NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
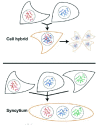
Despite the diversity of intercellular connections that are the subject of this book, most eukaryotic cells retain their distinct character as mononucleated compartments. Their membranes describe morphologically separate cytoplasms, while electrical connectivity and low-flux intercellular exchange of components occurs through small or selective channels between neighboring cell surfaces. However, in many instances throughout eukaryotes, pairs or groups of cells make a developmental decision to completely fuse their plasma membranes, allowing wholesale exchange and mixing of membranous, cytoplasmic and nuclear components. The products of these fusion events are either cell hybrids, in which chromosomes are combined into a single nucleus, or syncytia, wherein distinct nuclei are maintained within a single cytoplasm and plasma membrane (Fig. 1). While limited to very specific instances in the life cycle of any given organism, these precise cell fusions lead to a diverse set of dramatic developmental transitions: from formation of a new zygote, to construction of the musculoskeletal system, to refinement of the optical transparency of the developing eye. In addition, it appears possible to repair damaged cells, such as neurons, through the fusion of severed cellular fragments.1,2 This chapter will survey the various contexts for developmental cell fusion, examining the scant but growing knowledge of the molecules that initiate membrane permeability and removal of cell boundaries between merging partner cells. The understanding that is beginning to emerge suggests that cell-fusion channels or pores are transient affairs, both as structural antecedents of fully merged cell membranes, and possibly as replaceable molecular machines that were reinvented often through the course of evolution to drive a similar process by a variety of mechanisms.
The predominant case for formation of cell hybrids in nature is the fertilization or conjugation of haploid gametes to form diploid zygotes. This of course underlies the initiation of development by sexual reproduction in the vast majority of plant and animal species. It also plays a role in the development of many unicellular eukaryotes. Nonsexual somatic cell hybrids have been described as well, notably during the engraftment of stem cells into rodent hepatic and neural tissues after experimental cellular transfer.3-10 Chemically induced fusion of cells in vitro typically results in death for the majority of resulting hybrids, although viable cell hybrids are often selected experimentally and expanded as clones.11-13 It is unclear how prevalent the formation of somatic cell hybrids may be in the course of normal development and homeostasis. However, it has been reported that many cancer cell types are either fusogenic or are products of aberrant cell fusion,14,15 and the formation of rare yet viable cell hybrids can therefore have major impacts upon the health and lifespan of individual animals.
In contrast to cell hybrids, syncytia are most often formed during the terminal differentiation of specific cell types. In most cases, multinucleated giant cells are postmitotic and nonmotile, assuming a variety of roles in mature tissues. Reasoned teleologically, the functions performed by syncytial cells must be best accomplished by a single cellular compartment that is too large to be maintained by the gene-expression potential of a single diploid nucleus. Increases in gene copy-number can also be accomplished through endoreduplication of DNA.16- 18 However, multinucleation by cell fusion allows for establishment of pattern and form via the migration and proliferation of the mononucleate precursor cells before they fuse to form the final product. Fusion also allows the giant cell to be expanded or regenerated by later addition of new precursors to an existing syncytium. In addition, the distribution of individual nuclei through the giant cell permits local differences in gene expression to define sub-cellular specializations, such as the post-synaptic neuromuscular junction of skeletal muscle fibers.19,20
Below, several cases of developmental cell fusion are surveyed, and we summarize the current knowledge of molecules controlling actual plasma membrane merger and discuss the possibility that the same effect may be brought about by a variety of mechanisms. To focus on the process of membrane fusion, per se, we will avoid the discussion of the diverse regulatory pathways leading to the fusion-competent differentiated state in each of these cell types. In contrast to viral membrane-fusion pores and other forms of stable cell-cell channels covered in this book, very little is known about the biophysical properties of cell-fusion-initiating pores. We therefore also address the hypothetical involvement of initial cell-membrane fusion structures similar to those found in viral infection or specialized cell-cell channels between nonfusing cell types.
Fertilization (Mouse)
Sperm-egg fusion requires several steps prior to the actual fusion of gamete cell membranes, including binding of the sperm to the zona pelucida and initiation of the acrosome reaction in the sperm. Only sperm that have penetrated the zona, by secretion of degradative enzymes, and have extended an acrosomal process are competent to fuse with the egg. Izumo, an immunoglobulin-superfamily (IgSF) membrane glycoprotein, has been found newly presented on the surface of acrosome-reacted mouse sperm (Fig. 2A, Fig. 2B).21 When Izumo is either blocked by specific antibodies or deleted genetically, mouse sperm become completely incapable of fertilizing wild-type eggs, despite retaining their ability to penetrate the zona, elaborate a normal acrosomal process, and bind to the egg membrane. Furthermore, once experimentally introduced into an activated egg, Izumo-mutant sperm contribute normally to post-fusion zygotic development. Izumo, therefore, stands out among a collection of molecules previously implicated in fertilization,22-28 as it appears completely and specifically required for the fusion competence of sperm during fertilization.
On the egg plasma membrane, a tetraspanin molecule, CD9, is also absolutely required for fusion of the mouse egg membrane with wild-type sperm.29-31 Some peptide sequences in CD9 that are known to be required for sperm-egg fusion have also been shown to mediate binding of CD9 to IgSF molecules.32 Both CD9 and Izumo are encoded in the human genome, as well, suggesting that their role in sperm-egg fusion may be evolutionarily conserved in mammals other than mice. Yet, it is still a matter of conjecture whether the two molecules interact directly in trans to effect the membrane fusion reaction, or whether either molecule plays a direct role in the formation of a permeability pore between the two fusing cells.
Fertilization (Nematode)
Nematode sperm are unflagellated cells that move by amoeboid crawling. Current knowledge, via genetic studies of Caenorhabditis elegans fertility mutants, suggests that they also differ from mammalian sperm in their mechanism of sperm-egg fusion. Three different sperm-encoded proteins, SPE-9 (an EGF-repeat-containing membrane protein, TRP-3/SPE-41 (a TRPC-type calcium channel), and SPE-38 (a novel tetraspan membrane protein), are required specifically for sperm interaction and/or fusion with the egg.33-37 A recessive mutation in any of the three genes yields sperm that activate and migrate normally but fail to fertilize eggs. TRP-3 is interesting as it is perhaps the only ion-channel protein so far implicated specifically in a cell fusion event.35 SPE-38, although containing four membrane spanning domains, does not encode a homologue of the mammalian CD9, but it is structurally similar to the tetraspan protein PRM1 that regulates yeast cell fusion (see below).33 It is worth noting that EFF-1, while required for many tissue cell fusions in the worm (see below), is not required for sperm-egg fusion.38 This indicates two quite distinct mechanisms of membrane fusion in nematode gametes and somatic cells.
