NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
One strategy for the control of malaria and other vector-borne diseases relies on the ambitious goal of depleting natural vector populations' ability to transmit the pathogen through the introduction and spread of an engineered genetic construct. In this chapter, we assess whether the data accumulated so far on the population genetic structure of Anopheles gambiae, the major human malaria vector in Africa and the one studied most extensively, can be used to predict the spread of such genetic construct within and between wild populations. We conclude that available data offer good qualitative description of An. gambiae population structure, but do not provide the necessary information on the processes shaping this structure. We explore biological and methodological issues that prevented derivation of quantitative descriptions of these processes, focusing on the estimation of the effective population size and gene flow between populations. We discuss plans for bridging the gap between our present knowledge and where we should be, and outline a protocol for the direct estimation of relevant population genetics parameters and quantitative assessment of their interaction through a field population perturbation study. Finally, the epidemiological importance of other vector species in sustaining malaria transmission is highlighted as an additional roadblock that needs to be considered as part of any comprehensive vector control strategy designed to substantially lower the burden of malaria that overwhelms Africa.
Introduction
Novel approaches for the control of malaria transmission through genetic alteration of their mosquito vectors have received considerable attention in the past decade.1,2 They rely on the effective spread of transgene(s), i.e., gene(s) engineered to reduce vector competence such as by conferring refractoriness against the parasite,3-5 within natural vector mosquito populations. This suggests that the basis of the control, e.g., the transgene(s), will first be introduced (artificially) into the natural vector population(s) and that it will subsequently be transmitted to the offspring, to the extent that, within several generations, practically all individuals of the target species will express the refractory phenotype. Genetic drive mechanisms that should speed-up the process or improve efficient heritability are being developed and have received at least proof of principle.6-8 However, one fundamental assumption of this strategy is that mating occurs between individuals that carry the transgene(s) and individuals that do not.9 In other words, the target vector population is assumed to be a single, randomly mating unit, whereas assortative mating in wild mosquito populations has been demonstrated and could affect the spread of a transgene.10-14 Successful spread of such transgenes therefore depends on our ability to describe the basic reproductive units (demes, see Box 1) that compose the vector system responsible for malaria transmission in Africa, to understand their genetic and population dynamics, and determine the forces that shape it. Lessons from past genetic control programs demonstrated that the population structure and population dynamics of the target population(s) determine which, if any, genetic control approaches would be appropriate for addressing a specific problem.15 A critical part of this is obtaining a quantitative understanding of the spatial and temporal population structure of the mosquito vector. Such data are needed as input parameters for constructing predictive models for the prospects of different strategies to introduce genes into these populations. This constitutes the rationale for most population genetics studies aiming at unravelling the genetic structure of African malaria vectors.
In the following, we assess whether the available population genetics knowledge provides a solid basis for predicting the spread of a gene within and among natural malaria vector populations, with an emphasis on Anopheles gambiae, the most important vector throughout Africa and the most likely target for genetic control. As such, members of the An. gambiae complex have been extensively studied providing the most detailed information on their population biology and genetic structure. A number of reviews have been published recently on the knowledge gathered to date on this species complex (see for example refs. 9, 16-22). We do not attempt to duplicate this work. Rather, we assess whether these data can be used to predict the spread of an introduced gene within and between natural An. gambiae populations. Finally, we discuss the expected impact of the transgenic approach over malaria transmission in Africa.
Predicting the Spread of a Gene within and among Natural Vector Populations
Implementation of a novel public health control operation on a magnitude of a continent demands the highest and most rigorous preparation.2,23 The introduction and spread of genes into natural vector populations to interrupt disease transmission cannot be imagined without the capacity to predict, with sufficient accuracy, the outcome of a release effort. Prediction of changes in allele (gene) frequencies over time and space depends upon reasonable estimates of key parameters of the processes that determine such changes. The relevant outcomes are (i) the time until establishment of the introduced gene locally, within a single deme and (ii) the time for the gene establishment in other demes via natural spread. Establishment is defined as fixation (frequency = 1) or the frequency of stable equilibrium for the introduced gene. Such predictions require knowledge of contemporary migration between demes, selection, and drift as well as estimating the key parameters of these processes. Box 2 lists a minimal set of parameters, estimates of which are required to predict future changes in allele frequencies. Although not an exhaustive list, predictions based on fewer parameters may provide questionable results. Estimates of some of these parameters are found in the literature however, they suffer from serious flaws.
The Selective Value of the Transgene
In the absence of a genetic drive mechanism, establishment and further spread of a gene conferring refractoriness to malaria infection in wild mosquito population(s) will essentially rely on its net selective value, i.e., the balance between the fitness cost of phenotypic expression of the (introduced) gene and the overall benefit for the mosquito by escaping the detrimental effect of parasite infection and possibly, protection from other pathogens as well.24-26 Depending on the gene(s) involved, and their underlying expression dynamics, maintenance costs might be fixed (if the gene is to be expressed constitutively) or conditional (if the gene is expressed in certain conditions, e.g., in response to parasite infection in which case, the evolutionary cost of refractoriness is obviously sex specific, because only female anopheles are exposed to malaria parasites, and a function of the probability that the mosquito becomes infected). Furthermore, it is likely that environmental factors and the genetic background of differentially adapted vector populations will modulate the balance of evolutionary cost and benefit of refractoriness. The norm of reaction of any gene to be introduced in the genome of a vector species therefore needs to be assessed across the range of genetic variability the target species possesses and the diverse environments it experiences to assure that the phenotype (i.e., refractoriness) is predictable.23,27,28 This is a formidable challenge because relevant parameters of neither the natural environment (temperature, humidity, diet, crowding…) nor the relevant genetic variability (nucleotide polymorphism, genome structure, chromosomal inversions, cytological position…) are clearly defined. Although insights can be gained from cage experiments, whether these are conducted in a laboratory or in semi-field conditions will reflect at best only a parcel of the outcomes expected in a species like An. gambiae in nature. In this respect, the analysis of the spread of the Kdr mutation conferring insecticide resistance is very appealing because it represents the spread of a new gene under selection in natural settings. This single-nucleotide mutation was originally described from West African An. gambiae populations29 that are known to be genetically and ecologically differentiated subpopulations.9,16,19 Despite an apparently obvious fitness benefit in areas of intensive insecticide use, the Kdr allele was found only in populations of the S molecular form of An. gambiae and not in sympatric populations of the M form.30,31 It was subsequently found in the M form after an apparent introgression event from the S form,32 and is now spreading in this form as well (Etang J, Fondjo E, Simard F, unpublished).33,34 In the sibling species, An. arabiensis, the Kdr mutation apparently emerged as an independent mutation.35 The actual geographic distribution of the Kdr mutation in the An. gambiae complex suggests fluctuating balance between evolutionary costs and benefits that might favor its spread under certain ecological conditions only.32-34 It is likely that similar limitations applies to any mutation or gene with a strong phenotypic effect.
