NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The generic tendency of proteins to misassemble into nonfunctional, and sometimes cytotoxic, structures poses a universal problem for all types of cell. This problem is exacerbated by the high total concentration of macromolecules found within most intracellular compartments but it is solved by the actions of molecular chaperones. This review discusses some of the basic evidence and key concepts relating to this conclusion.
Introduction
A recent article in the journal Nature recommended authors to begin their review with a story, a story that is amusing but also sets the scene. My story concerns a professor of biology who decided to test the knowledge of his students at the end of his lecture course. He pointed to a young woman in the front row and said to her “There is an organ of the human body that under appropriate circumstances can increase in size by a factor of six-fold. What is that organ and what are the circumstances?” The young woman blushed and exclaimed that she found this question so offensive she would report the professor to the head of the university. The professor ignored this response and repeated the question to the next student “What is the organ of the body that can increase in size by six-fold?” This student replied that this organ is the iris of the human eye, which expands from a pinpoint in bright light to wide open in dim light. “Correct” said the professor, who then returned to the first student. “I have two things to say to you. Firstly, you clearly have not been paying sufficient attention to my lectures and secondly, I predict that at some point in the future you are going to be greatly disappointed!”
The point of this story is that the first student made an unwarranted assumption, unwarranted in the quantitative sense. This point of this article is to explain why those people who study the properties of isolated macromolecules in uncrowded buffers are similarly making unwarranted assumptions about the quantitative relevance of their measurements to the properties of these molecules inside the cell. They are thereby ignoring the universal cellular feature that explains the existence of molecular chaperones.
Inside the Cell
The term ‘crowded’ is a quick way of referring to the fact that the total concentration of macromolecules inside cells is very high, a phenomenon called macromolecular crowding. The cartoon in Figure 1A represents the densities and shapes of the macromolecules in part of the cytosol of an animal cell, as drawn by David Goodsell and it is obvious why the term ‘crowded’ is appropriate. Note that the ribosomes are connected by strands of messenger RNA to form polysomes. Figure 1B shows an actual polysome, isolated from the salivary gland of an insect and spread out on the grid of an electron microscope. You can see the newly synthesized polypeptide chains growing longer as each ribosome slides along the messenger RNA, and you will notice that these chains are close together—within touching distance. You can also see that, as the chains get longer, they show signs of folding into more compact structures whilst still attached to the ribosomes.
This proximity of identical, partly folded chains in the crowded environment of the cytosol creates a danger—a danger that these chains will not fold and assemble correctly into functional proteins but will bind to one another to form nonfunctional misassemblies. It follows that protein misassembly is a universal cellular problem created by the advantage of making several protein chains at the same time from one molecule of messenger RNA. But the misassembly problem is even wider than this, because it affects mature proteins as well, which can partly unfold as a result of environmental stresses. The most distressing consequences of protein misassembly are the human neurodegenerative diseases, characterised by the occurrence of protein aggregates called amyloid plaques in the brain. In these diseases misassembly does not take place at the stage of protein synthesis but when mature proteins partly unfold, but the principle is the same—misassembly occurs when partly folded identical chains are close to one another.
In the remainder of this article I shall discuss the reasons for believing that the dramatic stimulation of protein misassembly by macromolecular crowding accounts for the existence of proteins acting as molecular chaperones to combat this problem. This discussion will focus on the historical origins of the molecular chaperone concept because the fact that most computer databases rarely extend back more than about two decades is leading to ignorance of how this paradigm shift occurred.