Myoblasts (Mouse)
In arthropods and vertebrates, precursors of skeletal muscle tissue fuse together to form multinucleated myotubes (Fig. 3) and ultimately muscle fibers, each of which can contain thousands of nuclei. This is the most prevalent form of cell fusion in most animals, and it produces long tube-shaped cells in which a continuously reiterated lattice of contractile filaments can extend uninterrupted over many centimeters in length. In addition, the neurally stimulated excitation/ contraction response of this “spring” is controlled by one motor-neuron synapse at a single neuromuscular junction on the large cell membrane. Both of these properties of myofibers suggest selective advantages that concur with the universal incidence of multinucleated muscle in phyla having large muscles and jointed “lever-action” movements. Through the proliferation and fusion of normally quiescent satellite cells, muscle fibers can be repaired or regenerated, even late in life in mammals.39 Interestingly, a microtubule-binding compound, myoseverin, has been shown to induce the fission of mouse myotubes in vitro, producing mononucleated fragments that could resume proliferation.40 This suggests that even multinucleated terminally differentiated cells in mammals may have much of the capacity for regeneration exhibited by similar cells in amphibians.
Several IgSF, cadherin, and integrin proteins have been implicated in mouse myoblast fusion. However, each of the molecules tested directly by genetic deletion in mice has been found not to be required for multinucleation of muscle fibers.41-45 Three members of the ADAM family of proteins, meltrin-α, β and γ were found to be induced in differentiating myoblasts, and antibody or antisense inhibition of meltrin-α were reported to inhibit myoblast fusion in vitro.46 However, mouse knockouts of each of the meltrin genes (and several other ADAM-coding genes) show no apparent defects in myoblast fusion in vivo.47,48 Thus, the highly regulated and specific mechanism by which differentiating mammalian myoblasts fuse their membranes is still unknown.
Myoblasts (Fruit Fly)
Mutations in a variety of genes have been reported to block the fusion of myoblasts during formation of embryonic body-wall muscle fibers in Drosophila melanogaster. These include known components of the muscle contractile apparatus,49,50 as well as actin-associated, cytoplasmic, and integral membrane proteins (reviewed in Refs. 51, 52). Electron microscopy on staged embryos has led to a model for myoblast fusion involving trafficking of electron-dense vesicles, formation of electron-dense membrane plaques, and ultimate membrane fusion at the site of contact between fusing cells (Fig. 4).53 The hypothesis that these vesicles and plaques contain components required for fusion is supported to some degree by EM images of the fusion-arrested membranes of mutant embryos.53,54 However, none of the genetically-identified proteins has yet been reported localized to these structures, and the functional sequence and relationship between vesicle traffic, plaque, and pore formation has not been reported in live cells.
Three genes that are required for normal Drosophila myoblast fusion encode integral membrane IgSF proteins: Dumfounded/Kin of Irre (Duf/Kirre), Roughest/Irregular chiasm (Rst/IrreC), and Sticks and Stones (SNS).55-57 Duf/Kirre and Rst/IrreC are expressed in founder cells, early-differentiating myocytes that recruit fusion-competent myoblasts (fcm) for two rounds of developmentally regulated fusion to form mature muscle fibers. SNS, in contrast, is expressed only on the fcm cells. Models involving interaction of these receptors to produce chemoattractive and fusion-effecting signals are suggested by the reciprocal expression of these molecules on pairs of cells destined to fuse. However, while SNS can interact heterotypically with either Duf/Kirre or Rst/ IrreC to mediate efficient adhesion and aggregation of transfected Drosophila S2 cells in culture, these interactions cannot elicit the membrane fusion with which they are associated in vivo.58 At this point, no molecule or combination of molecules known to be required for Drosophila myogenic fusion has been shown to be sufficient for membrane fusion when ectopically expressed. This suggests either that the actual fusion mechanism involves fusogenic molecules not yet identified, or that it combines the action of the known players in a way that has not been reconstituted in heterologous cell types.
Placental Syncytiotrophoblast Cells (Human and Mouse)
During uterine implantation of the early embryo in placental mammals, the trophoblast cells of the conceptus proliferate and invade the maternal endometrium. Subsequently, many of these cells fuse to form multinucleated syncytiotrophoblast cells at the interface of the maturing placenta with maternal tissue.59 The syncytiotrophoblasts may serve as a more selective barrier or regulator of nutrient, metabolite, gas, waste, and immune interchange between the circulatory systems of mother and fetus than is possible with a typical epithelium of mononucleated cells. Interestingly, in some species, fusion of fetal and maternal cells has been observed by electron microscopy.60
Syncytin-1 and -2 are developmentally regulated genes encoding human genomic copies of retroviral envelope glycoproteins, identified through hybridization and expression screening of placental cDNA libraries and via in silico genomic analysis of human genome sequences.61-64 Syncytin expression is selectively induced during the development of the placenta, and forced overexpression of syncytins in cultured cells can induce fusion of normally mononucleated cells (Fig. 5). Thus syncytins appear individually sufficient to induce receptor-dependent cell membrane fusion, similar to homologous proteins acting during infection by enveloped viruses. However, syncytin-1 and -2 are encoded exclusively in a subset of recently evolved primate genomes.63,65 This has precluded genetic tests of their necessity in syncytiotrophoblast fusion. Furthermore, if syncytin-1 and -2 do underlie the fusion mechanism in primate placenta, the molecules must have been adapted to the task recently in evolution (apparently within the past 40 million years), and some different fusogen(s) must drive cell fusion in other mammal species with fusing placental cells.
Recently, two murine viral envelope genes have been discovered, syncytin-A and -B, that are evolutionarily distinct from primate syncytins, yet appear to serve the same function in the syncytiotrophoblasts of mice and related rodents.66 This suggests that placental cell fusion may involve a mechanism allowing fortuitous substitution of fusogenic proteins, including independent cooption of several different viral genes. In the mouse model system, the opportunity now exists to prove or disprove, through targeted gene knockouts, the necessity of syncytin genes in placental cell fusion. Given that the four known syncytins are not encoded in many species with fusing syncytiotrophoblast cells, the alternative possibility still remains tenable: that nonsyncytin molecules may comprise a more evolutionarily conserved cell fusion mechanism common to all placental mammals.
Lens Fiber Cells (Mouse and Other Vertebrates)
Cells of the crystalline lens also fuse extensively in the development of the amphibian, avian, and mammalian eye.67-70 Fusion appears to occur between hexagonal fiber cells that are already terminally differentiated, producing progressively more and more cytoplasmic communication between cells with increasing age (Fig. 6). However, fused cells maintain their form as individual hexagonal prisms over much of their length. The central core of the lens, where fusion is most frequent, is also the region where fiber cells are often enucleated. Thus, fusion may enable formation of a clarified syncytium, in which nucleated cells at the periphery of the central zone are able to sustain the viability of enucleated central fibers through free exchange of cytoplasm. Presumably, this overall structure of the lens enhances transmission and refraction that produce an image on the retina. As yet, no molecules involved in this process have been described.