However, in the case of genetically engineered mosquitoes, an efficient drive system should promote the spread of refractoriness allele(s), even in the face of unfavorable balance of evolutionary costs of refractoriness.24,25 Concerns about the stability of the genetic construct will need to be addressed separately,23,36,37 and the efficacy of the drive mechanism in promoting the spread of the transgene will need to be demonstrated under a variety of natural conditions.38 Indeed, although robust inferences were generated from theoretical work,6,24,25 experimental evidence for efficient drive mechanisms in mosquitoes has yet to be provided.37-40 Furthermore, as outlined above, the genetic structure of natural vector populations will mediate the spread of genes in space and time.
Limitations in the Knowledge of the Population Structure of African Malaria Vectors
Estimating Effective Population Size
The effective population size (Ne) reflects the degree to which a population is affected by random genetic drift.41 Genetic drift affects the stability of allele frequencies in populations over generations, such that large fluctuations in allele frequencies are expected in small populations, whereas small changes would occur in large populations.42,43 Hence, genetic drift influences the magnitude of genetic diversity within a population and the rate of differentiation between populations. Ne depends on demographic factors such as population density, dispersal, and the mating system. When population size varies among generations, Ne approximates the harmonic mean of the effective population sizes in each single generation, and hence is dominated by the smallest value.44,45 Episodes of small Ne (i.e., demographic and genetic bottlenecks) can be of great evolutionary significance because increased genetic drift during these periods can dramatically change allele frequencies and the distribution of the overall genetic variability within and between populations. In particular, a transient drop in Ne can favour the rise in frequency of alleles that otherwise would have been selected against due to fitness cost.
Several methods are available to estimate Ne from demographic or genetic data. They vary in the types of information they use, their sensitivity to various assumptions, and most importantly, they refer to somewhat different definitions of Ne.46-50 The most widely used genetic estimator derives Ne from the variance in allele frequencies between generations.42,44,51 The method has been used to estimate the effective population size of An. gambiae and An. arabiensis in a number of settings and using various genetic markers.43,48,51-54 Reported estimates of Ne were in the thousands for both species (but see ref. 53 for a geographically isolated An. arabiensis population) and significant differences in Ne were demonstrated between populations of An. gambiae.51 Overlooking such differences in Ne between populations leads to erroneous estimates of genetic differentiation, gene flow and divergence time.55,56 However, further interpretation of the results in a quantitative way had to be tentative, because the method relies on assumptions that do not hold true in populations of An. gambiae. These assumptions include random mating and equal reproductive potential across individuals, nonoverlapping generations, equal sex-ratio, and negligible selection, migration and mutation. Evidences showing that several of these assumptions are violated in natural vector populations have accumulated. Such violations can lead to severely biased Ne estimates. Further, Ne estimates derived through such moment-based estimators are biased upward.49 Often, confidence intervals around the estimated values of Ne were so wide that the estimate's biological significance was lost. Hence, although valuable to compare populations to one another, the available estimates of Ne are not suitable for use in predictive models of the spread of alleles within and between populations.
New methods are being developed to improve estimation of Ne, which appear robust over a wide range of realistic conditions due to relaxed assumptions.49 However, predicting the spread of an introduced gene in natural An. gambiae populations will require a detailed picture of the fluctuations, both in time and space, of the effective population size of target populations. Precise assessment of the number of reproductively active adults in a population is needed to plan the release effort, as well as the identification of the time and place where the natural population is most amenable to the genetic introgression of a new gene or allele. The geographical area associated with a deme and how this area varies in different environments may also be valuable for optimizing the release effort.
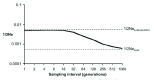
The above discussion assumes the existence of discrete demes as the building blocks of An. gambiae gene pool. However, some evidence suggests that the gene pool of An. gambiae in Africa is divided into few large subdivisions, within which isolation by distance applies.57-59 Under this model instead of discrete breeding units, there is a continuum where geographically closer populations are genetically more similar and reproductive adults disperse to all directions randomly with no barriers, except for their dispersal capacity and the limits of the subdivision or the species range.60-63 If the isolation by distance model accurately reflects the genetic structure of An. gambiae, then the values of Ne obtained so far do not refer to actual demes and are not useful. High rate of migration between populations acts as a buffer against genetic drift and results in estimate of Ne that increases as the period between the samples taken to estimate the variance in allele frequencies is longer, in sharp contrast to expectations if the estimate applies to a single deme (Fig. 1).64 Such results were obtained for an An. gambiae population from western Kenya (Lehmann et al unpublished) and for An. arabiensis in Cameroon.54 More studies are needed to determine if isolation by distance better describes the organization of the gene pool of An. gambiae and other malaria vectors in Africa before interpretation of the Ne estimates can be made.