The Principle of Protein Self-Assembly: Yesterday and Today
If we ask why protein chains fold as they do, the answer was provided by the classic refolding experiments initiated by Christian Anfinsen around 1956. In this type of experiment a pure native protein is dissolved in dilute buffer and denatured by a high concentration of a chaotrope such as urea. This treatment causes the protein to unfold and thereby lose its biological properties, but if the concentration of chaotrope is lowered by 10 to 50-fold, Anfinsen observed that many of the chains refold into their original functional conformations.1 This type of experiment has been repeated many times with many proteins, and it is clear that the majority of denatured proteins will refold into their original conformations in the absence of either an energy source or other macromolecules—in other words, proteins can self-assemble spontaneously by a process requiring only their primary structures. Despite the fact that the rates and yields of refolded proteins are often too small to meet biological requirements, as was pointed out by Anfinsen,2 this observation led to the assumption that spontaneous assembly also happens inside the cell. This assumption was reasonable because a basic principle of scientific enquiry is Occam's razor, which advises us not to complicate hypotheses unnecessarily, but a series of observations made in the 1980s challenged this view. Two reports appeared that newly synthesized polypeptide chains bind transiently to a preexisting protein before they fold.
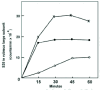
The first report concerns the synthesis of the photosynthetic enzyme rubisco in higher plants. Rubisco is a chloroplast enzyme, consisting of eight catalytic large subunits made by chloroplast ribosomes and eight structural small subunits made by cytosolic ribosomes. It is highly water-soluble, occurring at around 300g/l in the chloroplast stroma, but it is also one of the few proteins that fails to renature after dilution from a chaotrope. This failure stems from the extreme hydrophobicity of the large subunits, which aggregate with one another to form a water-insoluble precipitate. So we were surprised to find that when we allowed isolated chloroplasts to synthesize large subunits, these accumulate in a water-soluble form, despite the fact that they do not assemble into the holoenzyme until late in the incubation (Fig. 2). This solubility arises because newly synthesized large subunits bind transiently to another water-soluble protein before they assemble with small subunits.3 Sequencing of this large subunit binding protein revealed that it is 50% identical to the GroEL protein of Escherichia coli, a protein required for some bacteriophages to assemble in this bacterium but whose role in uninfected cells was unknown at that time. This identity was interpreted in terms of the molecular chaperone concept, and it was Sean Hemmingsen who coined the name ‘chaperonin’ for this family of molecular chaperone.4
The second report of a newly synthesized polypeptide binding to a preexisting protein before folding was found during experiments with a cultured cell line derived from lymphocyte precursors. These cells synthesize immunoglobin, an oligomeric protein composed of heavy and light chains. Haas and Wabl found that in a cell line of preB lymphocytes that are unable to make light chains, the heavy chains become bound to a preexisting small protein as they enter the lumen of the endoplasmic reticulum. They called this protein BiP and suggested that it is involved in the regulation of immunoglobulin assembly.5 BiP was later identified by Munro and Pelham as a member of the heat shock protein 70 family.6 We now know that both the chaperonins and the hsp70 chaperones assist the folding of many newly synthesized proteins in all types of cell.7
The Molecular Chaperone Concept
The term ‘molecular chaperone’ was coined by Ron Laskey in a Nature paper published in 1978 to describe a nuclear protein that solves a misassembly problem during the assembly of nucleosomes in amphibian eggs. Nucleosomes are octamers of basic histone protein bound to DNA by electrostatic interactions. Disruption of these interactions by high salt concentrations enables the histones to be separated from the DNA, but mixing these components together at physiological salt concentrations results in a failure of self-assembly—an insoluble precipitate forms instead of nucleosomes. Laskey and his coworkers discovered that this failure can be prevented by adding an abundant acidic nuclear protein called nucleoplasmin, which results in the correct assembly of nucleosome cores. The detailed mechanism is unclear but in general terms nucleoplasmin is thought to solve the misassembly problem by binding its acidic groups to the positively charged groups on the histones. This binding lowers their overall surface charge and allows the intrinsic self-assembly properties of the histones to predominate over the incorrect interactions favoured by the high densities of opposite charge. In the Discussion of this paper, the term ‘molecular chaperone’ is used for the first time.8
Control experiments show that nucleoplasmin does not provide steric information required for histones to bind correctly to DNA, nor is it a component of assembled nucleosomes. It is these latter two features that laid the basic foundation of our current general concept of the chaperone function, as I shall discuss later, but at this point I wish to emphasise that the role of nucleoplasmin is in the later stages of protein assembly, beyond the folding of newly synthesized histone chains, so the current common perception that chaperones are concerned solely with protein folding is incorrect and has been incorrect since this subject started. I suspect that this erroneous perception is retarding the search for chaperones that deal with the misassembly of folded subunits rather than with the problems posed by the folding of newly synthesized chains. It was only three years ago that a chaperone was found in red blood cells that mops up the excess of alpha subunits of haemoglobin that would otherwise misassemble and damage these cells.9 I predict that more examples of this type of subunit chaperone will be found in the future.