Macrophages/Osteoclasts (Mouse)
Cells of the mammalian monocyte-macrophage lineage form two major types of multinucleated cells: macrophage giant cells in many tissues and osteoclasts in bone.71 In each case, the physiological roles of these cells involves endocytosis and resorption of relatively large objects, including cell corpses, invading pathogens, foreign bodies, and chunks of mineralized bone. The increased size achieved through cell fusion presumably affords the extra membrane surface area and endosome/ lysosome volume to achieve these tasks. In addition, it is conceivable that a larger cell can more safely distance the nuclei and cell body from the noxious degradative cocktails used to dispose of objects within the engulfment apparatus.
MFR and CD44 are cell surface IgSF proteins implicated in the mechanism of macrophage and osteoclast fusion.72,73 The expression of each is induced transiently at the onset of fusion in macrophage cultures, and antibody blockade of MFR disrupts macrophage fusion in vitro. MFR is known to be a receptor for the constitutively expressed IgSF protein CD47,74 while CD44 has no known ligand. Despite their correlation with fusogenicity of macrophages, no genetic evidence for the requirement or sufficiency of these molecules in cell fusion has been reported. Recently, a seven-transmembrane receptor, DC-STAMP, was found to be required for macrophage/osteoclast fusion. Knockout mice lacking DC-STAMP fail to form multinucleated osteoclasts or macrophage-derived giant cells, and they display a mild osteopetrotic phenotype.75 Interestingly, the DC-STAMP mutation does not affect the mRNA levels of MFR, CD44, CD47, E-cadherin, or meltrin- α, all molecules hypothesized to contribute to the fusion mechanism. What the ligand for DC-STAMP is and what the actual components of the macrophage/osteoclast fusion mechanism might be are still unknown. It is interesting to note, however, that mononucleated DC-STAMP-/- osteoclasts appear to mediate reasonably normal bone development in the mouse, even without achieving the size of normal fused osteoclast giant cells.
Implanted Stem Cells (Mouse and Human)
Recent studies in mice have shown that implanted bone-marrow-derived cells undergo tissue-specific differentiation in host tissues via formation of cell-hybrids between donor and host cells.3, 5- 8, 10 Such donor-host fusion has even been reported in the neurons of human bone-marrow transplant recipients.9 In studies of hepatic disease in mice, where a selective advantage is conferred upon donor/host hybrids, fusion-based engraftment has been shown to rescue the viability of an entire organ.5,76 In a converse experiment, committed neuronal precursor cells cocultured with embryonic stem cells have been shown to produce cell-hybrids that regain pluripotent stem-cell character.4 The result of these cell-hybridizations in reprogramming gene expression is not unexpected, given previous evidence of the plasticity of nuclei in heterokaryons.77 But the cell fusions that are revealed between normally mononucleated cell types in the transplant studies is surprising. Although apparently quite rare events, these cell fusions might be hypothesized to proceed via similar mechanisms to the more robust developmentally programmed fusions in other tissues. Given that bone-marrow-derived lineages give rise to fusing macrophages and osteoclast giant cells, it has been reasonably hypothesized that the fusion mechanism responsible is conferred by a macrophage-like activity in the implanted bone-marrow cells. However, without further knowledge of the fusion mechanism of monocyte-derived cells (see above), it will be difficult to test this hypothesis directly.
Epithelia (Nematode)
One third of all somatic nuclei in adult Caenorhabditis elegans are found within multinucleated cells.78-82 Beginning halfway through embryogenesis, a stereotyped sequence of cell fusions between specific partner cells produces 44 adult syncytia ranging in size from 2 nuclei to 139. A number of tissues contain fused cells, including muscle cells in the pharynx (but not body wall), and cells of the somatic gonad and excretory system. The best-studied fusions occur among polarized epithelial cells in the epidermis and specialized organs derived from epidermal precursor cells, including the vulva, the male tail, and the lateral seam epidermis.78,83,84 Although apparently not required for viability of the animal in culture, these cell fusions are essential for achieving the sleek morphology and reproductive proficiency of the wild type.85 The invariance of these fusion events has allowed detailed observation of structural intermediates formed during the merger of two cells. Three-dimensional time-lapse imaging of epidermal fusions in the embryo indicates that the opening between fusing cells originates at a point on the apical edge of the lateral-membrane interface of two cells.86 The opening then widens as a single growing aperture, in part via vesiculation of the conjoined membranes, and displaces intercellular junctions that can remain intact even while the lipid bilayers retreat (Fig. 7). The initial permeabilization of the membrane to cytoplasmic diffusion precedes the visible widening of the opening by 5-10 minutes (W. Mohler, unpublished observations).
The integral membrane protein EFF-1 appears to lie at the heart of the membrane fusion mechanism in epidermal, vulval, and pharyngeal cells.38,85,87 Loss-of-function mutations in the eff-1 gene block essentially all epidermal cell fusions, without disrupting the ability of epidermal cells to function in other respects as an intact skin epithelium.85 Defects in the pharynx of eff-1 mutants87 also seem confined to cells' ability to fuse, as the organ forms and functions to feed the worm. In contrast, misexpression of EFF-1 in nonfusing cell types can be very lethal, causing inappropriate fusions that severely disrupt the normal course of development.85,87 Thus EFF-1 is both absolutely necessary for developmentally programmed fusions and quite sufficient to induce fusion in nonfusion-fated cells. This combination of attributes set it apart, for now, from all other molecules implicated in developmental cell fusion. But EFF-1 does not account for all somatic cell fusions in C. elegans. Neither sperm-egg, nor anchor cell-uterus, nor seam-seam cell fusion in the worm appears to require EFF-1, indicating that two or more distinct molecular mechanisms must be at work in producing all of the fused cell in the animal.
EFF-1 is also unique because its sequence does not match any known proteins or protein families in non-nematode organisms.38,85 Predicted to be a single-span transmembrane protein, EFF-1 has a large N-terminal domain and a small cytoplasmic C-terminus. Fluorescently tagged EFF-1 remains in cytoplasmic pools until a fusion-fated membrane contact forms between epidermal precursor cells. Once fusion-fated partners touch, EFF-1 rapidly accumulates at the point of cell contact. Furthermore, the pattern of this localization among groups of cells suggests that EFF-1 plasma-membrane localization is dependent upon EFF-1 expression in both partner cells; thus EFF-1 may interact homophilically between the surfaces of fusing membranes.38 In the extracellular domain of EFF-1 lies a short extracellular hydrophobic peptide that is hydropathically similar to fusion peptides and fusion loops in enveloped virus fusion proteins.88-94 Mutations in the EHP abrogate cell fusion activity, implicating this region as critical to EFF-1 function. However, EHP-mutant EFF-1 is not properly localized to cell-fusion-fated plasma membranes.38 Thus, it is unclear whether the EHP functions in targeting EFF-1 to the cell surface, in protein interactions that retain the protein at the membrane, or in formation of a membrane fusion pore itself (the function ascribed to fusion peptides in viral fusogens).88-94
No other genes have been discovered in C. elegans with the fusion-defective viable phenotype characteristic of loss of eff-1 function. Three possible scenarios explain this finding: 1) EFF-1 is the only required component of the fusion mechanism; 2) other components of the fusion mechanism function redundantly, and yield no phenotype when mutated singly; or 3) other nonredundant fusion components function pleiotropically in additional aspects of development, and therefore give lethal loss-of-function phenotypes. Interestingly, loss of subunits of the vacuolar ATPase (V-ATPase) complex causes ectopic cell fusions in C. elegans embryos, among other phenotypic defects.95 The V-ATPase complex is known to play critical roles in secretory pathways of mammalian and yeast cells.96 Extracellular barriers to fusion normally appear during the formation of tissues during C. elegans development,38 and V-ATPase function is known to affect extracellular protein activity.97 It is therefore possible that the promiscuous, delayed cell fusions caused by loss of the V-ATPase occur as a result of defective extracelluar matrix formation.
