NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The vertebrate nervous system is a major site of Hox gene expression and function. Studies on the patterns of expression, regulation and function of the vertebrate Hox gene family have played a key role in aiding our understanding of the basic ground plan of the CNS and processes that control how unique regional character is established and maintained in this complex organ system. This chapter will document the nature of the ordered patterns of Hox expression and link them with their regulation and functional roles in the nervous system.
Introduction
The adult vertebrate nervous system is a complex organ formed by a progressive series of intricately coordinated events that generate the components and circuitry essential for its function. While there is considerable diversity in the brain and CNS associated with higher order functions in vertebrates, underlying this final complexity there is a highly conserved basic genetic program that governs the fundamental patterns by which the central nervous system (CNS) is formed.
A central question is how this highly organized system arises in the body? For example, what ‘instructs’ a developing neuron to form in a particular location, correctly project to its targets or interact with other neurons? Part of the answer to these questions involves the products of the Hox transcription factor gene family that during development specify anterior-posterior (A-P) identity in different tissues, including the CNS. The purpose of this review is to highlight the expression, regulation and roles of Hox genes in patterning CNS development. A major focus will be placed on the hindbrain because of the considerable number of studies in different vertebrate models that demonstrate the important conserved role the Hox genes play in regulating the basic ground plan of this region of the CNS.
The Hox Gene Family
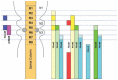
Initially identified and characterized in Drosophila as the genes associated with homeotic transformation, Hox genes were subsequently identified in vertebrates by virtue of their highly conserved homeobox sequences.1-5 From a concerted effort of many different laboratories, a total of 39 Hox genes have been identified in most vertebrates, except the ray-finned fishes. This large gene family is organized into four separate chromosomal clusters that can contain up to 11 genes with each complex (Fig. 1).5,6 Based on similarities between the putative translated sequences of the vertebrate Hox genes and the Drosophila Hox proteins, the Hox complex appear to have arisen by duplication and divergence from a common ancestor. The genes can be assigned into one of thirteen paralogous groups (Fig. 1). Thus, members of the labial (lab) subfamily, group 1 Hox genes, include Hoxa1, Hoxb1, and Hoxd1. Even though insect and vertebrates shared a common ancestor over hundreds of millions of year ago, their linear organization on the chromosome is maintain between complexes as well as conserved with the order found in Drosophila.7,8 However, unlike the Drosophila Hox complex, the vertebrate complexes are small (˜120 kb) and devoid of nonHox genes. In ray-finned fishes there has been a subsequent round of genome-wide duplication, generating up to 8 Hox clusters and many more Hox genes.9-12
Regardless of the species, one of the most fascinating features of Hox genes is the phenomenon known as colinearity.8,13-17 This term describes how the order in which each gene becomes activated matches their linear order along the chromosomal cluster. They are all transcribed in the same 5' to 3' orientation, with the 3' most genes expressed the earliest and with the most anterior borders of expression (Fig. 3). This process continues for each successive gene along the complex from the 3' to the 5' end. Consequently, the anterior borders of consecutively expressed genes are set more posteriorly than those of the earlier expressed genes, generating a nested and overlapping series of expression patterns. As a result of colinearity, unique combinations of Hox proteins are produced at different positions along the A-P axis of the developing embryo. This observation led to the proposal of a Hox Code whereby different combinations of Hox proteins generated by the nested domains of expression determine a cell's A-P placement and regional character within the developing hindbrain,16,18,19 limb,20-22 genitalia,23 and somites of the embryonic trunk.24,25
Hox proteins function as transcription factors to regulate other genes and hence play a pivotal role in the pathways that specify tissue identity. Each contains a homeodomain, translated from the sequence of the homeobox, which bears high amino acid sequence similarity to the homeodomain of the archetypical Hox member, Antennapedia. The homeodomain makes major groove contacts, via a helix-turn-helix motif, and minor groove contacts, via the N-terminal arm of the homeodomain, with DNA.26 From numerous loss-of-function and gain-of-function experiments in diverse vertebrate and invertebrate species, it has been shown that the anterior border of a Hox gene expression domain is important as perturbations of this border typically result in homeotic transformations and malformations.27,28 Here we review where these borders are set in the developing hindbrain of the CNS, what transcription factors and signaling pathways initiate the setting of these borders, and what are the consequences to CNS patterning when these borders are altered.
The Embryonic Vertebrate Nervous System
The central nervous system (CNS) begins as a thickened sheet of epithelial cells that forms the neural plate. Starting at the anterior end of the embryo, the lateral edges of the neural plate will roll upwards and fuse into the neural tube, while at the posterior end of the embryo the A-P axis is extended by cell proliferation, thereby elongating the trunk and neural plate. As the cells leave this proliferative region their A-P identity or regional character becomes determined. This process occurs in both the neural tube and the adjacent paraxial mesoderm as it differentiates into somites.
Since the development of the vertebrate embryo proceeds temporally in an anterior to posterior fashion, the rostral end of the embryo is more developmentally advanced than the caudal end. Following neural induction the neural tube becomes subdivided into morphologically distinct domains: the forebrain, midbrain, hindbrain, and spinal cord. Current models are consistent with the idea that the initial state of the neural tube is forebrain-like and that the more posterior regions (midbrain, hindbrain and spinal cord) arise through a progressive transformation to more posterior fates. This is referred to as the “activation and transformation” model of neural development.29-31 It is believed that distinct signaling pathways and processes govern each of these territories rather than a single common mechanism.32
The spinal cord can be further subdivided, according to the vertebra that will form to enclose it, into the cervical, thoracic, lumbar, and sacral domains. Within the cervical and thoracic domains, a third level of division can be identified as the brachial domain which spans vertebrae C5 to T1. These designations are important because different “columns” of MNs are generated in each domain. Hence in the brachial domain, the spinal cord will generate lateral column (LMC) neurons, whereas the remainder of the thoracic region will generate autonomic MNs.a The LMC neurons will send axons into the limb, whereas the autonomic MNs will project axons to the body wall muscles.33
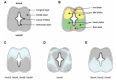
Within a transverse cross section of the spinal cord, there are differences in the dorsal-ventral (D-V) axis as different cell layers can be distinguished (Fig. 2A,B). The inner most layer, which lines the lumen of the neural canal, is the ventricular layer. This is composed of undifferentiated, proliferating cells. It is surrounding by the gray-colored mantle layer which contains differentiating neurons. The mantle layer is enclosed by the white-colored marginal layer that contains the nerve fibers. The mantle layer can be further subdivided into the dorsal alar and ventral basal plates. The alar plate forms the sensory area while the basal plate forms the motor area of the spinal cord.
So far we have described all of the major components of the CNS, whereas the peripheral nervous system (PNS) will arise from the neural crest. The neural crest is a transitory population of cells that delaminate from the border area located between the surface ectoderm and the neural plate as the neural tube is formed. After their induction, they undergo an epithelial-to-mesenchymal transition and migrate to give rise to numerous derivatives.34 In the head, the ‘first wave’ of neural crest migrate into the branchial arches while a ‘second wave’ migrates a short distance from the neural tube to form neural derivatives.