Ron Laskey did not use the term molecular chaperone to describe any other protein or develop the idea into a more general concept. This is where I enter the story. I read the Laskey paper in 1985 and it struck me that his observations with nucleoplasmin could be thought of in the same functional terms as the observations we had made about the rubisco binding protein, and others such as Hugh Pelham and Jim Rothman had made about hsp70. So I proposed in 1987 the existence of a new type of general cellular function, defined as ensuring that the folding of certain polypeptide chains and their assembly into oligomeric structures occur correctly.10
The term molecular chaperone rapidly caught on and the number of papers using this term contains to rise steadily, as you can see in Figure 3. The numbers of families of chaperone, as defined by sequence similarity, is now well over 30. Some of these families, but by no means all, are heat shock proteins and this reflects the fact that the need for chaperone function increases after proteins have been denatured by environmental stresses. Many chaperones are involved in protein folding but some are also assist disassembly processes, such as the remodelling of chromatin during fertilization and transcription, and the resolubilisation of insoluble aggregates that have escaped the attention of other chaperones. This accumulation of discoveries supports a new view of protein assembly. In this new view the principle of self-assembly is retained, but before chaperones, this assembly was thought to be spontaneous and require no energy expenditure, whereas now self-assembly is seen in many instances to require assistance by chaperones, some of which hydrolyse ATP. Thus the concept of spontaneous protein self-assembly has been replaced by the concept of assisted protein self-assembly. This new view was first articulated in a TiBS article in 1989 (see ref. 11), and has so far stood the test of time.
There is now a huge literature on the details of chaperone structure and function, but what I wish to concentrate on in this article is why chaperones exist. Why do cells need a chaperone function, given that most denatured proteins know how to fold correctly in the absence of other macromolecules?
The Problem of Protein Misassembly
There are two answers to this question. The fact that proteins are made by polysomes ensures that identical partly folded chains are close together, as I indicated earlier. Moreover, these chains grow vectorially so they cannot fold correctly until a complete folding domain has been made, raising the possibility that incomplete domains may misfold. In addition to these factors, polysomes function within a highly crowded compartment. All these features favour a process that competes with folding, the process of misassembly.
Misassembly is defined as the association of two or more polypeptide chains to form nonfunctional structures; these structures may be as small as dimers or large enough to be insoluble. This emphasis on function serves to distinguish misassembly from the formation of functional oligomers, which is thus termed oligomerization. Many authors use the term ‘aggregation’ to describe misassembly, and point to the amyloid fibrils found in the brains of people suffering from neurodegenerative disease as typical examples. The problem with this term is that there are increasing reports of amyloid structures that do have biological functions, so the essential distinction between aggregation and oligomerization has disappeared.12 Misassembly should be distinguished from misfolding, which I define as the formation of a conformation which cannot proceed to the functional conformation on a biologically relevant time scale. Misassemblies are by definition misfolded, but there are very few reports of renaturing proteins that misfold but remain monomeric. It seems likely that the vast majority of primary translation products are capable of folding correctly, provided they can avoid misassembly. It is for this reason that I suggest that the essential problem that chaperones have evolved to combat is misassembly rather than misfolding.