Primary Mesenchyme (Sea Urchin)
The primary mesenchyme cells of echinoderm embryos produce the pluteus larva's skeleton of calcified spicules. In doing so, they migrate to distinct positions in the blastocoel cavity, extend thin processes to one another, and fuse their plasma membranes to create a syncytial network (Fig. 8).98,99 Within the cytoplasm of this syncytium, mineralized calcium is deposited to form the calcified spicules.99 Cultured primary mesenchyme cells retain their ability to fuse.100 Yet they are unable to fuse with fusion-competent blastocoelar cells, suggesting that two distinct molecular mechanisms are at work in forming separate syncytial tissues simultaneously in the same same space.98 Molecular players in either of these mechanisms are still unknown.
Embryonic Blastomeres (Leech)
An exception to the rule of fusion as part of terminal differentiation appears in the early stages of embryogenesis in the leech Helobdella robusta.101 Here three large endodermal precursor cells undergo partial fusion to form a syncytium, which later gives rise to recellularized descendents lining the digestive tract. Experiments employing protein-synthesis inhibitors indicate that each cell in a fusion-pair must express new proteins for cell fusion to occur,102 but the molecules required for membrane fusion remain unknown.
Haploid Mating (Yeast)
Pheromone-induced conjugation of haploid Saccharomyces cerevisiae cells involves cell-wall remodeling in advance of plasma membrane fusion. Mutations in a number of single genes yield mating-defective yeast that undergo normal morphological changes but fail to fuse, as the cell wall continues to separate the cell membranes.103-105 These genes are apparently involved in degradation of the cell wall, and not in membrane fusion per se. Mutants in the PRM1 gene, in contrast, are often blocked at a step after cell-wall remodeling (Fig. 9).106 Still, Prm1 protein appears not to be completely required for fusion, because nearly half of prm1 null-mutant cell pairs fuse successfully. Recently, it was found that many prm1-mutant cells undergo lysis that is specifically associated with the membrane fusion step of mating.107 Thus, Prm1 appears to play a role in stabilizing the cell-fusion interface in the two cell membranes, permitting safe and efficient membrane merger by a mechanism whose components remain unknown.
Fungal Hyphae
Many fungi propagate as multinucleated hyphae, which can form anastomoses through fusion. In some species, it is hypothesized that fusion-dependent exchange of nuclei can stabilize the genome of such a multinucleated organism, even when the individual nuclei have varying genomic content, a phenonmenon termed heterokaryosis.108 In the genetically tractable species Neurospora crassa, the genes so/ham-1 and ham-2 have been implicated in the process of hyphal fusion,109,110 and the HAM-2 protein, which has orthologs in animals and other fungi, is predicted to contain three membrane-spanning domains.110 As in many other model systems, however, the nature of the membrane-fusion mechanism is still unclear.
Summary of Molecules Driving Cell Fusion: Obstacles to Their Discovery
To summarize the current understanding of molecules that drive cell membrane fusion in development, we know very little. Why is this? First, the fusion competent contact between cells is transient, and by definition self-destructive. This precludes biochemical preparation of membrane extracts specifically enriched in cell-membrane fusion activity. Second, the biology of each and every cell in an organism, including all nonfusing cell types, involves millions of intracellular membrane fusion events during a cell's lifetime. A fusion competent cell may fuse plasma membranes only once or twice with neighbors during development. Thus purifying and reconstituting a fusion mechanism, even from pure populations of fusing cells, is a “needle in a haystack” task. Third, although the fusion-fated cell must certainly change its membrane-protein content to become fusion competent, it typically changes its gene expression and proteome in many other ways while differentiating into a functioning myofiber, osteoclast, or lens fiber. This makes candidate-gene testing a more difficult task.
Loss-of-function genetics and expression-cloning, combined with functional-genome/proteome profiling have yielded the best current candidates for cell membrane fusogens, as well as several proteins that regulate the activities of still-unknown fusion proteins. However, forward genetic screening may be frustrated by either redundancy or pleiotropy of components of the fusion mechanism. Furthermore, confirmation of the importance of required proteins in a tightly regulated fusion machine ultimately will require reconstitution of that machine from its component parts. This work has only begun, and we can hope that lessons learned from the first apparent successes will guide the discovery of new cell fusion proteins either by similar screening approaches or by homology/analogy between confirmed fusogens and membrane proteins found in other systems.
Structural Origins of Cell Fusion Channels?
The best-understood examples of exoplasmic membrane fusion are those of enveloped viruses and virus-infected cells. Prevailing models, based on biochemical studies of the known fusogen molecules and electophysiological measurements of the early fusion pores, invoke a “fence” of oligomerized integral membrane proteins that span the intermembrane space and surround a lipid-lined pore with an inner diameter of 7-8 nm.111-116 This initial pore structure soon widens and eventually spreads to entirely join the two cytoplasms (Fig. 10). Synctyins, as clear orthologs of viral fusion proteins, fit implicitly within such models for pore formation. Lacking understanding of the key molecules in most other developmental fusions, we cannot yet address the structures of the pores in any detail. Nevertheless, time-lapse imaging in of EFF-1-induced fusions in C. elegans embryos indicates that (as in viral fusion) a small permeable pore forms to initiate the opening between cells, an aperture that can be seen visibly widening only several minutes later.38,86 Based on diffusion of fluorescently labeled globular proteins from the egg to sperm cytoplasm, it has been concluded that the mouse sperm-egg fusion pore is at least 8nm in diameter.117 However, like the identities of the pore forming molecules, an upper limit to the actual pore size has yet to be determined.
Another plausible model has arisen with the discovery of intercellular nanotubules (or nanotubes) in a variety of cultured animal cell types.118-120 These thin membrane bridges appear to form quite frequently between nonfusing cells, allowing exchange of organelles via active transport along cytoskeletal filaments, but preventing appreciable diffusion of cytoplasm between cells. If nanotubules are indeed as prevalent in vivo as the in vitro evidence would indicate, cell fusions could simply be instances in which a nanotubule dilates to fully merge the two connected cells. If this were the case, then the cell fusion mechanism may not involve specialized membrane fusion events at all, and could rely on ubiquitous nanotubule-forming fusogens. The activities specific to syncytium-forming cells could instead be dilation-promoting molecules that force this transition from a diffusion-impermeable tubule to a large fusion aperture. The example of EFF-1 makes it clear that developmental fusion can involve single molecules that are both required and sufficient to form syncytia. Whether they form virus-like pores, expand nanotubular channels, or employ some other mechanism involving lipases or other catalytic enaymes, fusing cell types are likely to express proteins that specifically enable a precise topological change that is unfavored in the constitutive composition of the plasma membrane.