The brain stem is a relatively small region just anterior to the spinal cord, yet its functional significance is very great. The brain stem includes the midbrain and the hindbrain. The Hox genes are central to events involved in specifying the hindbrain territory35 and furthermore they seem to be part of a highly conserved mechanism for regulating properties of this territory in vertebrates. During development, the vertebrate hindbrain undergoes a segmentation process which subdivides the neuroepithelium into a number of compartments or rhombomeres, along the anteroposterior (AP) axis.32,36,37 Early cellular partitioning of the hindbrain into segments represents an important mechanism by which neuronal organization and diversification are initially established. The hindbrain is densely packed with vital structures. It contains the nuclei and fibers of the cranial nerves which innervate the muscles of the head and neck, transmit sensory information on hearing, balance and taste and control the cardio-vascular and gastrointestinal systems. In addition, it contains the sensory tracts ascending from the spinal cord to the thalamus and cortex and the motor pathways descending from the forebrain to the brain stem and spinal cord. The reticular formation of the rhombencephalon contains higher order relay centers that control respiration and blood pressure as well as centers that mediate arousal and wakefulness. The distinct neuronal groups in the hindbrain are also the major source of noradrenergic and serotonergic inputs to most parts of the brain. Brain stem patterning appears to use common cellular and molecular mechanisms to generate these features because they are a shared feature of vertebrates. To understand how patterns of neuronal connectivity are established and maintained, we first need to know how a given neuronal precursor comes to occupy its particular position in the nervous system, how it acquires a specific identity appropriate to that position, and how this translates into its specific differentiated properties.
Overt and Covert Segmentation of the Nervous System
In Drosophila, Hox gene expression is associated with the visible segmentation of the embryo.38 Misexpression and mutational analysis of fly Hox genes result in the affected segments acquiring another segment's identity (homeotic transformation).5 One obvious example of segmentation in the vertebrate embryo is the formation of the somites from the paraxial mesoderm that will give rise to the axial skeleton, in addition to most of the musculature of the trunk. Early experiments with vertebrate Hox genes showed that their role in segmental identity was conserved when their loss or misexpression resulted in the transformation, and in some cases malformations, of elements of the axial skeleton.24,25,39-47
A similar level of serial segmentation is seen in the hindbrain which visibly subdivides into a series of seven to eight compartments called rhombomeres (r).32,36,37 Morphologically these compartments are short-lived, but each acts as a separate entity displaying very little cell mixing between adjacent compartments.37,48 Even though each rhombomere will generate a similar pool of neurons, their number and axonal projections are rhombomere specific.49,50 Patterns of expression in combination with extensive gain- and loss-of-function analyses in different vertebrates have revealed that the distinct cellular and molecular characteristics of rhombomeres are the result of different combinations of Hox genes being expressed in different rhombomeres; the so-called Hox Code.16,19,51-70
Superficially the developing spinal column appears to lack the ordered pattern of segmentation that is displayed by the adjacent somites or by the hindbrain. Work with the chick neural tube suggests that it indeed lacks any form of direct segmentation71,72 but that its local A-P character is probably determined by the adjacent somites hence there is some evidence for indirect segmentation through this route.73-75 However, others argue that the symmetrical pattern of axonal projections along its length suggest that it displays some level of intrinsic segmentation. Regardless of the arguments of whether or not it is molecularly segmented, the adjacent somites as well as their sclerotome derivatives (the prevertebrae) are often been used as landmarks to position the anterior boundaries of Hox gene expression in the developing spinal column (Fig. 4).
Hox Gene Expression in the CNS
Hox Genes from the Groups 1 to 4 Are Expressed in the Hindbrain
As a result of colinearity Hox genes expressed in the hindbrain are from paralog groups 1-4, where 10 of the 12 members display segmentally-restricted domains of expression (Fig. 3).13,19 Members from groups 5 to 13 have anterior boundaries of expression which map in the spinal cord (Fig. 4).35 Although earlier work documented individual expression patterns of vertebrate Hox genes as they were initially cloned (for example; Hoxb5,76 Hoxa1,77,78 Hoxb1- Hoxb3,16 Hunt and coworkers (1991) systematically cataloged the expression patterns of Hox genes that are expressed in the developing hindbrain, branchial arches, and cranial neural crest.19 This work highlighted several important observations. First, genes within the same paralogous group are generally expressed at the same rhombomere boundary although relative levels of expression in each segment vary between paralogs. Thus, members of the 1st, 3rd, and 4th groups have anterior boundaries that coincide with the r3/r4, r4/r5, and r6/r7 boundaries, respectively (Fig. 3). Second, the boundaries between the genes in the groups 1 to 4 occur with a two-rhombomere periodicity. Thus, the anterior border of early Hoxb1 expression is positioned two rhombomere lengths anterior to the border of Hoxb3, and four rhombomere lengths anterior to the border of Hoxb4.
Of course, there are some exceptions to these observations. First, Hoxa2 and Hoxb2 have different boundaries, such that Hoxa2 is expressed up to the r1/r2 boundary79 while Hoxb2 is expressed up to the r2/r3 boundary.16,19 Hence, Hoxa2 is the only Hox gene expressed in r2. Second, the early pattern of Hoxb1 later becomes restricted to r4, while Hoxa1 is no longer expressed in the hindbrain.16,77,78,80 Thirdly the anterior boundary of Hoxc4 expression starts approximately one rhombomere length posterior to that of Hoxd4.81 This border does not appear to correspond to any rhombomere boundary, but starts near the beginning of the spinal column.
Within each group, there are differences in terms of timing and levels of expression. For example, Hoxa1 expression proceeds Hoxb1 expression, such that at 9.5 days post-coitum (dpc), the former is no longer expressed in the hindbrain.16,77,78,80 Within the group 3 paralogs, Hoxa3 and Hoxb3 initially have identical patterns of expression but the anterior boundary of Hoxb3 regresses posteriorly later in development.82 They both display high levels of expression in r5 but have lower levels of expression in posterior domains. On the other hand, Hoxd3 has weak expression in r5 but stronger expression in the posterior domains.19 There are similar levels of variation within the group 4 paralogs. Although the anterior borders of Hoxa4, Hoxb4 and Hoxd4 are set at the r6/r7 boundary, their most anterior domain displays weaker expression, within r7, than their posterior domains of expression in the spinal column (Fig. 3).13,19 Between the groups, there are differences in how the anterior boundaries are set. The initial anterior border of Hoxb2 expression persists at later stages and corresponds to the r2/r3 border. However, the degree to which the majority of Hox genes maintain or modulate their anterior borders is uncertain.83,84 There is confusion because many have assumed that the expression domains represent a single pattern that is maintained or altered. However, regulatory analyses detailed below indicate that the overall patterns of Hox expression arise by the cumulative patterns mediated by a wide variety of individual local and global cis-regulatory elements and many of these individual sub-set of expression are highly variable.27 Therefore, it is best to think of the patterns of Hox expression as continually dynamic rather than static in any way.