Estimating the Level of Gene Flow between Populations
The principal malaria vectors in Africa (and typically elsewhere) are members of sibling (or cryptic) species complexes.65 Morphologically, the members of a complex are indistinguishable, reflecting that these species have diverged very recently. Some authors argued that speciation within the An. gambiae species complex, and most importantly lineage splitting between An. arabiensis and An. gambiae, occurred less than 4,000 years ago, as a byproduct of the development of agriculture in formerly unfavorable central African rainforest areas.16,17,66 As such, these species may retain substantial amounts of shared ancestral polymorphism because insufficient time has elapsed for reciprocal monophyly to establish.67 Post-mating reproductive barriers between members of the An. gambiae complex are incomplete, because only male hybrids are sterile but females are fertile, allowing some genetic exchange. Compelling evidence that such process occurred between An. arabiensis and An. gambiae in, at least parts of their genomes, has been provided in experimental as well as natural settings.68,69 As a result, discriminating between retention of ancestral polymorphism and genetic introgression proved problematic. Such discrimination has important evolutionary significance and implications for estimating the spread of a gene between vector species.
Retention of ancestral polymorphism also hindered interpretation of the description of the population structure of the major malaria vectors. Traditional population genetics inference is based on the analysis of variance in allele frequencies of putatively neutral markers. It relies upon a number of simplifying assumptions such as mutation-migration-drift equilibrium.70 Molecular signatures of recent demographic expansion have been detected in both An. arabiensis and An. gambiae and evidence suggests that neither of these species have reached equilibrium.57,71 Population expansion greatly reduces the rate of lineage sorting, resulting in inflated estimates of gene flow (Nm)72 by “historical” gene flow. Thus, high rate of gene flow do not necessarily reflects contemporary gene exchange between populations.73 Analysis of the population structure of vector species requires techniques that do not assume equilibrium and allow to distinguish between different models of gene flow and evolutionary scenarios explaining a given genetic structure (see ref. 74 and refs. therein).74
Population differentiation depends on the type of genetic markers used and the position of loci in the genome. DNA markers can exhibit dramatic variations in level of polymorphism due to locus-specific differences in the rate of mutation and to physical location in or near chromosomal inversions or loci under selection (a process that is known as ‘genetic hitchhiking’).75 Hence, results obtained from the same species using different types of markers or different sets of loci will not necessarily agree21,76,77 and summary statistics representing genome-wide trends must exclude outlier/deviant loci.59 Distinguishing locus-specific from genome-wide effects is a prerequisite for a correct description of population structure. Furthermore, estimates of genetic differentiation between populations depend on the analytic method used and the (evolutionary and demographic) model assumed.77 As no consensus has yet been reached, comparison across studies remains problematic.
With these limitations, it is not surprising that the low level of genetic divergence typically observed between natural populations of An. gambiae led to largely inconclusive results as far as contemporary gene flow is concerned.19,59,78 Similar finding seems to emerge from recent analyses conducted in the other major human malaria vector, An. funestus.79-81 However, few consistent trends have emerged providing a good qualitative description of the patterns of gene flow between An. gambiae populations. In the face of shallow geographical population structure between neighboring populations, recent studies revealed strong, if incomplete, barriers to gene flow between the molecular forms M and S of An. gambiae.18-20,59,82 Because both forms have extensively overlapping geographical and temporal distributions and are widespread throughout the continent,20 such findings suggest that genes might spread over large geographical areas, within one molecular form, before potentially invading the other form.14 This is reminiscent of the Kdr gene situation described above. However, the degree of differentiation between molecular forms appears very low over most of the genome, but is remarkably high in few small genomic regions not only because of paracentric inversions.59,77,83,84 Such semipermeable barriers to gene flow in a mosaic genome prompts further studies to identify regions of the genomes with different abilities to introgress between molecular forms and species within the An. gambiae complex.
Proposed Plan for Bridging the Gap
Population genetics studies produced robust description of the population structure, but they failed to quantify the processes that have shaped this structure. As stated by Gould & Schliekelman15 “Researchers working with classical genetic manipulations learned over and over again that there is no substitute for examining behavior of a genetically manipulated strain under local field conditions. This will not change in the future”. We echo their view and advocate that, the ultimate approach to estimate contemporary gene flow and derive robust estimates of all key parameters is by tracking new multiple mutations (genetic markers) that are experimentally introduced into natural populations by small scale release experiments.
Tracking new genetic markers and the lineages harboring them will provide a clear, complete, and nearly “assumption free” information to address the spread of a new gene over time and space in natural settings. Among population genetics approaches, direct tracking of genes under natural conditions has unparallel power to resolve alternative hypotheses, but its technical demands throughout its development and application as well as its ethical implications cannot be justified in every case. Here we outline the basic components, prominent advantages, and main challenges of this approach because, in our view, natural vector populations perturbation studies are indispensable for the development of every genetic control strategy, and will have to be implemented prior to the introduction of a functional gene(s) to alter the vectorial capacity phenotype of the vector.
The development phase of the experimental release of new makers (thereafter, ERNM) involves (a) colonization of mosquitoes from the region where experimental release is planned, (b) inducing multiple mutations spread throughout the genome by low intensity irradiation or chemical mutagenesis (or by inserting stable genetic tags using molecular methods) across the genome of a number of specimens, (c) derive a few iso-female lines from specimens carrying induced mutations by inbreeding over ca. twenty generations (desirable range) to produce practically homozygous lines and insure removal of most severely deleterious mutations, (d) after the lines have been inbred for several generations (i.e., successful breeding for over ca. 7 generations in outbred organisms would ensure overcoming the inbreeding depression that causes small colonies to crash), a few dozens of the newly induced mutations are identified and (e) molecular assays are developed for genotyping of field collected specimens. Efficient genome scanning tools (e.g., DNA chips) will allow identifying and later monitoring dozens or even hundreds of these genetic markers, thus maximizing the number of “loci” and minimizing the number of mosquitoes to be released and analyzed. The derived lines are ready for experimental release in the region where they originated. The release may require only few hundreds of mosquitoes per line, so no mass production is required. The application phase involves (a) identifying three release sites ca. 60-100 km apart and coordinating the release with all the relevant parties, (b) removing the same (or larger) number of females to be released prior to the release date and releasing the set numbers from one to three lines in each release site, (c) large samples of adults will be taken periodically from every release site for genotyping to determine the markers frequencies, (d) adult sampling of nearby populations will follow findings showing that some of the new markers have reached set frequencies at the release sites. Monitoring will involve genotyping of mosquitoes collected by a flexible sampling scheme that increases in the area surrounding the release site based on the data from previous dates.