Protein misassembly has not been widely studied but two important aspects are established. It has been known for many years that misassembly competes with folding in an Anfinsen-type refolding experiment (Fig. 4A). Since misassembly is a high order process, it is very sensitive to the concentration of the unfolded chains, so as this concentration rises, misassembly outcompetes folding. The molecular basis for this competition is simply that polypeptide chains do not distinguish between inter- and intra-molecular interactions. Protein chemists traditionally solve this problem by lowering the concentration of the chains, and/or the temperature, but this solution is not open to cells.
A more surprising feature of misassembly is that in many cases it is highly specific - chains misassemble only if they are identical or very similar. This observation is interpreted to mean that the aggregating species have a degree of secondary or tertiary structure - in other words they are partly folded. This point was first established by adding total crude extracts of E. coli cells to the 8M urea in a standard Anfinsen refolding experiment applied to tryptophanase (Fig. 4B). The numbers marked with asterisks in the box indicate the enzymic activity of the tryptophanase chains that have refolded after dilution of the urea, and you can see that this activity is unaffected by the addition of 100 times as much total protein from E. coli to the urea-containing buffer. Thus the several thousand different unfolded polypeptide chains in the E. coli extract do not affect the competition between correct folding and misassembly.13 What does affect this competition however, is the phenomenon of macromolecular crowding.
Macromolecular Crowding
We use the term ‘crowded’ rather than ‘concentrated’ because in general no single macromolecular species occurs at a high concentration, but taken together, macromolecules occupy a significant fraction of the total volume. This fraction is in the range 8-40%. That is, 8-40% of the total volume is physically occupied by macromolecules and therefore is unavailable to other molecules, just as in a football crowd most of the space is occupied by people and is unavailable to other people—other people are sterically excluded. This steric exclusion of part of the volume generates considerable energetic consequences, whose magnitude is not generally appreciated because it is so counter-intuitive—8-40% reduction in available volume does not sound very much. You may think that it simply means that the concentration of macromolecules is 8-40% larger than it appears to be, but in reality this steric exclusion produces large highly nonlinear effects on the effective concentration of macromolecules. This nonlinearity results in an exquisite sensitivity of macromolecular properties to changes in their environment.
There exists a quantitative theory of crowding developed largely by Allen Minton, a biophysicist at the National Institutes of Health in America.14-16 This theory predicts two major consequences of crowding. The first prediction is that crowding will reduce the diffusion rates of both large and small molecules. There is good experimental evidence that diffusion is reduced in the range three-four fold in eukaryotic cells and eleven-fold in E. coli relative to the rate in water; prokaryotic cells appear to be more crowded than eukaryotic cells.17 Much more important with respect to misassembly however is the large increase in association constants for macromolecules. For example, let us consider the association of two 40 kDa monomers into a dimer. Suppose that the association constant for this oligomerization in water is 1.0. Crowding theory predicts that the value of this constant inside a cell of E. coli will be between 8 and 40, depending on what specific volume of the protein is assumed. If we suppose that the dimers can form a homotetramer, the effect is even larger—the association constant is now 10,000 (see ref. 18).
This dramatic consequence arises because crowding increases the effective concentration of macromolecules—in other words, it increases their thermodynamic activity. This increase in activity is produced by the reduction in excluded volume when molecules bind to one another. In fact macromolecular crowding is more precisely called ‘the excluded volume effect’. This term emphasises the fact that crowding is a purely physical nonspecific effect based solely on steric exclusion. As the number and the size of molecules in a solution increase, the less randomly they can be distributed. So as the concentration rises, the free energy of the solution also rises, because the entropy of each molecule becomes less. But if these molecules bind to one another, the volume available to them increases, their entropy therefore is greater and the total free energy of the solution decreases. In other words, the most favoured state is the state that excludes the least volume to other macromolecules.