Evolution and Cell Fusion
Has the mechanism of cell fusion been reinvented multiple times during evolution? To date, none of the membrane proteins implicated in the fusion of one cell type have been found to function in another system. This fact may simply reflect a dearth of knowledge, as molecules verifiably essential to the fusion mechanism are unknown in most examples of cell fusion (see above). Then again, the syncytins and EFF-1 are the only two cases where current data strongly support the model of a developmentally regulated fusogenic (sufficient) membrane protein, and each of these molecules appears restricted phyletically to only a few million years of evolution between closely related animal genomes. The syncytins, as clear orthologs of retroviral envelope proteins, present a ready explanation for this novelty, since their coding sequences may have simply arrived as mobile genetic elements that fell under the control of placenta-specific cis-regulatory DNA sequences. EFF-1, in contrast, is not obviously related to any known viral proteins. However, its tight sequence conservation within the nematodes and apparent absence in other phyla suggest a sudden origin in the worm genome, possibly also by transfer from a viral or other transposable source. If so, then it is remarkable how the various cis regulatory sequences controlling eff-1 expression have been tuned to drive transcription in the precise and complex spatiotemporal pattern that occurs during development.85,121
It is possible that developmental cell fusogens have been adopted in specific cases over only short spans of evolution, and that the fusion mechanisms of different cell types are radically different, even within the same organism. Mismatched fusion machines in distinct cell types - as are indicated in nematodes and echinoderms - may permit the formation of separate syncytial tissues without promiscuous cross-fusion of cell types. Cell fusion, a cellular function that is not required for cellular viability and is quite specialized to discrete developmental contexts, might be more tolerably lost and replaced than other more highly conserved molecular systems: at least in C. elegans, fusing not at all is safer than fusing too much.38,85,87 Moreover, the task of inducing membrane-fusion, without building a stable intercellular connection, could require somewhat less molecular specificity than other cell-cell channels, allowing selection from a variety of protein types that can do the job. Still, we might expect that long-conserved syncytial structures, such as skeletal muscle, that have become integral to the body plans of multiple phyla, may be formed by mechanisms that have remained little-changed during evolution.
References
- 1.
- Deriemer SA, Elliott EJ, Macagno ER. et al. Morphological evidence that regenerating axons can fuse with severed axon segments. Brain Res. 1983;272(1):157–161. [PubMed: 6616192]
- 2.
- Shi R, Borgens RB, Blight AR. Functional reconnection of severed mammalian spinal cord axons with polyethylene glycol. J Neurotrauma. 1999;16(8):727–738. [PubMed: 10511246]
- 3.
- Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM. et al. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature. 2003;425(6961):968–973. [PubMed: 14555960]
- 4.
- Ying QL, Nichols J, Evans EP. et al. Changing potency by spontaneous fusion. Nature. 2002;416(6880):545–548. [PubMed: 11932748]
- 5.
- Wang X, Willenbring H, Akkari Y. et al. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422(6934):897–901. [PubMed: 12665832]
- 6.
- Terada N, Hamazaki T, Oka M. et al. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature. 2002;416(6880):542–545. [PubMed: 11932747]
- 7.
- Wurmser AE, Gage FH. Stem cells: Cell fusion causes confusion. Nature. 2002;416(6880):485–487. [PubMed: 11932725]
- 8.
- Weimann JM, Johansson CB, Trejo A. et al. Stable reprogrammed heterokaryons form spontaneously in Purkinje neurons after bone marrow transplant. Nat Cell Biol. 2003;5(11):959–966. [PubMed: 14562057]
- 9.
- Weimann JM, Charlton CA, Brazelton TR. et al. Contribution of transplanted bone marrow cells to Purkinje neurons in human adult brains. Proc Natl Acad Sci USA. 2003;100(4):2088–2093. [PMC free article: PMC149963] [PubMed: 12576546]
- 10.
- Medvinsky A, Smith A. Stem cells: Fusion brings down barriers. Nature. 2003;422(6934):823–825. [PubMed: 12712184]
- 11.
- Wang HS, Niewczas V, de SNHR. et al. Cytogenetic characteristics of 26 polyethylene glycol-induced human-hamster hybrid cell lines. Cytogenet Cell Genet. 1979;24(4):233–244. [PubMed: 509993]
- 12.
- Chu EH, Powell SS. Selective systems in somatic cell genetics. Adv Hum Genet. 1976;7:189–258. [PubMed: 797245]
- 13.
- Antczak DF. Monoclonal antibodies: Technology and potential use. J Am Vet Med Assoc. 1982;181(10):1005–1010. [PubMed: 6757209]
- 14.
- Hart IR. Tumor cell hybridization and neoplastic progression. Symp Fundam Cancer Res. 1983;36:133–143. [PubMed: 6382503]
- 15.
- Duelli D, Lazebnik Y. Cell fusion: A hidden enemy? Cancer Cell. 2003;3(5):445–448. [PubMed: 12781362]
- 16.
- Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol. 1985;107(1):128–133. [PubMed: 2578115]
- 17.
- Zybina EV, Zybina TG. Polytene chromosomes in mammalian cells. Int Rev Cytol. 1996;165:53–119. [PubMed: 8900957]
- 18.
- Royzman I, Orr-Weaver TL. S phase and differential DNA replication during Drosophila oogenesis. Genes Cells. 1998;3(12):767–776. [PubMed: 10096018]
- 19.
- Moscoso LM, Merlie JP, Sanes JR. N-CAM, 43K-rapsyn, and S-laminin mRNAs are concentrated at synaptic sites in muscle fibers. Mol Cell Neurosci. 1995;6(1):80–89. [PubMed: 7599960]
- 20.
- Schaeffer L, de Kerchove d'Exaerde A, Changeux JP. Targeting transcription to the neuromuscular synapse. Neuron. 2001;31(1):15–22. [PubMed: 11498047]
- 21.
- Inoue N, Ikawa M, Isotani A. et al. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434(7030):234–238. [PubMed: 15759005]
- 22.
- Manandhar G, Toshimori K. Exposure of sperm head equatorin after acrosome reaction and its fate after fertilization in mice. Biol Reprod. 2001;65(5):1425–1436. [PubMed: 11673259]
- 23.
- Nishimura H, Cho C, Branciforte DR. et al. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev Biol. 2001;233(1):204–213. [PubMed: 11319869]
- 24.