Hox Genes from Groups 5 to 13 Begin Their Expression in the Developing Spinal Cord
As with the Hox genes that begin their expression in the hindbrain, those that begin their expression in the spinal column display some general trends. The anterior border of most genes in groups 5 to 9 arise in the cervical region of the spinal cord while the anterior borders of groups 10 to 13 map to the lumbar region (Fig. 4).85 There are slight differences reported between the clusters. HoxA genes 10 through 13 have slightly more anterior borders than paralogs 10 to 13 from the HoxC and HoxD clusters (Fig. 4).85 HoxB members 5 through 9 have anterior borders that are more rostrally located in the cervical region of the spinal cord than their paralogs in the HoxA, HoxC, and HoxD clusters.14,85 On the other hand, the anterior border of Hoxb13 is more caudally located compared to group 13 paralogs from the other clusters.86
Group 1 to 4 Hox genes are also expressed in the spinal column, but their expression tends to be weaker than their expression in the hindbrain. For example, Hoxb1, Hoxb2 and Hoxb3 display higher levels of expression in the hindbrain but lower levels of expression in the spinal cord.16 Hoxb4 and Hoxd4 have a similar level of expression throughout the spinal cord with slightly elevated levels in the dorsal part.13 On the other hand, except for its most anterior domain, the expression of Hoxa4 appears to be low throughout most of the spinal cord.13
Many early papers reported the domains of Hox gene expression at a uniform level throughout the spinal cord. However, more detailed studies have shown a variety of differences in both the D-V and A-P axes whereby discontinuous domains and varying levels occur in distinct regions of the spinal cord. For example, the expression pattern of Hoxc4 gene displays a cyclic distribution pattern in the 12.5 dpc mouse CNS. It is strongly expressed from its anterior border near the end of the hindbrain down through the spinal column to an axial level that coincides with the adjacent developing 6th prevertebrae (pv). Hoxc4 expression is relatively lower between pv6 and pv21, but again is detected at a higher level from pv21-pv29/pv30 before then decreasing in more posterior regions.81 Another pattern had high levels in an anterior domain followed by a uniform lower level in more posterior regions. This is seen for both Hoxa7 and Hoxc8 expression (Fig. 4). The CNS domain of Hoxa7 expression starts adjacent to pv3 and quickly fades at ˜pv19 and is maintained at this lower level in the more posterior neural tube (Fig. 4).87 Similarly, the spinal cord domain of Hoxc8 expression starts at the level of pv4 and abruptly drops to low levels in the neural tube adjacent to pv14 (Fig. 4).88 Hence, Hox genes can be expressed in a continuous domain that continues to the caudal end of the neural tube, in a restricted domain that begins and ends within the neural tube, and/or display areas of higher and lower expression along its length.
The anterior borders of Hox expression are not statically fixed in the spinal cord following their initial activation. This is particularly true for Hox members from groups 5 and 6 whose expression begins in the spinal column but spread forward at later stages to reside near or within the hindbrain by 12.5 dpc (Fig. 4). For example, from 9.5 to 11.5 dpc, the anterior borders of Hoxb5, Hoxb6 and Hoxb8 expand toward the hindbrain before reaching their anterior-most border.89 At 11.5 dpc, Hoxb5's anterior border resides within the posterior hindbrain while the anterior borders of Hoxb6 and Hoxb8 are level with the somite (s) boundaries s2/s3 and s4/s5, respectively.89 Less dramatic changes have been reported for several group 9 to 13 members (Abd-B type). For example, the Hoxc11 anterior border moves rostrally from the level of the second lumbar (L2) vertebrae at 12.5 dpc to reside adjacent to the level of L1 at 13.5 dpc.90 So in addition to displaying dynamic levels of expression within A-P restricted domains, the initial anterior border of expression of many Hox genes may move more rostral as CNS development proceeds.
This dynamic repositioning of the anterior borders of expression in the developing neural tube helps to explain why there are discrepancies in the reporting of these borders. For example, the anterior border of Hoxc9 in the 12.5 dpc mouse neural tube is reported to be at the level of pv10 or pv6.88,91 Similarly, the anterior border of Hoxc10 is either at the level of pv20 or pv18.88,90b Furthermore, the shifts in A-P boundaries mean that assigning precise borders at any given stage is also complicated by the need for accurate staging of embryos. Another interesting complication in determining A-P boundaries of expression is that many Hox genes produce alternate spliced products. Therefore, differences in the probes used for a given gene can detect different mRNA species with varying boundaries. This is observed for Hoxa5 and Hoxb3 in the spinal cord.92,93
While most studies on Hox expression have concentrated on RNA levels, the distribution of the encoded proteins is the actual domain of functional activity. It is interesting that post-translational regulation has shown differences in the posterior aspects of Hox expression as revealed by RNA in situ hybridization. In general anterior borders of expression as determined by RNA or antibody approaches are in agreement for any given gene. However, there often appears to be a sharp posterior border not seen with the probes for RNA. For example, the initial distribution of Hoxc8 protein in the spinal cord was examined at different stages by raising antibodies against the protein's unconserved amino-terminus.94 In general, its protein's distribution occupied a smaller subset of the larger, relatively diffuse posterior distribution of its mRNA, being localized in the region that corresponds to the brachial domain of the spinal column. A more thorough examination of HoxC protein localization in the spinal cord was performed with antibodies against Hoxc5, Hoxc6, Hoxc8, Hoxc9 and Hoxc10.95 Jessell and colleagues found that each Hox protein displayed discrete staggered and overlapping domains within the developing neural tube of the chick. In general, the protein distribution of the various HoxC proteins corresponds to the anterior end of their mRNA profile. Thus, the domain of Hoxc5 protein was the most anteriorly located, in the cervical region, while Hoxc10 protein was the most posteriorly located in the lumbar region, while Hoxc6, Hoxc8 and Hoxc9 proteins occupied distinct domains in between. An implication of these studies is that overlapping RNAs for HoxC genes does not necessarily correlate with overlapping protein expression, which has implications for understanding posterior prevalence.
The specific distribution of HoxC proteins is crucial for specifying “columns” of motor neurons produced at different regions along the spinal cord96 in line with their dynamic D-V domains of restricted expression (see below). Thus Hoxc6 protein in the brachial region specifies LMC neurons that will go on to innervate the forelimb muscles, while Hoxc9 protein in the thoracic region specifies autonomic motor neurons that will innervate targets in the flank.96 Misexpression of either gene in the other's domain of expression results in the down-regulation of the endogenous gene with a corresponding switch in MN columnar identity.96 This is consistent with the idea that auto and cross-regulatory influences among the Hox genes are important for modulating their activity. Members from the other Hox clusters are also expressed in the spinal column, but only a handful of papers have looked at the effects that a single mutant,97,98 compound mutants,99-101 or misexpression102,103 have on MN projections.
Dorsal-Ventral (D-V) Patterning
The dynamic patterning of Hox gene expression in the A-P axis of the developing CNS is extended to the D-V axis as well. This is interesting as there is a precise ordered series of events associated with the birth of the major classes of neurons in the CNS.104 As development proceeds over time from anterior to posterior the anterior regions of the spinal cord are more developmentally advanced than the posterior domains. In a comprehensive analysis of the D-V patterns of HoxB expression it was observed that all of the genes in the cluster are initially activated throughout the entire D-V domain, but as development proceeds they become dorsally-restricted in a manner that correlates with the formation of commissural and sensory neurons.105 For example, at 10.5 dpc Hoxb3 to Hoxb9 are uniformly expressed throughout the D-V axis of the spinal cord.105 But by 11.5 dpc, their expression intensifies in the lateral regions of the spinal cord where the relay neurons are being formed (Fig. 2C). The expression pattern of Hoxb3, Hoxb6, Hoxb8 and Hoxb9 then intensifies in the alar plates, with little or no expression of these genes in the basal plates. This strong dorsal expression forms an ‘M’ shaped-pattern, where the central domain of HoxB expression drops slightly ventrally into the ventricular layer past the level of the sulcus limitans. By 14.5 dpc, expression of Hoxb8 and Hoxb6 increases in the basal plates, particularly in the anterior portion of the neural tube, and matches the strong expression seen in the alar plates. But as one moves further posterior within the spinal cord, this ventral domain of expression fades, while the strong expression in the alar plates persists.