ERNM can provide direct information on contemporary gene flow of alleles with various selective values (expected to vary between neutral and mildly deleterious) across geographic distance and various putative barriers to gene flow such as that separating the molecular forms of An. gambiae. A central element in ERNM is the replication in three independent sites in the same region, that together with the change over generations, facilitates separating systematic change in allele frequency due to selection from stochastic change due to drift, hence, providing means to estimate the selective value of each marker (assuming similar marker's selective values and drift in the three sites). Thus, the effects of chromosomal location and the selective value of the marker on gene flow will be estimated. The data can also provide accurate estimates of the effective population size and the deme's geographical area, without being confounded by migration. The variation between populations in these parameters will be obtained. The experimental release will provide comprehensive and direct information on all key parameters required for prediction of the outcomes of different genetic control strategies. Apart from providing additional population genetics (e.g., recombination rates under natural conditions in relation to the chromosomal position and inversions) and ecological (e.g., dispersal, longevity) parameters, it will provide practical information on the behavior and viability of the released mosquitoes and the effectiveness of various release strategies. Finally, release experiments in West, Central, and East Africa will facilitate comparison of results from different geographical and ecological regions.
The value of the experimental release for genetic control programs cannot be overestimated despite its logistical demands (e.g., above), but it also involves ethical challenges that must be addressed. The most important is the possible increased risk of disease transmission and personal irritation due to (1) a larger number of mosquitoes in the released area, and (2) a higher threat associated with mutagenized mosquitoes. Unlike typical genetic control programs, the experimental release aims at a partial and temporary introduction of a fraction of the markers (mutations) into populations. Thus, a single release of up to several thousand mosquitoes is required. The overall number of females in the area will not increase since the number released will be matched by the same or larger number of females removed (prior to the release). Further, cumulative sampling for monitoring the change in markers frequencies definitely will reduce the number of vectors in the area. Unlike introducing a new functional gene with expected phenotypic effects, ERNM uses randomly “sprinkled” mutations induced by irradiation or chemical mutagenesis, or by inserting a stable marker into multiple sites throughout the genome. Such mutations are expected to consist primarily of deleterious, slightly deleterious, and neutral mutations and therefore present a safe material for release. Notably, released mosquitoes originated from an area within 100 km of the release site, thus the risk of introduction of adaptive genes into the release area is negligible.
Clearly, the possibility of introducing a beneficial mutation (for the mosquito) can not be ruled out, but we stress that it is a very remote possibility and making the mosquito a more dangerous disease vector is even more unlikely. However, this point needs to be further evaluated and weighed against the risk of every intervention. In the case of developing a genetic control strategy using a functional gene attached to a genetic drive mechanism, the benefit of ERNM appears to outweigh its risks. Finally there is the possibility to release males that carry new markers on the Y chromosome only, thereby “disconnecting” the marker from the female phenotype. While informative in its unique way, it will not address many of the issues addressed using markers spread throughout the genome. Nevertheless it can be a starting point. Although developed to meet the needs of a genetic control program, ERNM can revolutionize population genetic research, especially if it provides different results from those derived based on classical population genetics approach.
Overall Impact on Malaria Transmission Intensity and Disease Burden
The successful introduction of a transgene into An. gambiae across Africa does not imply removal of malaria from the top of public health priorities in the continent. In fact, the expected impact of a successful spread of a transgene on malaria transmission is not clear. Epidemiological models dating back to the classical model of Macdonald-Ross85 have shown that considerable reduction in human exposure to infective mosquito bites is needed to achieve substantial impact on malaria morbidity and mortality in most parts of tropical Africa.86,87 With this in mind and using a simple population genetical and epidemiological model, Boëte and Koella24,25 demonstrated that even in conditions that allow the allele conferring refractoriness to reach fixation in the local vector population, the efficacy of refractoriness should be almost 100% (i.e., assuming no parasite escape from the refractory phenotype of its vector) for a significant effect on malaria prevalence.
Unlike classical means for vector control such as insecticide impregnated bednets or intra-domiciliary spraying that are directed to reduce exposure of peoples to infective bites by targeting anthropophilic and endophilic mosquitoes regardless of species, transgenesis-based methods target a single species. Hence, even if natural populations of An. gambiae became completely refractory to Plasmodium parasites Africa-wide (including all its chromosomal and molecular forms, and even extending this to the sibling An. arabiensis as well), other anophelines species will maintain transmission of malaria in large areas.65,88 The importance of these ‘neglected’ vector species in contributing to the overall malaria transmission must be considered when the question of the benefits expected from the release of transgenic mosquitoes is discussed.
In addition to the members of the An. gambiae complex, at least three species are considered as vectors of epidemiological importance in Africa: An. funestus, An. nili and An. moucheti. In certain areas, these vector species may contribute more to disease transmission than the members of the An. gambiae species complex.89-94 This is particularly the case in the humid savannas and forests of Central Africa, which remain largely unexplored.95-97 One example of this situation that demonstrates how little we know on malaria vectors in Central Africa is the recent description, based on morphological and molecular evidences, of a new species, member of the An. nili group.98 This newly described species appears to be the major malaria vector along rivers in South Cameroon. In such highly malaria endemic areas, eliminating malaria transmission by An. gambiae would change little the epidemiology of the disease and may even trigger unexpected worsening effects through insufficient decrease in transmission intensity.99,100 Only if the transgenic approach proved successful in An. gambiae and is extended to the other vectors, then this strategy could realize its outmost impact on disease prevalence.
