892. [PMC free article: PMC107514] [PubMed: 9733692]
- 77.
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:277–301. [PMC free article: PMC156469] [PubMed: 12794193]
- 78.
- Belhocine K, Plante I, Cousineau B. Conjugation mediates transfer of the LI.LtrB group II intron between different bacterial species. Mol Microbiol. 2004;51:1459–1469. [PubMed: 14982638]
- 79.
- Clewell DB. Movable genetic elements and antibiotic resistance in enterococci. Eur J Clin Microbiol Infect Dis. 1990;9:90–102. [PubMed: 2156704]
- 80.
- Schaberg DR, Zervos MJ. Intergeneric and interspecies gene exchange in gram-positive cocci. Antimicrob Agents Chemother. 1986;30:817–822. [PMC free article: PMC180600] [PubMed: 3028249]
- 81.
- Berg T, Firth N, Apisiridej S. et al. Complete nucleotide sequence of pSK41: Evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol. 1998;180:4350–4359. [PMC free article: PMC107441] [PubMed: 9721269]
- 82.
- Firth N, Ridgway KP, Byrne ME. et al. Analysis of a transfer region from the staphylococcal conjugative plasmid pSK41. Gene. 1993;136:13–25. [PubMed: 8293996]
- 83.
- Dougherty BA, Hill C, Weidmann JF. et al. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol Microbiol. 1998;29:1029–1038. [PubMed: 9767571]
- 84.
- Kurenbach B, Bohn C, Prabhu J. et al. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid. 2003;50:86–93. [PubMed: 12826062]
- 85.
- Thompson JK, Collins MA. Completed sequence of plasmid pIP501 and origin of spontaneous deletion derivatives. Plasmid. 2003;50:28–35. [PubMed: 12826055]
- 86.
- Morton TM, Eaton DM, Johnston JL. et al. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J Bacteriol. 1993;175:4436–4447. [PMC free article: PMC204884] [PubMed: 7687249]
- 87.
- Sharma VK, Johnston JL, Morton TM. et al. Transcriptional regulation by TrsN of conjugative transfer genes on staphylococcal plasmid pGO1. J Bacteriol. 1994;1769:3445–3454. [PMC free article: PMC205530] [PubMed: 8206820]
- 88.
- Tanimoto K, Ike Y. Analysis of the conjugal transfer system of the pheromone-independent highly transferable Enterococcus plasmid pMG1: Identification of a tra gene (traA) up-regulated during conjugation. J Bacteriol. 2002;184:5800–5804. [PMC free article: PMC139602] [PubMed: 12270839]
- 89.
- Kurenbach B, Grothe D, Farías ME. et al. The tra region of the conjugative plasmid pIP501 is organized in an operon with the first gene encoding the relaxase. J Bacteriol. 2002;184:1801–1805. [PMC free article: PMC134879] [PubMed: 11872736]
- 90.
- Salmond GPC. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu Rev Phytopathol. 1994;32:181–200.
- 91.
- Llosa M, de la Cruz F. Bacterial conjugation: A potential tool for genomic engineering. Res Microbiol. 2005;156:1–6. [PubMed: 15636742]
- 92.
- Christie PJ. Type IV secretion: Intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol Microbiol. 2001;40:294–305. [PMC free article: PMC3922410] [PubMed: 11309113]
- 93.
- Bateman A, Rawlings ND. The CHAP domain: A large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci. 2003;28:234–237. [PubMed: 12765834]
- 94.
- Rigden DJ, Jedrzejas MJ, Galperin MY. Amidase domains from bacterial and phage autolysins define a family of γ-D,L-glutamate-specific amidohydrolases. Trends Biochem Sci. 2003;28:230–234. [PubMed: 12765833]
- 95.
- Climo MW, Sharma VK, Archer GL. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol. 1996;178:4975–4983. [PMC free article: PMC178282] [PubMed: 8759863]
- Mating Cell-Cell Channels in Conjugating Bacteria - Madame Curie Bioscience Data...Mating Cell-Cell Channels in Conjugating Bacteria - Madame Curie Bioscience Database
- FROM STRUCTURE TO FUNCTION OF RNA BINDING DOMAINS - Madame Curie Bioscience Data...FROM STRUCTURE TO FUNCTION OF RNA BINDING DOMAINS - Madame Curie Bioscience Database
- Regulation and Coordination of Intracellular Trafficking: An Overview - Madame C...Regulation and Coordination of Intracellular Trafficking: An Overview - Madame Curie Bioscience Database
- Pre-Ribosomal RNA Processing in Multicellular Organisms - Madame Curie Bioscienc...Pre-Ribosomal RNA Processing in Multicellular Organisms - Madame Curie Bioscience Database
- Regulation of T Cell Immunity by OX40 and OX40L - Madame Curie Bioscience Databa...Regulation of T Cell Immunity by OX40 and OX40L - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...