- Blobel CP, Wolfsberg TG, Turck CW. et al. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356(6366):248–252. [PubMed: 1552944]
- 25.
- Hao Z, Wolkowicz MJ, Shetty J. et al. SAMP32, a testis-specific, isoantigenic sperm acrosomal membrane-associated protein. Biol Reprod. 2002;66(3):735–744. [PubMed: 11870081]
- 26.
- Ilayperuma I. Identification of the 48-kDa G11 protein from guinea pig testes as sperad. J Exp Zool. 2002;293(6):617–623. [PubMed: 12410611]
- 27.
- Anderson DJ, Abbott AF, Jack RM. The role of complement component C3b and its receptors in sperm-oocyte interaction. Proc Natl Acad Sci USA. 1993;90(21):10051–10055. [PMC free article: PMC47711] [PubMed: 8234255]
- 28.
- Rochwerger L, Cohen DJ, Cuasnicu PS. Mammalian sperm-egg fusion: The rat egg has complementary sites for a sperm protein that mediates gamete fusion. Dev Biol. 1992;153(1):83–90. [PubMed: 1516754]
- 29.
- Kaji K, Oda S, Shikano T. et al. The gamete fusion process is defective in eggs of CD9-deficient mice. Nat Genet. 2000;24(3):279–282. [PubMed: 10700183]
- 30.
- Le Naour F, Rubinstein E, Jasmin C. et al. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287(5451):319–321. [PubMed: 10634790]
- 31.
- Miyado K, Yamada G, Yamada S. et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287(5451):321–324. [PubMed: 10634791]
- 32.
- Ellerman DA, Ha C, Primakoff P. et al. Direct binding of the ligand PSG17 to CD9 requires a CD9 site essential for sperm-egg fusion. Mol Biol Cell. 2003;14(12):5098–5103. [PMC free article: PMC284811] [PubMed: 14528020]
- 33.
- Chatterjee I, Richmond A, Putiri E. et al. The Caenorhabditis elegans spe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development. 2005;132(12):2795–2808. [PubMed: 15930110]
- 34.
- Putiri E, Zannoni S, Kadandale P. et al. Functional domains and temperature - sensitive mutations in SPE-9, an EGF repeat-containing protein required for fertility in Caenorhabditis elegans. Dev Biol. 2004;272(2):448–459. [PubMed: 15282160]
- 35.
- Xu XZ, Sternberg PW. A C. elegans sperm TRP protein required for sperm-egg interactions during fertilization. Cell. 2003;114(3):285–297. [PubMed: 12914694]
- 36.
- Singson A, Mercer KB, L'Hernault SW. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 1998;93(1):71–79. [PubMed: 9546393]
- 37.
- Zannoni S, L'Hernault SW, Singson AW. Dynamic localization of SPE-9 in sperm: A protein required for sperm-oocyte interactions in Caenorhabditis elegans. BMC Dev Biol. 2003;3(1):10. [PMC free article: PMC305347] [PubMed: 14653860]
- 38.
- del Campo JJ, Opoku-Serebuoh E, Isaacson AB. et al. Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Curr Biol. 2005;15(5):413–423. [PubMed: 15753035]
- 39.
- Collins CA, Olsen I, Zammit PS. et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122(2):289–301. [PubMed: 16051152]
- 40.
- Rosania GR, Chang YT, Perez O. et al. Myoseverin, a microtubule-binding molecule with novel cellular effects. Nat Biotechnol. 2000;18(3):304–308. [PubMed: 10700146]
- 41.
- Taverna D, Disatnik MH, Rayburn H. et al. Dystrophic muscle in mice chimeric for expression of alpha5 integrin. J Cell Biol. 1998;143(3):849–859. [PMC free article: PMC2148145] [PubMed: 9813102]
- 42.
- Yang JT, Rando TA, Mohler WA. et al. Genetic analysis of alpha 4 integrin functions in the development of mouse skeletal muscle. J Cell Biol. 1996;135(3):829–835. [PMC free article: PMC2121061] [PubMed: 8909554]
- 43.
- Charlton CA, Mohler WA, Radice GL. et al. Fusion competence of myoblasts rendered genetically null for N-cadherin in culture. J Cell Biol. 1997;138(2):331–336. [PMC free article: PMC2138190] [PubMed: 9230075]
- 44.
- Charlton CA, Mohler WA, Blau HM. Neural cell adhesion molecule (NCAM) and myoblast fusion. Dev Biol. 2000;221(1):112–119. [PubMed: 10772795]
- 45.
- Hollnagel A, Grund C, Franke WW. et al. The cell adhesion molecule M-cadherin is not essential for muscle development and regeneration. Mol Cell Biol. 2002;22(13):4760–4770. [PMC free article: PMC133893] [PubMed: 12052883]
- 46.
- Yagami-Hiromasa T, Sato T, Kurisaki T. et al. A metalloprotease-disintegrin participating in myoblast fusion. Nature. 1995;377(6550):652–656. [PubMed: 7566181]
- 47.
- Kurohara K, Komatsu K, Kurisaki T. et al. Essential roles of Meltrin beta (ADAM19) in heart development. Dev Biol. 2004;267(1):14–28. [PubMed: 14975714]
- 48.
- Kurisaki T, Masuda A, Sudo K. et al. Phenotypic analysis of Meltrin alpha (ADAM12)-deficient mice: Involvement of Meltrin alpha in adipogenesis and myogenesis. Mol Cell Biol. 2003;23(1):55–61. [PMC free article: PMC140658] [PubMed: 12482960]
- 49.
- Zhang Y, Featherstone D, Davis W. et al. Drosophila D-titin is required for myoblast fusion and skeletal muscle striation. J Cell Sci. 2000;113(Pt 17):3103–3115. [PubMed: 10934048]
- 50.
- Menon SD, Chia W. Drosophila rolling pebbles: A multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev Cell. 2001;1(5):691–703. [PubMed: 11709189]
- 51.
- Chen EH, Olson EN. Towards a molecular pathway for myoblast fusion in Drosophila. Trends Cell Biol. 2004;14(8):452–460. [PubMed: 15308212]
- 52.
- Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308(5720):369–373. [PubMed: 15831748]
- 53.
- Doberstein SK, Fetter RD, Mehta AY. et al. Genetic analysis of myoblast fusion: Blown fuse is required for progression beyond the prefusion complex. J Cell Biol. 1997;136(6):1249–1261. [PMC free article: PMC2132517] [PubMed: 9087441]
- 54.
- Schroter RH, Lier S, Holz A. et al. Kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development. 2004;131(18):4501–4509. [PubMed: 15342475]
- 55.
- Ruiz-Gomez M, Coutts N, Price A. et al. Drosophila dumbfounded: A myoblast attractant essential for fusion. Cell. 2000;102(2):189–198. [PubMed: 10943839]
- 56.
- Bour BA, Chakravarti M, West JM. et al. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000;14(12):1498–1511. [PMC free article: PMC316690] [PubMed: 10859168]
- 57.