When Graham and coworkers (1991) compared the expression pattern of Hoxb9 versus Hoxc8 in 12.5 dpc mouse neural tube, they found that while Hoxb9 exhibited the ‘M’ shaped pattern of strong dorsally restricted expression, Hoxc8 had an entirely different pattern of expression stronger in ventral domains. It was excluded from a central ‘V’ shaped domain of expression within the transverse section of the neural tube, instead displaying a strong middle stripe of expression projecting laterally away from the border of the ventricular zone with lower levels of expression within the ventricular zone (Fig. 2D). This correlates with the birth of motor neurons and is in agreement with the more recent studies of Jessell and colleagues showing the important functional role of HoxC genes in specification of motorneuron fates.33
Although this was only a comparison between two genes from different clusters, it suggested that each complex might have a distinct D-V pattern of expression unique from the other Hox clusters, thereby forming a D-V combinatorial code for cell fates specification.105, 106 Further support for this idea come from additional expression studies. Unlike the strong, lateral stripe of Hoxc8 expression, Hoxc4,81 Hoxc5 and Hoxc6107 display robust expression throughout the basal plates of the neural tube, with a dorsal extension of this domain in the central region of the mantle layer into the alar plates. Except for some overlap in the D-V border, this pattern of expression is complementary to the pattern of expression observed for HoxB members in the spinal cord's D-V axis.105 Knowing that the specific distribution of HoxC proteins along the A-P axis95 specifies which ‘column’ of MNs will form from the ventral domain,96 the distribution of HoxB protein in the dorsal domain may play an analogous role in determining the organization of the dorsal sensory area. Interestingly the targeted disruption of the Hoxb8 gene results in mutant mice that display excessive, pathological grooming.108 In trying to determine the mechanism of this behavior, Greer and Capecchi examined normal Hoxb8 expression in the adult mouse and found that it is expressed in regions of the CNS known as the obsessive-compulsive disorder (OCD) circuit in humans. Loss of Hoxb8 may have ‘short-circuited’ this neuronal network involved in innate behavior. It will be interesting to see what the effects that the loss of other HoxB members expressed within the spinal column has on behavioral-neuronal networks.
Regulation of Neural Expression
Given that their temporal and spatial expression must be precisely controlled, the regulation of Hox gene expression is understandably complex. It can be broken down into three stages: (1) initiation, (2) establishment, and (3) maintenance. Many cis-acting elements that control the initiation and establishment of Hox gene expression have been identified.27,109 The majority of these involve the regulation of the members from the first four paralog group because of interest in understanding how expression is regulated and coupled to hindbrain segmentation. Regulatory analyses have revealed that there are multiple rhombomeric enhancers that act as independent regulatory elements. They direct expression in specific-subsets of hindbrain segments and function outside of the Hox complexes on heterologous promoters linked to reporter genes.82,110-129 A variety of other regulatory elements that mediate neural expression in other domains have also been found.89,92,112,116-118,129-137
While these enhancers can display regulatory activity outside of the complex there is evidence that within a cluster some elements and Hox enhancers for other tissues may also be shared or competed for between adjacent genes.93,130,136,138-140 An interesting but open question is the extent to which these enhancers may work more globally within a complex. In addition to these local transcriptional control elements, Hox expression can be controlled post-transcriptionally by the recently discovered Hox specific miRNAs.141,142
Local Regulators
The identification of local cis-regulatory enhancers capable of regulating segmental expression in the hindbrain opened the way for further analyses to identify upstream factors that serve to direct these patterns of express and help build a picture of the upstream regulatory network of Hox genes in hindbrain segmentation. Here, we will focus on progress made in identifying upstream regulators that initiate segmental patterns of Hox gene expression in the hindbrain and craniofacial development.
Activating Protein-2 (AP-2) Regulates Expression of Hoxa2 in the Cranial Neural Crest
Ap-2 genes code for transcription factors that contain a proline-rich trans-activation domain, a dimerization domain, and a basic helix DNA-binding domain.143 They bind to DNA as homodimers and heterodimers. Initially, Ap-2 is expressed in extra-embryonic tissue,144 and subsequently becomes expressed in premigratory neural crest cell, remaining actively expressed in these cells during their migration and differentiation.144-146 Since AP-2 and Hox genes are both expressed in cranial neural crest cells, it is interesting to find that the expression of Hoxa2 in neural crest cells migrating into the second branchial arch is dependent upon an enhancer that contains and requires Ap-2 binding sites.120 Ap-2 is expressed in crest cells that do not express Hoxa2 indicating that mechanisms restricting its activity on the Hoxa2 enhancer must be working in head development. To date AP-2 binding sites have not been characterized for other Hox genes that are expressed in cranial neural crest.