This is a general conclusion—it applies not just to associating macromolecules, but to all processes where a change in excluded volume occurs. So it applies to the folding of newly synthesized polypeptide chains, the folding of nucleic acid chains, the unfolding of mature proteins as a result of environmental stresses such as high temperature, the condensation of DNA in chromosomes, the operation of motile systems such as actin filaments and microtubules, and, of particular relevance here, to the formation of protein misassemblies.
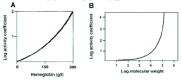
It is important to grasp that the effect of crowding on thermodynamic activity is highly nonlinear with respect to the concentration of the crowding agent. The term ‘crowding agent’ is used to describe the molecules that cause the crowding. Figure 5A shows how the activity coefficient, that is the ratio of the effective concentration to the actual concentration, varies with the concentration of haemoglobin.19 Note that this is a log/linear plot and that the activity coefficient rises in a nonlinear fashion with respect to the actual concentration. The concentration of hemoglobin in red blood cells is about 340 g/l, so you can see that the activity of haemoglobin inside the cell is more than two orders of magnitude greater than it is in the dilute buffer where its properties are commonly studied.
Figure 5B plots the activity coefficient against molecular weight for a test molecule placed in a background of hemoglobin at 300 g/l. So in this experiment the crowding agent is haemoglobin and we are asking how the activity of another molecule placed in that solution depends on the molecular weight of that molecule. Note that this is a log/log plot. You can see that the effect of crowding on activity becomes significant only after about 10,000 molecular weight.20 This is why we use the term ‘macromolecular crowding’ because the effect on the activity of small molecules such as metabolites and inorganic ions is small by comparison.
I have so far talked about the effect of crowding on thermodynamics. What about effects on kinetics? Figure 6 summarises the effects of crowding on reaction rates for a bimolecular association reaction.21 The vertical axis of the graph is the log of the association rate constant. We can consider two situations in turn. If the rate-limiting step is the encounter rate of the two components A and B, then the reaction is diffusion-limited. But crowding reduces diffusion so in this case the reaction rate will decrease as the concentration of crowding agent rises. However in the other situation, the rate-limiting step is the conversion of the activated complex (or transition state) to the dimer, so in this case the rate depends on the concentration of this complex. But crowding increases association, so the equilibrium between A + B and the activated complex is displaced to the right. Thus the reaction rate will rise as the concentration of crowding reagent increases. However, if you think about it, the absolute upper limit of any bimolecular reaction must be ultimately set by the encounter rate of the two molecules, so even for transition state-limited reactions, the rate must eventually come down as the concentration of the crowding agent gets high enough. So we have a resultant curve between two opposing effects.
Thus we see that the effect of crowding on reaction rate is complex; it depends on the nature of the reaction and where you are on the concentration axis. The sad fact however, is that although these dramatic effects of crowding have been known for at least 25 years, the vast majority of studies on isolated macromolecules, including those on protein folding, protein misassembly and molecular chaperones, continue to be done in uncrowded buffers. In my view this is a mistake. Until the effect of crowding agents becomes as routine to study as the effects of pH or redox potential, the extrapolation of interpretations made from studies of isolated systems to the intact cell must have a question mark hanging over them.22
Stimulation of Misassembly by Crowding Agents
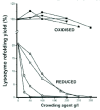
The first demonstration that crowding agents increase the misassembly of refolding chains was obtained with hen lysozyme.23,24 Oxidised lysozyme refolds impeccably, achieving almost complete recovery of enzymic activity in one or two seconds, but when the two four disulfide bonds are reduced, the chains misassemble in the refolding buffer, the extent depending on their concentration. Misassembly occurs because at least two disulfide bonds have to form by slow air oxidation before the chains collapse and during this time the chains have a chance to misassemble with one another. When several different crowding agents are added to the refolding buffer, all the reduced chains misassemble but the refolding of oxidised chains is unaffected (Fig. 7). Missasembly can be prevented by adding protein disulfide isomerase (PDI) to the refolding buffer. Low concentrations of PDI act by speeding up the rate of disufide formation, which stabilises the correctly folded chains, while higher concentrations act in a chaperone fashion. Further examples of the stimulation of protein misassembly by crowding are discussed in reference 25.