Finally, we point out that unlike conventional means of control such as insecticides, drugs or vaccines, we can do nothing to halt the spread of an undesirable effect brought about by the transgene spread in the vector populations. Designing a “recall mechanism”, allowing halting the spread and possibly reversing it, would greatly improve the prospects and acceptability of the genetic control strategy.
Altogether, this discussion highlights serious limitations of our current ability to apply the genetic control strategy for malaria control in Africa. Current knowledge of vector populations and the epidemiology of malaria in Africa has lagged behind and its limitations call for caution when assessing the expected outcomes of a release of genetically altered vectors into the wild. However, the impressive progress in our understanding of the genetics and molecular biology of Plasmodium falciparum, its vectors and their interactions suggests that addressing these limitations is not beyond our reach.
References
- 1.
- Collins FH. Prospects for malaria control through the genetic manipulation of its vectors. Parasitol Today. 1994;10(10):3170–371. [PubMed: 15275536]
- 2.
- Varmus H, Klausner R, Zerhouni E. et al. Public health. Grand Challenge in Global Health. Science. 2003;302(5644):398–399. [PMC free article: PMC243493] [PubMed: 14563993]
- 3.
- James AA, Beerntsen BT, Capurro ML. et al. Controlling malaria transmission with genetically-engineered, Plasmodium-resistant mosquitoes: Milestones in a model system. Parassitologia. 1999;41:461–471. [PubMed: 10697903]
- 4.
- Coluzzi M, Costantini C. An alternative focus in strategic research on disease vectors: The potential of genetically modified nonbiting mosquitoes. Parassitologia. 2002;44:131–135. [PubMed: 12701373]
- 5.
- Besansky NJ, Hill CA, Costantini C. No accounting for taste: Host preference in malaria vectors. Trends Parasitol. 2004;20(6):249–251. [PubMed: 15147668]
- 6.
- Ribeiro JMC, Kidwell MG. Transposable elements as population drive mechanisms: Specification of critical parameters values. J Med Entomol. 1994;31:10–15. [PubMed: 8158612]
- 7.
- Davis S, Bax N, Grewe P. Engineered underdominance allows efficient and economical introgression of traits into pest populations. J theor Biol. 2001;212:83–98. [PubMed: 11527447]
- 8.
- Sinkins SP, Godfray HC. Use of Wolbachia to drive nuclear transgenes through insect populations. Proc Biol Sci. 2004;271(1546):1421–1426. [PMC free article: PMC1691734] [PubMed: 15306342]
- 9.
- Lanzaro G, Tripet F. Gene flow among populations of Anopheles gambiae: A critical review. In: Takken W, Scott T, eds. Ecological aspects for application of genetically modified mosquitoes. Wageningen Frontis Series. Vol. 2. Dodrecht, The Netherlands: Kluwer Academic Press. 2003:109–132.
- 10.
- Okanda FM, Dao A, Njiru BN. et al. Behavioural determinants of gene flow in malaria vector populations: Anopheles gambiae males select large females as mates. Malar J. 2002;1:10. [PMC free article: PMC140138] [PubMed: 12296972]
- 11.
- Charlwood JD, Pinto J, Sousa CA. et al. ‘A mate or a meal’ – Pregravid behaviour of female Anopheles gambiae from the islands of Sao Tomé and Principe, West Africa. Malar J. 2003;2:9. [PMC free article: PMC161788] [PubMed: 12801421]
- 12.
- Tripet F, Touré YT, Taylor CE. et al. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae. Mol Ecol. 2001;10:1725–1732. [PubMed: 11472539]
- 13.
- Diabate A, Baldet T, Brengues C. et al. Natural swarming behaviour in the molecular M form of Anopheles gambiae. Trans R Soc Trop Med Hyg. 2003;97:1–4. [PubMed: 16117970]
- 14.
- Tripet F, Dolo G, Lanzaro GC. Multilevel analysis of genetic differentiation in Anopheles gambiae s.s. reveal patterns of gene flow important for malaria-fighting projects. Genetics. 2005;169:313–324. [PMC free article: PMC1448890] [PubMed: 15677750]
- 15.
- Gould F, Schliekelman P. Population genetics of autocidal control and strain replacement. Annu Rev Entomol. 2004;49:193–217. [PubMed: 14651462]
- 16.
- Coluzzi M, Sabatini A, della Torre A. et al. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. [PubMed: 12364623]
- 17.
- Ayala FJ, Coluzzi M. Chromosome speciation: Humans, drosophila, and mosquitoes. Proc Natl Acad Sci USA. 2005;102(1):6535–6542. [PMC free article: PMC1131864] [PubMed: 15851677]
- 18.
- della Torre A, Fanello C, Akogbeto M. et al. Cytogenetic and molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10(1):9–18. [PubMed: 11240632]
- 19.
- della Torre A, Costantini C, Besansky NJ. et al. Molecular and ecological aspects of incipient speciation within Anopheles gambiae: The glass is half full. Science. 2002;298:115–117. [PubMed: 12364784]
- 20.
- della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol. 2005;35:755–769. [PubMed: 15894192]
- 21.
- Krzywinski J, Besansky NJ. Molecular systematics of Anopheles: From subgenera to subpopulations. Annu Rev Entomol. 2003;48:111–139. [PubMed: 12208816]
- 22.
- Powell JR, Petrarca V, della Torre A. et al. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia. 1999;41:101–114. [PubMed: 10697841]
- 23.
- Tabachnick WJ. Reflections on the Anopheles gambiae genome sequence, transgenic mosquitoes and the prospect for controlling malaria and other vector borne diseases. J Med Entomol. 2003;40(5):597–606. [PubMed: 14596272]
- 24.
- Boëte C, Koella JC. A theoretical approach to predicting the success of genetic manipulation of malaria mosquitoes in malaria control. Malaria Journal. 2002;1:3–9. [PMC free article: PMC111501] [PubMed: 12057019]
- 25.
- Boëte C, Koella JC. Evolutionary ideas about genetically manipulated mosquitoes and malaria control. Trends Parasitol. 2003;19(1):32–38. [PubMed: 12488224]
- 26.