kreisler (kr)
kreisler (kr) is a classic mouse mutation generated in an X-ray mutagenesis screen.147 Affected mice have inner ear defects accompanied by posterior hindbrain malformations in the r5 and r6 territories.148,149 The locus responsible for the kr phenotype was identified and it was found that the mutation was the result of a chromosomal inversion of the kr gene.150 The kr gene encodes the KRML1 protein, a member of the Maf B/Zip protein family.150 The expression of this gene normally starts at 8.0 dpc and rapidly decreases after 9.5 dpc.150 It is strongly expressed in r5 and r6, with a sharp rostral border at r4/r5 boundary and a more diffuse caudal border in the vicinity of r6/r7. In addition, kr is expressed in the cranial neural crest derived from the caudal hindbrain but its expression is not detected in the otic vesicle that lies adjacent to r5 and r6. This pattern of expression correlates with the morphological defects seen in r5 and r6; however, patterning defects are also present in r3 and r4 which are outside the normal expression domains of kr.115,151,152 A mutation in the zebrafish version of kreisler (valentino) also has been shown to have a conserved role in regulating hindbrain segmentation.53,55,153
In kr embryos, the caudal border of Hoxb1 expression, normally at r4/r5 is fuzzy and extends more posteriorly. Similarly, the anterior borders of Hoxb3 and Hoxb4, normally at r4/r5 and r6/r7 respectively, are poorly defined. Consequently, there are missing cranial nerves and the affected patterns of gene expression in the presumptive r5/r6 area suggests that these specific rhombomeres fail to form.149,151 In the absence of this gene this territory adopts characteristics of r4 and r5 fails to form, r6 is present.115,154
A direct role for KRML1 in Hox gene expression was first shown for Hoxb3 where enhancers in both mouse and chick responsible for r5-restricted expression of this gene were identified.113 Subsequently, an r5/r6 enhancer for Hoxa3 was also identified and found to be under direct regulatory control by KRML1.115 Additional experimental evidence for a direct role of KRML1 in the regulation of group 3 paralogs in r5 and r6 was provided by the ectopic expression of kr in r3.155 This induced expression of Hoxa3 and Hoxb3 in r3 thereby transforming it into an r5-like segment.155
Krox20
Krox20 encodes a zinc finger DNA-binding protein that is homologous to the Drosophila gap gene Krüppel. In mice, chickens, fish and Xenopus embryos, Krox20 is transiently expressed in presumptive r3 and r5.54,156,157 After the rhombomeres have formed, Krox20 is sequentially down-regulated in r3 and then r5.156,157 Targeted inactivation of Krox20 causes mutant mice to die shortly after birth.158,159 Histological examination reveals that nerves derived from the embryonic hindbrain are fused and disorganized. This defect is associated with a failure to maintain early r3 character and a failure to specify r5.158-160
Functional Krox20 binding sites have been identified in enhancers of murine Hoxa2, Hoxb2, and Hoxb3.114,124,125,128,161 The Krox20-dependent enhancers of Hoxa2 and Hoxb2 drive expression of these genes in r3 and r5125,128 and are conserved in both chicken and pufferfish.123,124 Regulation of Hoxb3 expression in r5 requires both Krox20 and KRML1.114 Krox20 binding sites have also been identified in the promoter regions of Hoxa4,162 and human HOXA7 and HOXA9.163,164 However, none of these genes are expressed in r3 and r5. These Krox20 binding sites may regulate expression of these genes in other tissues where Krox20 is expressed such as in the myelinating Schwann cells, chondrocytes, or osteoblasts. Krox20 has also been found to regulate the segmental expression of EphA4 in r3 and r5 of the hindbrain.165
Nuclear Factor Y (NFY) and Ying-Yang 1 (YY1)
NFY is a ubiquitously expressed transcription factor166,167 that interacts with the histone acetylase P/CAF168 and the coactivators p300/CBP.169 Hence, it acts as a transcriptional activator. On the other hand, YY1 is a zinc finger transcription factor that displays a dual nature, being capable of recruiting either histone acetyltransferases (HATs) or histone deacetylases (HDACs).170 Since HATs acetylate amino-terminal histone tails thereby reducing the interactions between neighboring nucleosomes, they make DNA more accessible for transcription factors. On the other hand, HDACs deacetylate histone tails, making DNA more compact and less accessible. Together, these two factors, NFY and YY1, have been found to be important for a subdomain of Hoxb4 expression in the neural tube.171 More specifically, NFY binds to a region within Hoxb4's Region C117 and requires YY1 to stabilize its binding to the NFY binding site within this element.171 Given the central role of HATs in gene activation and NFY's ubiquitous expression pattern, one would predict that other NFY binding sites exist in other Hox regulatory elements, but so far additional function sites have not been reported.
Sox and Oct
More than twenty Sox genes are found in vertebrates, the prototype of which is the Sry gene.172 These genes code for proteins that bind to DNA through a high mobility group (HMG) domain. This domain interacts with the minor groove and induces a dramatic bend in the DNA. On its own, the binding of Sox to naturally occurring enhancers is often not enough to exert regulatory activity because many of these family members lack an activation domain. Therefore to recruit coactivators there is frequently an adjacent DNA binding site. A common copartner for Sox appears to be Oct family members.173-175 This Sox-Oct partnership appears to be relevant in the regulation of Hoxb1 as there is an enhancer that mediates r4-restricted expression (the Hoxb1 autoregulatory element, ARE)110 and Sox and Oct cooperatively bind to a highly conserved bipartite binding site found within this element.176 Mutation of this SOX-OCT binding site reduces reporter activity in P19 embryonal carcinoma (EC) cells but does not abolish r4-restricted expression of the transgene in mouse embryos.176 Instead, the loss of this bipartite site alters the ability of the ARE to respond to retinoic acid.176
Global Regulators
Global regulators are factors or mechanistic features that simultaneously affect the expression of many Hox genes within a complex. In many instances, evidence for their direct action on Hox gene expression (i.e., the identification of cis-acting elements) has not been demonstrated. Two types of global regulators are major embryonic signaling pathways: the retinoic acid (RA) and fibroblast growth factor (FGF) signaling pathways. In a variety of tissues these signaling pathways act in an antagonistic fashion and it has been found that they differentially regulate Hox gene expression in the hindbrain and spinal cord. Their ability to regulate neural patterning can depend upon integration by a third global regulator, Cdx, a transcription factor whose expression is modulated by both of these signaling pathways.
Cdx Mutants Result in Posteriorization of Hox Anterior Expression Borders
Caudal (cad) was initially identified in Drosophila177 where it is expressed in the fly embryo in a gradient that peaks at the posterior pole.178 Using the Drosophila cad cDNA as a probe, a murine caudal ortholog, Cdx1 was identified.179 Subsequently, a total of three Cdx genes have been identified in vertebrates.c These genes code for proteins that have a highly conserved homeodomain and thus they function as transcription factors.
During early mouse embryogenesis, Cdx2 and Cdx4 are first expressed in extra-embryonic tissues,180,181 whereas expression of Cdx1 is first detected in the ectoderm and mesoderm of the primitive streak.182 By 8.5 dpc, all three genes are expressed in an overlapping pattern in the neural tube and mesoderm. As the trunk of the embryo elongates and somitogenesis proceeds, their anterior borders of expression recede posteriorly. This dynamic pattern of expression in the trunk is conserved amongst the vertebrates [chick,183Xenopus,184 zebrafish,185,186].
A role for Cdx in Hox gene regulation in the mouse was revealed by the targeted disruption of Cdx1.187 These mutant mice display vertebral homeotic transformations that coincide with a posterior shift in the anterior borders of several Hox genes in the prevertebrae. In the homozygous state targeted disruption of Cdx2 is embryonic lethal; however, heterozygous Cdx2 mutants display homeotic transformations of the axial skeleton.188 Products from both these genes were shown to have a synergistic role in setting Hox anterior borders when these two mouse mutants were crossed. Thus, compound mutants (Cdx1-/-, Cdx2+/-) have greater axial skeletal defects and greater posterior shifts in Hox gene expression than those present in single mutants.189
Work in Xenopus shows that over-expression of different Xenopus cad genes affects different subsets of Hox genes. For example, over-expression of Xcad2 results in microencephaly and shortening of the A-P axis.190 These morphological changes are accompanied by repression of Hoxd1, Hoxb1, Hoxb3 and Hoxb4 expression, and an anterior shift in the expression domains of Hoxc6 and Hoxb9. On the other hand, over-expression of Xcad3 does not affect Hoxb1 and Hoxb3 expression but does stimulate expression of Hoxc6, Hoxa7, Hoxb7 and Hoxb9.191 However, as with over-expression of Xcad2, it does anteriorize the borders of Hoxa7 and Hoxb9 resulting in embryos with anterior truncations.191 When a dominant negative form of Xcad3 is over-expressed in Xenopus embryos, expression of Hoxc6, Hoxa7, Hoxb7 and Hoxb9 are blocked.191 These studies show that there is a correlation between altered Cdx expression and Hox gene expression but do not establish a direct interaction.