- Strunkelnberg M, Bonengel B, Moda LM. et al. Rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128(21):4229–4239. [PubMed: 11684659]
- 58.
- Galletta BJ, Chakravarti M, Banerjee R. et al. SNS: Adhesive properties, localization requirements and ectodomain dependence in S2 cells and embryonic myoblasts. Mech Dev. 2004;121(12):1455–1468. [PubMed: 15511638]
- 59.
- Robertson WB. Pathology of the pregnant uterus. In: Fox H, ed. Obstetrical and Gynaecological Pathology. 3rd ed. London: Churchill Livingstone. 1987:1149–1176.
- 60.
- Wooding FBP. Role of binucleate cells in fetomaternal cell fusion at implantation in the sheep. American Journal of Anatomy. 1984;170:233–250. [PubMed: 6465051]
- 61.
- Blond JL, Beseme F, Duret L. et al. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol. 1999;73(2):1175–1185. [PMC free article: PMC103938] [PubMed: 9882319]
- 62.
- Blond JL, Lavillette D, Cheynet V. et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74(7):3321–3329. [PMC free article: PMC111833] [PubMed: 10708449]
- 63.
- Blaise S, de Parseval N, Benit L. et al. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100(22):13013–13018. [PMC free article: PMC240736] [PubMed: 14557543]
- 64.
- Mi S, Lee X, Li X-p. et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. [PubMed: 10693809]
- 65.
- Mallet F, Bouton O, Prudhomme S. et al. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci USA. 2004;101(6):1731–1736. [PMC free article: PMC341840] [PubMed: 14757826]
- 66.
- Dupressoir A, Marceau G, Vernochet C. et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102(3):725–730. [PMC free article: PMC545540] [PubMed: 15644441]
- 67.
- Kuszak JR, Macsai MS, Bloom KJ. et al. Cell-to-cell fusion of lens fiber cells in situ: Correlative light, scanning electron microscopic, and freeze-fracture studies. J Ultrastruct Res. 1985;93(3):144–160. [PubMed: 3879764]
- 68.
- Kuszak JR, Ennesser CA, Bertram BA. et al. The contribution of cell-to-cell fusion to the ordered structure of the crystalline lens. Lens Eye Toxic Res. 1989;6(4):639–673. [PubMed: 2487276]
- 69.
- Shestopalov VI, Bassnett S. Expression of autofluorescent proteins reveals a novel protein permeable pathway between cells in the lens core. J Cell Sci. 2000;113(Pt 11):1913–1921. [PubMed: 10806102]
- 70.
- Shestopalov VI, Bassnett S. Development of a macromolecular diffusion pathway in the lens. J Cell Sci. 2003;116(Pt 20):4191–4199. [PMC free article: PMC1401505] [PubMed: 12953070]
- 71.
- Sutton JS, Weiss L. Transformation of monocytes in tissue culture into macrophages, epithelioid cells, and multinucleated giant cells. An electron microscope study. J Cell Biol. 1966;28(2):303–332. [PMC free article: PMC2106921] [PubMed: 5914695]
- 72.
- Saginario C, Sterling H, Beckers C. et al. MFR, a putative receptor mediating the fusion of macrophages. Mol Cell Biol. 1998;18(11):6213–6223. [PMC free article: PMC109208] [PubMed: 9774638]
- 73.
- Sterling H, Saginario C, Vignery A. CD44 occupancy prevents macrophage multinucleation. J Cell Biol. 1998;143(3):837–847. [PMC free article: PMC2148144] [PubMed: 9813101]
- 74.
- Han X, Sterling H, Chen Y. et al. CD47, a ligand for the macrophage fusion receptor, participates in macrophage multinucleation. J Biol Chem. 2000;275(48):37984–37992. [PubMed: 10964914]
- 75.
- Yagi M, Miyamoto T, Sawatani Y. et al. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med. 2005;202(3):345–351. [PMC free article: PMC2213087] [PubMed: 16061724]
- 76.
- Lagasse E, Connors H, Al-Dhalimy M. et al. Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat Med. 2000;6(11):1229–1234. [PubMed: 11062533]
- 77.
- Blau HM, Pavlath GK, Hardeman EC. et al. Plasticity of the differentiated state. Science. 1985;230(4727):758–766. [PubMed: 2414846]
- 78.
- Podbilewicz B, White JG. Cell fusions in the developing epithelial of C.elegans. Dev Biol. 1994;161(2):408–424. [PubMed: 8313992]
- 79.
- Sulston JE, Schierenberg E, White JG. et al. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100(1):64–119. [PubMed: 6684600]
- 80.
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56(1):110–156. [PubMed: 838129]
- 81.
- Shemer G, Podbilewicz B. Fusomorphogenesis: Cell fusion in organ formation. Dev Dyn. 2000;218(1):30–51. [PubMed: 10822258]
- 82.
- Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149(3):1323–1334. [PMC free article: PMC1460238] [PubMed: 9649523]
- 83.
- Nguyen CQ, Hall DH, Yang Y. et al. Morphogenesis of the Caenorhabditis elegans male tail tip. Dev Biol. 1999;207(1):86–106. [PubMed: 10049567]
- 84.
- Sharma-Kishore R, White JG, Southgate E. et al. Formation of the vulva in Caenorhabditis elegans: A paradigm for organogenesis. Development. 1999;126(4):691–699. [PubMed: 9895317]
- 85.
- Mohler WA, Shemer G, del Campo JJ. et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2(3):355–362. [PubMed: 11879640]
- 86.
- Mohler WA, Simske JS, Williams-Masson EM. et al. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr Biol. 1998;8(19):1087–1090. [PubMed: 9768364]
- 87.
- Shemer G, Suissa M, Kolotuev I. et al. EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr Biol. 2004;14(17):1587–1591. [PubMed: 15341747]
- 88.
- Gamblin SJ, Haire LF, Russell RJ. et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303(5665):1838–1842. [PubMed: 14764886]
- 89.
- Gibbons DL, Vaney MC, Roussel A. et al. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427(6972):320–325. [PubMed: 14737160]
- 90.
- Stevens J, Corper AL, Basler CF. et al. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303(5665):1866–1870. [PubMed: 14764887]
- 91.
- Modis Y, Ogata S, Clements D. et al. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. [PubMed: 14737159]
- 92.
- Bullough PA, Hughson FM, Skehel JJ. et al. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. [PubMed: 8072525]
- 93.
- Nieva JL, Agirre A. Are fusion peptides a good model to study viral cell fusion? Biochim Biophys Acta. 2003;1614(1):104–115. [PubMed: 12873771]
- 94.
- Harter C, James P, Bächi T. et al. Hydrophobic binding of the ectodomain of influenza hemagglutinin to membranes occurs through the “fusion peptide” J Biol Chem. 1989;264:6459–6464. [PubMed: 2703499]
- 95.
- Kontani K, Moskowitz IP, Rothman JH. Repression of cell-cell fusion by components of the C. elegans vacuolar ATPase complex. Dev Cell. 2005;8(5):787–794. [PubMed: 15866168]
- 96.