How do Chaperones Combat Misassembly?
If we survey the literature on the roles of many different chaperones in preventing the misassembly of newly synthesized polypeptides, the picture looks complex, but I suggest that the available data can be encompassed within just two basic principles of chaperone operation.7
It is useful to divide chaperones into two classes on the basis of size. Small chaperones are defined as those less than 200 kDa in size and include hsp70, hsp40, PDI and the very recently discovered cases of membrane chaperones i.e., proteins that occur inside membranes and prevent the aggregation of other transmembrane proteins.26 Small chaperones bind transiently to short hydrophobic sequences as these appear on nascent or just released polypeptide chains, and prevent them from both folding prematurely or misassembling together by binding to these sequences for a time. These chaperones do not appear to affect protein conformation, but function essentially by reducing the time potentially interactive surfaces are exposed by cycling on and off these surfaces until they are buried by folding. Such a simple mechanism can be thought of as analogous to tossing a hot potato from hand-to-hand until it has cooled, an analogy suggested by Ulrich Hartl.
Large chaperones, exemplified by the chaperonins such as GroEL, function by a much more sophisticated mechanism that uses what I call the Anfinsen cage principle.27 The chaperonins function essentially by providing a molecular cage made of one oligomer of GroEL capped by one oligomer of GroES. Single partly folded chains are encapsulated one at a time inside this cage. The enclosed chain continues to fold in the absence of other folded chains until the hydrophobic surfaces that cause misassembly are buried within the final folded structure. The time of folding inside this cage is set by the slow ATPase activity of the GroEL subunits. Completion of ATP hydrolysis allows ATP to bind to the opposite end of the GroEL/ ES complex from the enclosed protein, and this binding triggers via allosteric interactions the release of the folded chain into the cytosol.7
But suppose the released chain has not managed to bury its hydrophobic regions during the time it is enclosed? It would be dangerous to release such misassembly-prone chains into the cytosol but this problem is prevented by the crowded state of the cytosol. Experiment with crowding agents added to isolated GroEL/ES complexes by Jorg Martin and Ulrich Hartl show that crowding ensures that any such released, but partly folded chains, bind back to the same GroEL molecule from which they have just been released.28,29 So we reach the pleasing conclusion that the chaperonins use the crowded state of the cytosol to combat the problem of protein aggregation that has been created by crowding in the first place.
The Molecular Chaperone Function
My current definition of molecular chaperones is that they are a large and diverse group of proteins that share the property of assisting the noncovalent assembly and/or disassembly of other macromolecular structures, but which are not permanent components of these structures when these are performing their normal biological functions.30 It is important to note that this definition is functional and not structural, but it contains no constraints on the mechanisms by which different chaperones may act; this is the reason for the use of the imprecise term ‘assist’. Thus molecular chaperones are not defined by a common mechanism or by sequence similarity. In my view only two criteria need be satisfied to designate a macromolecule a molecular chaperone. Firstly, it must in some sense assist the noncovalent assembly or disassembly of some other macromolecular structure, the mechanism being irrelevant, and secondly, it must not be a component of these structures when they are performing their normal biological function. Note that the term ‘assembly’ is used in this definition in a very broad sense, and includes the folding of newly synthesized or stress-denatured proteins, the unfolding of proteins during transport across membranes, the association of monomers into oligomers and macromolecular disassembly processes.
All these considerations can be reduced to a simple unifying concept of the chaperone function, defined as the prevention and reversal of incorrect interactions that may occur when potentially interactive surfaces are exposed to the crowded intracellular environment. These surfaces occur on nascent and newly synthesized polypeptide chains, on mature proteins unfolded by environmental stresses, and also on folded proteins in near-native conformations, as in signalling proteins such as the steroid receptor. Thus the term ‘molecular chaperone’ is not a metaphor, nor an example of academic whimsy, but a precise description. The word ‘chaperone’ is appropriate because the traditional role of the human chaperone is to prevent incorrect interactions between pairs of people without either providing the steric information required for their correct interaction, or being present during married life - but often reappearing during divorce and remarriage!