- Moreira LA, Wang J, Collins FH. et al. Fitness of anopheline mosquitoes expressing transgenes that inhibit Plasmodium development. Genetics. 2004;166:1337–1341. [PMC free article: PMC1470781] [PubMed: 15082552]
- 27.
- Boëte C. Malaria parasites in mosquitoes: Laboratory models, evolutionary temptation and the real world. Trends Parasitol. 2005;21(10):445–447. [PubMed: 16099724]
- 28.
- Lambrechts L, Halbert J, Durand P. et al. Host genotype by parasite genotype interactions underlying the resistance of anopheline mosquitoes to Plasmodium falciparum. Malar J. 2005;4:3. [PMC free article: PMC548507] [PubMed: 15644136]
- 29.
- Martinez-Torres D, Chandre F, Williamson MS. et al. Molecular characterization of pyrethroid knock-down resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol Biol. 1998;7:179–184. [PubMed: 9535162]
- 30.
- Chandre F, Manguin S, Brengues C. et al. Current distribution of pyrethroid resistance gene (kdr) in Anopheles gambiae complex from West Africa and further evidence for reproductive isolation of the Mopti form. Parassitologia. 1999;41:319–322. [PubMed: 10697876]
- 31.
- Fanello C, Petrarca V, della Torre A. et al. The pyrethroid knock-down resistance gene in the Anopheles gambiae complex in Mali and further indication of incipient speciation within An. gambiae s.s. Insect Mol Biol. 2003;12:241–245. [PubMed: 12752657]
- 32.
- Weill M, Chandre F, Brengues C. et al. The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Mol Biol. 2000;9:451–455. [PubMed: 11029663]
- 33.
- Diabate A, Baldet T, Chandre F. et al. Kdr mutation, a genetic marker to assess events of introgression between the molecular M and S forms of Anopheles gambiae (Diptera: Culicidae) in the tropical savannah area of West Africa. J Med Entomol. 2003;40(2):195–198. [PubMed: 12693848]
- 34.
- Yawson AE, McCall PJ, Wilson MD. et al. Species abundance and insecticide resistance of Anopheles gambiae in selected areas of Ghana and Burkina Faso. Med Vet Entomol. 2004;18:372–377. [PubMed: 15642004]
- 35.
- Diabate A, Baldet T, Chandre F. et al. First report of a kdr mutation in Anopheles arabiensis from Burkina Faso, West Africa. J Am Mosq Control Assoc. 2004;20(2):195–196. [PubMed: 15264630]
- 36.
- Meister GA, Grigliatti TA. Rapid spread of a P element/Adh gene construct through experimental populations of Drosophila melanogaster. Genome. 1993;36:1169–1175. [PubMed: 8112576]
- 37.
- Brown EB, Bugeon L, Crisanti A. et al. Stable and heritable gene silencing in the malaria vector Anopheles stephensi. Nucleic Acids Res. 2003;31(15):e85. [PMC free article: PMC169974] [PubMed: 12888537]
- 38.
- Curtis C, Coleman PG, Kelly DW. et al. Advantages and limitations of transgenic vector control: Sterile males versus gene drivers In: Boëte C, ed. Genetically modified mosquitoes for malaria control. Georgetown: Eurekah/Landes Bioscience 2005 . (http//wwweurekahcom/ abstractphp?chapid=2731&bookid=188&catid=38)
- 39.
- Cateruccia F, Nolan T, Loukeris TG. et al. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature. 2000;405:959–962. [PubMed: 10879538]
- 40.
- Cateruccia F, Godfray HCJ, Crisanti A. Impact of genetic manipulation on the fitness of Anopheles stephensi mosquitoes. Science. 2003;299:1225–1227. [PubMed: 12595691]
- 41.
- Crow JF, Kimura M. Introduction to population genetics theory. New York:Harper and Row. 1970
- 42.
- Waples RS. Genetic methods for estimating the effective population size of cetacean populations. In: Hoelzel AR, ed. Genetic ecology of whales and dolphins. Cambridge, UK: International Whaling Commission (Special Issue nº13). 1991:279–300.
- 43.
- Taylor CE, Touré YT, Coluzzi M. et al. Effective population size and persistence of Anopheles arabiensis during the dry season in West Africa. Med Vet Entomol. 1993;7:351–357. [PubMed: 8268490]
- 44.
- Nei M, Tajima F. Genetic drift and estimation of effective population size. Genetics. 1981;98:625–640. [PMC free article: PMC1214463] [PubMed: 17249104]
- 45.
- Pollak E. A new method for estimating the effective population size from allele frequency changes. Genetics. 1983;104:531–548. [PMC free article: PMC1202093] [PubMed: 17246147]
- 46.
- Caballero A. Developments in the prediction of effective population size. Heredity. 1994;73:657–679. [PubMed: 7814264]
- 47.
- Berthier P, Beaumont MA, Cornuet JM. et al. Likelihood-based estimation of the effective population size using temporal changes in allele frequencies: A genealogical approach. Genetics. 2002;160:741–51. [PMC free article: PMC1461962] [PubMed: 11861575]
- 48.
- Taylor CE, Manoukis NC. Effective population size in relation to genetic modification of Anopheles gambiae sensu stricto. In: Takken W, Scott T, eds. Ecological aspects for application of genetically modified mosquitoes. Wageningen Frontis Series. Vol. 2. Dodrecht, The Netherlands: Kluwer Academic Press. 2003:133–146.
- 49.
- Tallmon DA, Luikart G, Beaumont MA. Comparative evaluation of a new effective population size estimator based on approximate bayesian computation. Genetics. 2004;167:977–88. [PMC free article: PMC1470910] [PubMed: 15238546]
- 50.
- Waples RS. Genetic estimates of contemporary effective population size: To what time periods do the estimates apply? Mol Ecol. 2005;14(11):3335–3352. [PubMed: 16156807]
- 51.