- Nishi T, Forgac M. The vacuolar (H+)-ATPases—nature's most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3(2):94–103. [PubMed: 11836511]
- 97.
- Maquoi E, Peyrollier K, Noel A. et al. Regulation of membrane-type 1 matrix metalloproteinase activity by vacuolar H+-ATPases. Biochem J. 2003;373(Pt 1):19–24. [PMC free article: PMC1223473] [PubMed: 12667140]
- 98.
- Hodor PG, Ettensohn CA. The dynamics and regulation of mesenchymal cell fusion in the sea urchin embryo. Dev Biol. 1998;199(1):111–124. [PubMed: 9676196]
- 99.
- Gibbins JR, Tilney LG, Porter KR. Microtubules in the formation and development of the primary mesenchyme in Arbacia punctulata. I. The distribution of microtubules. J Cell Biol. 1969;41(1):201–226. [PMC free article: PMC2107725] [PubMed: 5775786]
- 100.
- Karp GC, Solursh M. In vitro fusion and separation of sea urchin primary mesenchyme cells. Exp Cell Res. 1985;158(2):554–557. [PubMed: 4007067]
- 101.
- Liu NL, Isaksen DE, Smith CM. et al. Movements and stepwise fusion of endodermal precursor cells in leech. Dev Genes Evol. 1998;208(3):117–127. [PubMed: 9601984]
- 102.
- Isaksen DE, Liu NJ, Weisblat DA. Inductive regulation of cell fusion in leech. Development. 1999;126(15):3381–3390. [PubMed: 10393117]
- 103.
- White JM, Rose MD. Yeast mating: Getting close to membrane merger. Curr Biol. 2001;11(1):R16–20. [PubMed: 11166190]
- 104.
- McCaffrey G, Clay FJ, Kelsay K. et al. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7(8):2680–2690. [PMC free article: PMC367884] [PubMed: 3313002]
- 105.
- Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: Evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7(7):2316–2328. [PMC free article: PMC365362] [PubMed: 3302672]
- 106.
- Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151(3):719–730. [PMC free article: PMC2185589] [PubMed: 11062271]
- 107.
- Jin H, Carlile C, Nolan S. et al. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot Cell. 2004;3(6):1664–1673. [PMC free article: PMC539027] [PubMed: 15590839]
- 108.
- Bever JD, Wang M. Arbuscular mycorrhizal fungi: Hyphal fusion and multigenomic structure Nature 2005433(7022)E34 (discussion E4) [PubMed: 15650700]
- 109.
- Fleissner A, Sarkar S, Jacobson DJ. et al. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot Cell. 2005;4(5):920–930. [PMC free article: PMC1140088] [PubMed: 15879526]
- 110.
- Xiang Q, Rasmussen C, Glass NL. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics. 2002;160(1):169–180. [PMC free article: PMC1461943] [PubMed: 11805054]
- 111.
- Leikina E, Chernomordik LV. Reversible merger of membranes at the early stage of influenza hemagglutinin-mediated fusion. Mol Biol Cell. 2000;11:2359–2371. [PMC free article: PMC14925] [PubMed: 10888674]
- 112.
- Chernomordik LV, Frolov VA, Leikina E. et al. The pathway of membrane fusion catalyzed by influenza hemagglutinin: Restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol. 1998;140:1369–1382. [PMC free article: PMC2132678] [PubMed: 9508770]
- 113.
- Zimmerberg J, Blumenthal R, Sarkar DP. et al. Restricted movement of lipid and aqueous dyes through pores formed by influenza hemagglutinin during cell fusion. J Cell Biol. 1994;127(6 Pt 2):1885–1894. [PMC free article: PMC2120276] [PubMed: 7806567]
- 114.
- Plonsky I, Zimmerberg J. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. J Cell Biol. 1996;135(6 Pt 2):1831–1839. [PMC free article: PMC2133954] [PubMed: 8991094]
- 115.
- Bonnafous P, Stegmann T. Membrane perturbation and fusion pore formation in influenza hemagglutinin-mediated membrane fusion. A new model for fusion. J Biol Chem. 2000;275(9):6160–6166. [PubMed: 10692407]
- 116.
- 117.
- Jones KT, Soeller C, Cannell MB. The passage of Ca2+ and fluorescent markers between the sperm and egg after fusion in the mouse. Development. 1998;125(23):4627–4635. [PubMed: 9806912]
- 118.
- Rustom A, Saffrich R, Markovic I. et al. Nanotubular highways for intercellular organelle transport. Science. 2004;303(5660):1007–1010. [PubMed: 14963329]
- 119.
- Onfelt B, Davis DM. Can membrane nanotubes facilitate communication between immune cells? Biochem Soc Trans. 2004;32(Pt 5):676–678. [PubMed: 15493985]
- 120.
- Onfelt B, Nedvetzki S, Yanagi K. et al. Cutting edge: Membrane nanotubes connect immune cells. J Immunol. 2004;173(3):1511–1513. [PubMed: 15265877]
- 121.
- Opoku-Serebuoh E. Transcriptional regulation of the eff-1 gene [Ph.D.] Farmington, CT: Genetics and Developmental Biology, University of Connecticut Health Center. 2005
- Fertilization (Mouse)
- Fertilization (Nematode)
- Myoblasts (Mouse)
- Myoblasts (Fruit Fly)
- Placental Syncytiotrophoblast Cells (Human and Mouse)
- Lens Fiber Cells (Mouse and Other Vertebrates)
- Macrophages/Osteoclasts (Mouse)
- Implanted Stem Cells (Mouse and Human)
- Epithelia (Nematode)
- Primary Mesenchyme (Sea Urchin)
- Embryonic Blastomeres (Leech)
- Haploid Mating (Yeast)
- Fungal Hyphae
- Summary of Molecules Driving Cell Fusion: Obstacles to Their Discovery
- Structural Origins of Cell Fusion Channels?
- Evolution and Cell Fusion
- References
- Cell-Cell Fusion: Transient Channels Leading to Plasma Membrane Merger - Madame ...Cell-Cell Fusion: Transient Channels Leading to Plasma Membrane Merger - Madame Curie Bioscience Database
- Replacement of Specific Neuronal Populations in the Spinal Cord - Madame Curie B...Replacement of Specific Neuronal Populations in the Spinal Cord - Madame Curie Bioscience Database
- The End in Sight: Poly(A), Translation and mRNA Stability in Eukaryotes - Madame...The End in Sight: Poly(A), Translation and mRNA Stability in Eukaryotes - Madame Curie Bioscience Database
- Origin and Evolution of DNA and DNA Replication Machineries - Madame Curie Biosc...Origin and Evolution of DNA and DNA Replication Machineries - Madame Curie Bioscience Database
- Polyomavirus Allograft Nephropathy: Clinico-Pathological Correlations - Madame C...Polyomavirus Allograft Nephropathy: Clinico-Pathological Correlations - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...