Acknowledgements
I thank Robert Freedman for his generous provision of Departmental facilities during my retirement and Allen Minton for correcting my misconceptions about crowding theory.
References
- 1.
- Anfinsen CB. Principles that govern the folding of polypeptide chains. Science. 1973;181:223–230. [PubMed: 4124164]
- 2.
- Epstein CJ, Goldberg RF, Anfinsen CB. The genetic control of tertiary protein structure: Studies with model systems. Cold Spring Harbor Symp Quant Biol. 1963;28:439–448.
- 3.
- Barraclough R, Ellis RJ. Protein synthesis in chloroplasts. IX. Assembly of newly synthesized large subunits into ribulose bisphosphate carboxylase in isolated intact chloroplasts. Biochim Biophys Acta. 1980;608:19–31. [PubMed: 7388030]
- 4.
- Hemmingsen SM, Woolford C, van der Vies SM. et al. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. [PubMed: 2897629]
- 5.
- Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. [PubMed: 6417546]
- 6.
- Munro S, Pelham SRB. An hsp70-like protein in the ER: Identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. [PubMed: 3087629]
- 7.
- Young JC, Agashe VR, Siegers K. et al. Pathways of chaperone-mediated folding in the cytosol. Nature Rev Mol Cell Biol. 2004;5:781–791. [PubMed: 15459659]
- 8.
- Laskey RA, Honda BM, Mills AD. et al. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. [PubMed: 692721]
- 9.
- Kihm AJ, Kong YI, Hong W. et al. An abundant erythroid protein that stabilizes free alpha-hemoglobin. Nature. 2002;417:758–767. [PubMed: 12066189]
- 10.
- Ellis RJ. Proteins as molecular chaperones. Nature. 1987;328:378–379. [PubMed: 3112578]
- 11.
- Ellis RJ, Hemmingsen SM. Molecular chaperones: Proteins essential for the biogenesis of some macromolecular structures. Trends Biochem Sci. 1989;14:339–342. [PubMed: 2572080]
- 12.
- Fowler DM, Koulov AV, Alory-Jost C. Functional amyloid formation in mammalian tissue. PloS Biol. 2006;4:0001–0008. [PMC free article: PMC1288039] [PubMed: 16300414]
- 13.
- Zettlmeiss G, Rudolph R, Jaenicke R. Reconstitution of lactic dehydrogenase. Noncovalent aggregation vs. reactivation. 1. Physical properties and kinetics of aggregation. Biochemistry. 1979;18:5567–5571. [PubMed: 518855]
- 14.
- Minton AP. Molecular crowding: Analysis of effects of high concentrations of inert cosolutes on biochemical equilibria and rates in terms of volume exclusion. Methods Enzym. 1988;295:127–149. [PubMed: 9750217]
- 15.
- Minton AP. Implications of macromolecular crowding for protein assembly. Curr Opin Struct Biol. 2000;10:34–39. [PubMed: 10679465]
- 16.
- Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276:10577–10580. [PubMed: 11279227]
- 17.
- Ellis RJ. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr Opin Struct Biol. 2001;11:114–119. [PubMed: 11179900]
- 18.
- Zimmerman SB, Trach SO. Estimation of macromolecular concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. [PubMed: 1748995]
- 19.
- Minton AP. The effect of volume occupancy upon the thermodynamic activity of proteins: Some biochemical consequences. Mol Cell Biochem. 1983;55:119–140. [PubMed: 6633513]
- 20.
- Minton AP, Colclasure GC, Parker JC. Model for the role of macromolecular crowding in regulation of cellular volume. Proc Natl Acad Sci USA. 1992;89:10504–10506. [PMC free article: PMC50367] [PubMed: 1332050]
- 21.