- Lehmann T, Hawley WA, Grebert H. et al. The effective population size of Anopheles gambiae in Kenya: Implications for population structure. Mol Biol Evol. 1998;15(3):264–276. [PubMed: 9501493]
- 52.
- Simard F, Lehmann T, Lemasson JJ. et al. Persistence of Anopheles arabiensis during the severe dry season conditions in Senegal: An indirect approach using microsatellite loci. Insect Mol Biol. 2000;9(5):467–479. [PubMed: 11029665]
- 53.
- Morlais I, Girod R, Hunt R. et al. Population structure of Anopheles arabiensis in La Reunion island, Indian Ocean Am J Trop Med Hyg 2005 . (in press) [PubMed: 16354815]
- 54.
- Wondji C, Simard F, Lehmann T. et al. Impact of insecticide treated bed nets implementation on the genetic structure of Anopheles arabiensis in an area of irrigated rice fields in the Sahelian region of Cameroon. Mol Ecol. 2005;14:3683–3693. [PubMed: 16202089]
- 55.
- Rousset F. Genetic differentiation within and between two habitats. Genetics. 1999;151:397–407. [PMC free article: PMC1460448] [PubMed: 9872976]
- 56.
- Nei M, Chesser RK. Estimation of fixation indices and gene diversities. Ann Hum Genet. 1983;47:253–259. [PubMed: 6614868]
- 57.
- Donnelly MJ, Simard F, Lehmann T. Evolutionary studies of malaria vectors. Trends Parasitol. 2002;18:75–80. [PubMed: 11832298]
- 58.
- Lehmann T, Hawley WA, Grebert H. et al. The Rift valley complex as a barrier to gene flow for Anopheles gambiae in Kenya. J Hered. 1999;90(6):613–621. [PubMed: 10589511]
- 59.
- Lehmann T, Licht M, Elissa N. et al. Population structure of Anopheles gambiae in Africa. J Hered. 2003;94(2):133–147. [PubMed: 12721225]
- 60.
- Wright S. Isolation by distance. Genetics. 1943;28:139–156. [PMC free article: PMC1209196] [PubMed: 17247074]
- 61.
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. [PMC free article: PMC1207888] [PubMed: 9093870]
- 62.
- Rousset F. Genetic differentiation between individuals. J Evol Biol. 2000;13:58–62.
- 63.
- Slatkin M. Isolation by distance in equilibrium and nonequilibrium populations. Evolution. 1993;47:264–279. [PubMed: 28568097]
- 64.
- Wang J, Whitlock MC. Estimating population size and migration rates from genetic samples over space and time. Genetics. 2003;163:429–446. [PMC free article: PMC1462406] [PubMed: 12586728]
- 65.
- Fontenille D, Simard F. Unraveling complexities in human malaria transmission dynamics in Africa through a comprehensive knowledge of vector populations. Comp Immunol Microbiol Infectious Diseases. 2004;27(5):357–375. [PubMed: 15225985]
- 66.
- Coluzzi M. The clay feet of the malaria giant and its African roots: Hypotheses and inferences about origin, spread and control of Plasmodium falciparum. Parassitologia. 1999;41:277–283. [PubMed: 10697869]
- 67.
- Avise JC. Phylogeography: The history and formation of species. Cambridge, MA: Harvard University Press. 2000
- 68.
- della Torre A, Merzagora L, Powell JR. et al. Selective introgression of paracentric inversions between two sibling species of the Anopheles gambiae complex. Genetics. 1997;146:239–244. [PMC free article: PMC1207938] [PubMed: 9136013]
- 69.
- Besansky NJ, Krzywinski J, Lehmann T. et al. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: Evidence from multilocus DNA sequence variation. Proc Natl Acad Sci USA. 2003;100(19):10818–10823. [PMC free article: PMC196886] [PubMed: 12947038]
- 70.
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration: Fst?1/(4Nm+1). Heredity. 1999;82:117–125. [PubMed: 10098262]
- 71.
- Donnelly MJ, Licht MC, Lehmann T. Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles gambiae. Mol Biol Evol. 2001;18:1353–1364. [PubMed: 11420373]
- 72.
- Wright S. Evolution and the genetics of populations: Variability within and among natural populations. Chicago: University of Chicago Press. 1978
- 73.
- Balloux F, Lugon-Moulin N. The estimation of population differentiation with microsatellite markers. Mol Ecol. 2002;11:155–165. [PubMed: 11856418]
- 74.
- Excoffier L. Special issue: Analytical methods in phylogeography and genetic structure. Mol Ecol. 2004;13:727. [PubMed: 15012751]
- 75.
- Barton N. Genetic hitchhiking. Phil Trans R Soc Lond B Biol Sci. 2000;355:1553–1562. [PMC free article: PMC1692896] [PubMed: 11127900]
- 76.
- Lanzaro GC, Touré YT, Carnahan J. et al. Complexities in the genetic structure of Anopheles gambiae populations in West Africa as revealed by microsatellite DNA analysis. Proc Natl Acad Sci USA. 1998;95:14260–14285. [PMC free article: PMC24361] [PubMed: 9826688]
- 77.
- Wang R, Zheng L, Touré YT. et al. When genetic distance matters: Measuring genetic differentiation at microsatellite loci in whole-genome scans of recent and incipient mosquito species. Proc Natl Acad Sci USA. 2001;98(19):10769–10774. [PMC free article: PMC58550] [PubMed: 11553812]
- 78.
- Gentile G, della Torre A, Maegga B. et al. Genetic differentiation in the African malaria vector, Anopheles gambiae s.s., and the problem of taxonomic status. Genetics. 2002;161:1561–1578. [PMC free article: PMC1462204] [PubMed: 12196401]
- 79.
- Costantini C, Sagnon NF, Ilboudo-Sanogo E. et al. Chromosomal and bionomic heterogeneities suggest incipient speciation in Anopheles funestus from Burkina Faso. Parassitologia. 1999;41:595–611. [PubMed: 10870569]
- 80.