In the mouse functional Cdx sites have been characterized in the mesodermal enhancers of Hoxa5,192Hoxa7,193,194Hoxb8,195 and Hoxc8196 thus arguing that Cdx directly affects Hox gene expression. A scan of the genomic sequences of many Hox genes reveals that they can contain one to four putative Cdx1 binding sites.187 Although many of these putative Cdx sites may not be functional, the resulting shifts in the anterior borders of Hox genes in the Cdx mutants suggest that Cdx plays a central role in fine-tuning the establishment of these borders. The degree to which Cdx regulates neural expression is not clear. Most of the putative Cdx sites described above reside in mesodermal regulatory elements. However there is evidence in chick that Cdx may play a global role in regulating neural expression in the context of RA and FGF signaling.197 We will further highlight the central role that Cdx plays as an integrator of FGF and RA signaling in section 5.3.
Retinoic Acid (RA) Signaling
Early experiments in which pregnant rats were maintained on vitamin A-deficient (VAD) diets resulted in fetuses with malformations in their visual, respiratory, vascular and urogenital systems.198- 200 These defects could be reversed by supplementing the VAD diet with vitamin A. However, the timing of its administration is important to prevent a given malformation.200 At the time, these results suggested that vitamin A is important for early developmental events. We now know that retinoic acid (RA) is the active derivative of vitamin A.
Too much or too little RA results in embryological defects. When excess RA is applied during gastrulation and neurulation, embryos display abnormal brain, head and vertebral development.24,201-209 These abnormalities include anterior displacement of the otic vesicles, expansion of the anterior hindbrain and spinal cord, and the formation of the somites at more anterior locations. Conversely, reducing or eliminating RA signaling results in the partial transformation of the rhombomeres or the complete loss of the posterior hindbrain and anterior spinal cord depending on the severity of the perturbation to the RA signaling pathway.210-215 In either case, the resulting abnormal patterning of the hindbrain in these embryos coincided with misexpression of the 3' Hox genes in this structure.202,213,216,217
Early cell culture experiments showed that Hox genes display another form of colinearity associated with RA signaling: genes at the 3' end of the complex respond rapidly to RA while genes positioned in more 5' positions respond slowly.218-220 For several of these Hox genes, their response to RA is directly mediated through retinoic acid response elements (RAREs) identified in their adjacent sequences. Functional RAREs have been identified for group 1,111,112,221-223 group 4,116,129,131,224,225 and recently for group 5 Hox genes.226 In transgenic mice assays the individual mutation of several of these RAREs reduces or eliminates neural expression of the reporter,112,116,224,225 arguing that they play a pivotal role in the endogenous gene's RA-mediated CNS expression. Unexpectedly, the genetic deletion or mutation of these neural-specific RAREs only delays and diminishes their cognate Hox gene's expression producing mice that are viable and fertile.70,227 Even the generation of the compound mutant of these altered RAREs produces only mild facial motor nerve defects and results in low perinatal lethality amongst the homozygous compound mutants.67 One possible explanation for the discrepancy between the RARE transgenic reporter results and their mutation within the endogenous locus is that other cis-acting elements such as auto- and cross-regulatory enhancers compensate for the loss of these individual RAREs. RA excess or depletion can produce the gross rearrangements of the hindbrain with altered Hox gene expression profiles because these compensatory mechanisms are over-ridden when all Hox genes are misregulated.
Although initial in vitro work suggested that all Hox genes are up-regulated by RA, in the embryo 5' members may be actually negatively regulated by RA signaling.95 In early chick neural tube explants, adding RA to the explants did not induce Hoxc6, Hoxc8, Hoxc9 nor Hoxc10 expression in the developing MNs or adjacent cells. However, when these explants are exposed to FGF-signaling and subsequent RA-signaling, Hoxc6 expression is elevated by 2 folds whereas Hoxc8 and Hoxc9 expression are decreased by 3 and 6 folds, respectively. The authors concluded that RA signaling (from the paraxial mesoderm) refines the pattern of HoxC expression induced in the MNs by FGF signaling.95 Similar differences in the response of 3' and 5' HoxB genes to RA in the CNS have been reported.197 Hence, RA signaling regulates the anterior border in a positive fashion in the hindbrain, whereas in the spinal column it appears to function in an opposite manner, limiting the anterior expansion of more posteriorly expressed Hox genes. The presence of RAREs clearly indicates that many of the influences of RA may be direct on the Hox genes via their RAR and RXR nuclear receptors.
Fibroblast Growth Factor (FGF) Signaling
For more than a decade, it has been known that exogenously added FGF can up-regulate expression of posteriorly expressed Hox genes in the mesoderm and neural tissues of Xenopus embryos.228-234 Conversely, blocking FGF signaling in Xenopus embryos, by expressing a dominant-negative FGF receptor, results in repression or posteriorization of Hox gene expression. 234,235 These observations suggest that endogenous FGF signaling controls A-P patterning in part through regulation of Hox gene expression. A growing body of work now shows that this is a common mechanism during vertebrate development.
The fibroblast growth factor (FGFs) family contains twenty-two secreted heparin-binding molecules that interact with one of four FGF receptors (FGFRs).236 This signaling pathway is further complicated by the observations that three of FGFRs have various isoforms, and that these isoforms display different preferences for FGF ligands.237 Most of the Fgf genes have been mutated in mouse gene targeting experiments and result in a variety of phenotypes.238 Of these, the targeted deletion of Fgf2, Fgf3, Fgf8, and Fgf14 have been reported to affect some aspect of CNS development.
In the early zebrafish hindbrain, both Fgf3 and Fgf8 are transiently expressed. Their expression precedes the expression of kreisler and Krox20 and becomes refined to the region that will form r4.239,240 When the rhombomeres form, the expression of Fgf3 persists in r4 while Fgf8 expression in this region disappears. Expression of Fgf8 is subsequently detected at the midbrain/hindbrain border.241,242 The patterns of Fgf expression vary among vertebrates suggesting that different members of the family may have distinct roles in each species. Inhibition of FGF signaling in the early chick and fish hindbrain reduces and blocks expression of kreisler and Krox20.240,243 In the zebrafish, this inhibition leads to the failure of Hoxa2 expression while the initial expression of Hoxb1 becomes down-regulated at later stages. These changes result in a zebrafish hindbrain in which the axonal organization is severely affected.240 Conversely, exogenous applied FGF, or its ectopic expression, in the vertebrate hindbrain induces ectopic expression of kreisler and Krox20.239,243 The expression of Fgf in the presumptive r4 of zebrafish has been used to suggest that it may act as a signaling center for hindbrain patterning by regulating the upstream effectors of Hox gene expression but this pattern is not conserved amongst all vertebrates.244-246
In the early hindbrain FGF signaling positively regulates Hox gene expression, but at later stages, its role is reversed, as Fgf8 expression from the isthmus represses Hox gene expression thereby limiting the anterior border of Hoxa2 expression at the r1/r2 border.247 However, at the opposite end of the embryo, FGF signaling from the regressing node activates group 5 to group 13 Hox genes in the spinal cord.197 This apparent paradox is the result of Cdx availability which is no longer present in the hindbrain when these genes are active.197
In chick neural tube explants, increasing concentrations of FGF induce expression of more 5' located HoxC genes.95 At low concentrations of Fgf8 or Fgf2, chick neural plate explants express Hoxc6. Higher concentrations of Fgf8 or Fgf2 induce Hoxc8 and Hoxc9 (in addition to Hoxc6), and at the highest concentration they also induce Hoxc10 expression. Conversely, blocking FGF signaling with SU5402, an inhibitor of FGFR1 activity, prevent the expression of Hoxc8, Hoxc9 and Hoxc10 in chick neural explants. Furthermore, the presence of SU5402 is able to block the HoxC inducing activities of Hensen node that were cocultured with the chick explants. Higher concentrations of SU5402 are needed to inhibit expression of Hoxc6. Thus the expression of Hoxc6 to Hoxc10 in MNs relies on FGF signaling provided by Hensen's node. Similarly it has been shown that FGF signaling is required for expression of 5' HoxB genes in the neural tube.197 These data suggest that FGF signaling acts in a reciprocal fashion to RA signaling by promoting expression of Hox genes in the spinal column.