- Zimmerman SB, Minton AP. Macromolecular crowding: Biochemical, biophysical and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. [PubMed: 7688609]
- 22.
- Ellis RJ. Macromolecular crowding: Obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. [PubMed: 11590012]
- 23.
- van den Berg B, Ellis RJ, Dobson CM. Effects of macromolecular crowding on protein folding and aggregation. EMBO J. 1999;18:6927–6933. [PMC free article: PMC1171756] [PubMed: 10601015]
- 24.
- van den Berg B, Wain R, Dobson CM. et al. Macromolecular crowding perturbs protein refolding kinetics: Implications for folding inside the cell. EMBO J. 2000;19:3870–3875. [PMC free article: PMC306593] [PubMed: 10921869]
- 25.
- Ellis RJ, Minton AP. Protein aggregation in crowded environments Biol Chem 2006 . (in press) [PubMed: 16740119]
- 26.
- Kota J, Ljungdahl PO. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J Cell Biol. 2005;168:79–88. [PMC free article: PMC2171667] [PubMed: 15623581]
- 27.
- Ellis RJ. Revisiting the Anfinsen cage. Folding and Design. 1996;1:R9–R15. [PubMed: 9162143]
- 28.
- Martin J, Hartl FU. The effect of macromolecular crowding on chaperonin-mediated protein folding. Proc Natl Acad Sci USA. 94:1107–1112. [PMC free article: PMC19752] [PubMed: 9037014]
- 29.
- Martin J. Chaperonin function - Effects of crowding and confinement. J Mol Recog. 2004;17:465–472. [PubMed: 15362106]
- 30.
- Ellis RJ. Chaperone function: The orthodox view. In: Henderson B, Pockley AG, eds. Molecular Chaperones and Cell Signalling. Cambridge University Press. 2005:3–41.
- 31.
- Goodsell DS. The Machinery of Life. Springer-Verlag. 1992;68
- 32.
- Kiseleva EV. Secretory protein synthesis in Chironomus salivary gland cells is not coupled with protein translocation across endoplasmic reticulum membranes. Electron microscopic evidence. FEBS Lett. 1989;257:251–253. [PubMed: 2583269]
- 33.
- Musgrove JE, Ellis RJ. The Rubisco large subunit binding protein. Phil Trans R Soc London B. 1986;313:419–428.
- 34.
- London J, Skrzynia C, Goldberg ME. Renaturation of E. coli tryptophanase after exposure to 8 M urea. Evidence for the existence of nucleation centers. Eur J Biochem. 1974;47:409–415. [PubMed: 4607014]
- Introduction
- Inside the Cell
- The Principle of Protein Self-Assembly: Yesterday and Today
- The Molecular Chaperone Concept
- The Problem of Protein Misassembly
- Macromolecular Crowding
- Stimulation of Misassembly by Crowding Agents
- How do Chaperones Combat Misassembly?
- The Molecular Chaperone Function
- Acknowledgements
- References
- Protein Misassembly: Macromolecular Crowding and Molecular Chaperones - Madame C...Protein Misassembly: Macromolecular Crowding and Molecular Chaperones - Madame Curie Bioscience Database
- p53's Dilemma in Transcription: Analysis by Microarrays - Madame Curie Bioscienc...p53's Dilemma in Transcription: Analysis by Microarrays - Madame Curie Bioscience Database
- Genetics, Mutations, and Polymorphisms - Madame Curie Bioscience DatabaseGenetics, Mutations, and Polymorphisms - Madame Curie Bioscience Database
- Tumor-Initiating Cells, Cancer Metastasis and Therapeutic Implications - Madame ...Tumor-Initiating Cells, Cancer Metastasis and Therapeutic Implications - Madame Curie Bioscience Database
- Dynamic Kinetic Modelling of Mitochondrial Energy Metabolism - Madame Curie Bios...Dynamic Kinetic Modelling of Mitochondrial Energy Metabolism - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...