- Cohuet A, Dia I, Simard F. et al. Gene flow between chromosomal forms of the malaria vector Anopheles funestus in Cameroon, Central Africa, and its relevance in malaria fighting. Genetics. 2005;169:301–311. [PMC free article: PMC1448888] [PubMed: 15677749]
- 81.
- Michel AP, Guelbeogo WM, Grushko O. et al. Molecular differentiation between chromosomally defined incipient species of Anopheles funestus. Insect Mol Biol. 2005;14(4):375–387. [PubMed: 16033431]
- 82.
- Wondji C, Simard F, Fontenille D. Evidence for genetic differentiation between the molecular forms M and S within the Forest chromosomal form of Anopheles gambiae in an area of sympatry. Insect Mol Biol. 2002;11(1):11–19. [PubMed: 11841498]
- 83.
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3(9):e85. [PMC free article: PMC1182689] [PubMed: 16076241]
- 84.
- Stump AD, Shoener JA, Costantini C. et al. Sex-linked differentiation between incipient species of Anopheles gambiae. Genetics. 2005;169:1509–1519. [PMC free article: PMC1449544] [PubMed: 15654109]
- 85.
- Macdonald G. The epidemiology and control of malaria. London: Oxford University Press. 1957
- 86.
- Smith TA, Leuenberger R, Lengeler C. Child mortality and malaria transmission intensity in Africa. Trends Parasitol. 2001;17:145–149. [PubMed: 11286800]
- 87.
- Trape JF, Pison G, Spiegel A. et al. Combating malaria in Africa. Trends Parasitol. 2002;18:224–230. [PubMed: 11983604]
- 88.
- Fontenille D, Lochouarn L. The complexity of the malaria vectorial system in Africa. Parassitologia. 1999;41:267–271. [PubMed: 10697867]
- 89.
- Antonio-Nkondjio C, Awono-Ambene P, Toto JC. et al. High malaria transmission intensity in a village close to Yaounde, the capital city of Cameroon. J Med Entomol. 2002;39:350–355. [PubMed: 11931035]
- 90.
- Antonio-Nkondjio C, Simard F, Awono-Ambene P. et al. Malaria vectors and urbanization in the equatorial forest region of south Cameroon. Trans R Soc Trop Med Hyg. 2005;99:347–354. [PubMed: 15780341]
- 91.
- Carnevale P, Le Goff G, Toto JC. et al. Anopheles nili as the main vector of human malaria in villages of southern Cameroon. Med Vet Entomol. 1992;6:135–138. [PubMed: 1421483]
- 92.
- Cohuet A, Simard F, Wondji C. et al. High malaria transmission intensity due to Anopheles funestus in a village of Savannah-Forest transition area in Cameroon. J Med Entomol. 2004;41(5):901–905. [PubMed: 15535619]
- 93.
- Dia I, Diop T, Rakotoarivony I. et al. Bionomics of Anopheles gambiae Giles, An. arabiensis Patton, An. funestus Giles and An. nili (Theobald) (Diptera: Culicidae) and transmission of Plasmodium falciparum in a Sudano-Guinean zone (Ngari, Senegal). J Med Entomol. 2003;40(3):279–283. [PubMed: 12943105]
- 94.
- Mendis C, Jacobsen JL, Gamage-Mendis A. et al. Anopheles arabiensis and Anopheles funestus are equally important vectors of malaria in Matola coastal suburb of Maputo, southern Mozambique. Med Vet Entomol. 2000;14:171–180. [PubMed: 10872861]
- 95.
- Hay SI, Rogers DJ, Toomer JF. et al. Annual Plasmodium falciparum entomological inoculation rates (EIR) across Africa: Literature survey, internet access and review. Trans R Soc Trop Med Hyg. 2000;94:113–127. [PMC free article: PMC3204456] [PubMed: 10897348]
- 96.
- Coetzee M, Craig M, le Sueur D. Distribution of African malaria mosquitoes belonging to the Anopheles gambiae complex. Parasitology Today. 2000;16(2):74–77. [PubMed: 10652493]
- 97.
- Levine RS, Townsend Peterson A, Benedict MQ. Geographic and ecologic distributions of the Anopheles gambiae complex predicted using genetic algorithm. Am J Trop Med Hyg. 2004;70(2):105–109. [PubMed: 14993618]
- 98.
- Awono-Ambene P, Kengne P, Simard F. et al. Description and bionomics of Anopheles (Cellia) ovengensis (Diptera: Culicidae), a new malaria vector species of the Anopheles nili group from South Cameroon. J Med Entomol. 2004;41(4):561–568. [PubMed: 15311444]
- 99.
- Trape JF, Rogier C. Combating malaria morbidity and mortality by reducing transmission. Parasitol Today. 1996;12:236–240. [PubMed: 15275204]
- 100.
- Reyburn H, Drakeley C. The epidemiological consequences of reducing the transmission intensity of P. falciparum In: Boëte C, ed. Genetically modified mosquitoes for malaria control. Georgetown: Eurekah/Landes Bioscience 2005. (http//wwweurekahcom/ abstractphp?chapid=2654&bookid=188&catid=38)
- Predicting the Spread of a Transgene in African Malaria Vector Populations: Curr...Predicting the Spread of a Transgene in African Malaria Vector Populations: Current Knowledge and Limitations - Madame Curie Bioscience Database
- Coronin 1 in Innate Immunity - Madame Curie Bioscience DatabaseCoronin 1 in Innate Immunity - Madame Curie Bioscience Database
- CTLA-4 in Type 1 Diabetes Mellitus - Madame Curie Bioscience DatabaseCTLA-4 in Type 1 Diabetes Mellitus - Madame Curie Bioscience Database
- Cell Adhesion Molecules - Madame Curie Bioscience DatabaseCell Adhesion Molecules - Madame Curie Bioscience Database
- Coronin Structure and Implications - Madame Curie Bioscience DatabaseCoronin Structure and Implications - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...