Tying It All Iogether: RA and FGF Signaling Regulate Hox Gene Expression in the CNS
So far, we have reviewed how RA signaling can initiate expression of the 3' Hox genes (from groups 1 to 5) while FGF signaling can induce expression of the 5' Hox genes (from groups 6 to 13) in the developing vertebrate CNS. At the boundary between these two groups, group 6 Hox members can be induced by both signaling pathways 95,197. Recent work has highlighted how these two signaling pathways play a pivotal role in promoting neuron formation in the developing spinal column and how they act in a reciprocal and antagonistic process to do so.248,249 RA signaling from the somitic mesoderm represses Fgf8 expression in the developing spinal cord thus restricting its expression to the caudal end of the embryo. This repressive interaction is necessary for neuronal differentiation to proceed within the neural tube adjacent to the forming somites.248 Paradoxically FGF signaling initiates Raldh2 expression in the paraxial mesoderm, resulting in the somites that flank the neural tube to produce RA.249 In both Xenopus and chick, caudal genes are downstream targets of FGF signaling191,197,234 and in the chick neural tube Cdx is required to anteriorize the FGF response of 5' HoxB members into the hindbrain.197 Moreover, RA signaling can directly regulate the expression of Cdx genes.250 Hence if FGFs and RA themselves global regulators can modulate another global regulator, Cdx, there is a complex network that regulates the balance of these opposing pathways. This might help explain their roles in neurogenesis and the differential responsiveness of the 3' Hox genes and 5' Hox genes to RA and FGF signaling. There is also evidence that Cdx genes may integrate signals from the Wnt pathway, which regulates A-P patterning.251-255 By simultaneously linking the establishment of A-P character in both the paraxial mesoderm (from which the muscles are generated) and neural tube, there is a coordinated system whereby the muscles and neurons that innervate them have the same molecular code.
Expression and Neuronal Phenotypes
The majority of Hox loss-of-function and gain-of-function studies describe the effects that these mutations have on axial and limb skeleton patterning and formation. In terms of what effects these mutations have on CNS development, more work has been produced looking at the defects in the organization of the hindbrain and its cranial nerves; whereas, a smaller body of work deals with the alterations and deletions of the MNs projecting from the spinal column into the limbs or trunk.
In the hindbrain targeted inactivation of the Hoxa1 gene results in loss of r5 and reduction in the size of r4.65,256 However, the loss of Hoxb1 function is mild in comparison. Initially r4 character is triggered but it is not maintained and there is a transformation to an r2-like character. 69,257 The generation of compound group 1 mutants leads to more severe phenotypes. Hence in Hoxa1 and Hoxb1 double homozygous mutants r4 and r5 fail to be specified and are missing along with derivatives of the second branchial arch which are generated from neural crest cells that migrate from r4.61,67,70 As a consequence of their gain or loss of expression in the hindbrain, Hoxb1 and Hoxa1 affect the projections of the cranial MNs from the hindbrain.59,60,66,69,258 These studies have shown that Hoxb1 is essential for the formation, migration and projections of the facial MN.59,69,257,259 Studies with the Hoxa1, Hoxb1 and Hoxb2 mutants also show later effects of these genes on neurogenesis.68,260 Similar work with group 3 paralogs have highlighted their important role in the formation of the abducens and hypoglossal neurons.63,261
Forcing expression of Hox genes in more anterior domains of the hindbrain typically leads to rhombomere transformations. Ectopic expression of Hoxa1 results in transformation of r2 into r4,56,60 and similar results are produced when Hoxa1 and Hoxb1 expression is anteriorized in the hindbrain by excess RA treatment.206,217 These changes in rhombomere identity have profound effects on the subsequently formed CNS structures, such as the cerebellum that is derived primarily from r1.262 Loss of Hoxa2 causes a caudal expansion of the cerebellum,66 which is further extended into r2 and r3 territory when both paralogous Hoxa2 and Hoxb2 are absent.260
The products of Hox genes also determine the axonal projection patterns from the spinal column. Targeted inactivation of HoxA, HoxC and HoxD genes results in alterations and/or reductions of the MNs axonal projections to muscle targets in the limbs.97-100,263 In general, members from groups 6 and 8 are important for specifying brachial MNs that innervate the forelimbs whereas members from groups 9, 10 and 11 have been shown to control MN projections into the hindlimbs. Interestingly, mutation of the Hoxb13 gene does not produce rearrangements or deletions of elements derived from the CNS but results in the overgrowth of the posterior spinal cord and tail vertebrae.264 This suggests that, unlike the other Abd-B like Hox proteins, Hoxb13 functions as a repressor of neuronal and caudal vertebral proliferation.
Studies in which the gene is mis-expressed in the target area, or in tissue through which the neurons project, have highlighted the importance of coordinating the simultaneous expression of these genes in both the paraxial mesoderm and neuroectoderm. Viral misexpression of Hoxc6 in the chick cervical paraxial mesoderm disrupts the spinal nerve's projection, causing it to prematurely halt is migration.102 This suggests that the signals from the mesoderm through which the axon migrates are important to direct their outgrowth.102 Similarly, the viral misexpression of Hoxb1 in the first branchial arches causes Hoxb1-specified neurons to alter their migration patterns so that they project axons into the first branchial arch rather than their normal targets located in the second branchial arch.59 This link between the local A-P character of the neural tube and the flanking mesoderm tissue is missed in gene-targeting experiments because such experiments eliminated the targeted gene's product from both compartments.
The Roles of Auto- and Cross-Regulatory Interactions between Hox Genes
An important consideration in both the regulation and function of Hox genes in the nervous system is the degree to which they cross-regulate each other. A common feature from the cis-regulatory studies on segmental expression of Hox genes in the hindbrain is that early signals (RA, FGFs) and transcription factors (kreisler, Krox20) are transient. Hence, other mechanisms are required to maintain or stabilize their domains of expression critical for later expression and function in rhombomeric segments. By analogy to Drosophila, mechanisms involving Polycomb and trithorax mediated epigenetic changes in chromatin, have been widely postulated to be important in vertebrate Hox regulation. In support of this, mouse mutants for members of the PcG and trxG have been shown to alter Hox expression, primarily examined in the mesoderm due to the prevalence of skeletal defects.265 However, the regulatory analyses of rhombomeric enhancers has uncovered a surprising degree of auto and cross-regulation between the Hox genes themselves, as an important mechanism for maintaining segmentally-restricted expression.70,82,110,116,122,123,140,266-268 During the establishment and maintenance stages, Hox proteins can directly cross-regulate the expression of other Hox genes,82,122,140,269-271 or perpetuate their own expression through auto-regulatory elements.82,110,267,271 Hence, following initiation by transiently expressed upstream factors or signals, Hox response elements serve to generate feedback loops that stabilize and perpetuate segmental expression. These lock-down or feedback loops have important implications in functional studies because mutation of one Hox gene can alter the regulation of other family members, generating a more complex phenotype.
One of the best examples of such loops from regulatory and mutant analyses relates to rhombomere 4. It appears that there is a pathway for specifying r4 character and later neuronal identities dependent upon cross-talk between Hox genes.268 Early retinoid signaling triggers both Hoxb1 and Hoxa1 expression directly via RAREs located at the 3' end of these genes.56,112,217,227 In combination with Pbx and Prep/Meis cofactors, both Hoxa1 and Hoxb1 then bind to the auto-regulatory enhancer of Hoxb1 to maintain its expression. 110,272-278 Hoxb1 in turn directly cross regulates both Hoxb2 and Hoxa2 specific expression in r4 and Hoxb2 feedbacks upon Hoxb1 to support its expression.110,112,122,123,268 By these loops it is clear that Hoxb1 plays a central role in specifying and maintaining r4 identity and the mutation of any of the individual Hox genes in this these cross-talk loops directly correlates with the neuronal phenotypes observed.268
Further evidence for the importance of these feedback loops in other vertebrates is provided by loss and gain-of-function experiments in zebrafish where Hox, Pbx and Meis mutations all show primary defects in hindbrain patterning and alterations of Hox expression.51,55,58,279-283 There is also emerging evidence that both positive and negative feedback loops among Hox genes is important in the spinal cord.33,96
In conclusion, regulatory analyses seeking to understand the cis-regulation of Hox genes themselves and the upstream cascade have surprisingly provided novel insight into potential Hox target genes. By identifying a series of known in vivo relevant target elements it will be possible to define the cis-regulatory code of a Hox response element and use it to predict down-stream target genes identified by genomic and microarray analysis. Therefore, a major area of interest in future Hox research will be to understand how different Hox proteins bindtheir target sites, what cofactors they use and how the output of activation or repression is achieved. This will be important in predicting and understanding their interactions on down-stream target genes. This work should also be highly relevant to Hox gene expression, regulation and function in tissues outside the nervous system.
References
- 1.
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. [PubMed: 103000]
- 2.
- McGinnis W, Garber RL, Wirz J. et al. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984;37:403–408. [PubMed: 6327065]
- 3.
- Carrasco AE, McGinnis W, Gehring WJ. et al. Cloning of an X. laevis gene expressed during early embryogenesis coding for a peptide region homologous to Drosophila homeotic genes. Cell. 1984;37:409–414. [PubMed: 6327066]
- 4.
- Scott M, Weiner A, Hazelrigg T. et al. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983;35:763–776. [PubMed: 6418389]
- 5.
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. [PubMed: 1346368]
- 6.
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. [PubMed: 7913880]
- 7.
- Graham A, Papalopulu N, Hunt P. et al. The murine and Drosophila homeobox clusters are derived from a common ancestor based on similarities in structure and expression. In: Capecchi M, ed. Molecular Genetics of Early Drosophila and Mouse Development. Cold Spring Harbor: Cold Spring Harpor Press. 1989:85–90.
- 8.
- Duboule D, Dollé P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8(5):1497–1505. [PMC free article: PMC400980] [PubMed: 2569969]
- 9.
- Amores A, Force A, Yan YL. et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. [PubMed: 9831563]
- 10.
- Amores A, Suzuki T, Yan YL. et al. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004;14(1):1–10. [PMC free article: PMC314266] [PubMed: 14707165]
- 11.
- Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends Genet. 2005;21:421–424. [PubMed: 15967537]
- 12.
- Aparicio S, Hawker K, Cottage A. et al. Organization of the Fugu rubripes Hox clusters, evidence for continuing evolution of vertebrate Hox complexes. Nat Gen. 1997;16:79–84. [PubMed: 9140399]
- 13.
- Gaunt SJ, Krumlauf R, Duboule D. Mouse homeo-genes within a subfamily, Hox-1.4, -2.6 and -5.1, display similar anteroposterior domains of expression in the embryo, but show stage- and tissue-dependent differences in their regulation. Development. 1989;107(1):131–141. [PubMed: 2576400]
- 14.
- Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. [PubMed: 2566383]
- 15.
- Dollé P, Izpisùa-Belmonte JC, Falkenstein H. et al. Coordinate expression of the murine Hox-5 complex homeobox-containing genes during limb pattern formation. Nature. 1989;342(6251):767–772. [PubMed: 2574828]
- 16.
- Wilkinson DG, Bhatt S, Cook M. et al. Segmental expression of Hox-2 homeobox-containing genes in the developing mouse hindbrain. Nature. 1989;341:405–409. [PubMed: 2571936]
- 17.
- Izpisua-Belmonte JC, Falkenstein H, Dollé P. et al. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991;10:2279–2289. [PMC free article: PMC452918] [PubMed: 1676674]
- 18.
- Hunt P, Wilkinson D, Krumlauf R. Patterning the vertebrate head: Murine Hox 2 genes mark distinct subpopulations of premigratory and migrating neural crest. Development. 1991;112:43–51. [PubMed: 1685117]
- 19.
- Hunt P, Gulisano M, Cook M. et al. A distinct Hox code for the branchial region of the vertebrate head. Nature. 1991;353(6347):861–864. [PubMed: 1682814]
- 20.
- Dollé P, Izpisua-Belmonte JC, Falkenstein H. et al. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. [PubMed: 2574828]
- 21.
- Izpisua-Belmonte J, Falkenstein H, Dollé P. et al. Murine genes related to the Drosophila AbdB homeotic gene are sequentially expressed during development of the posterior part of the body. EMBO J. 1991;10:2279–2289. [PMC free article: PMC452918] [PubMed: 1676674]
- 22.
- Nohno T, Noji S, Koyama E. et al. Involvement of the Chox-4 chicken homeobox genes in determination of anteroposterior axial polarity during limb development. Cell. 1991;64:1197–1205. [PubMed: 1672266]
- 23.
- Dollé P, Izpisùa-Belmonte JC, Brown JM. et al. Hox-4 genes and the morphogenesis of mammalian genitalia. Genes Dev. 1991;5:1767–1776. [PubMed: 1680771]
- 24.
- Kessel M, Gruss P. Homeotic transformations of murine prevertebrae and concommitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. [PubMed: 1680565]
- 25.
- Kessel M, Balling R, Gruss P. Variations of cervical vertebrae after expression of a Hox 1.1 transgene in mice. Cell. 1990;61:301–308. [PubMed: 1970515]
- 26.
- Gehring WJ, Qian YQ, Billeter M. et al. Homeodomain-DNA recognition. Cell. 1994;78:211–223. [PubMed: 8044836]
- 27.
- Maconochie M, Nonchev S, Morrison A. et al. Paralogous Hox genes: Function and regulation. Ann Rev Genet. 1996;30:529–556. [PubMed: 8982464]
- 28.
- Favier B, Dolle P. Developmental functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115–131. [PubMed: 9239717]
- 29.
- Slack JMW, Tannahill D. Mechanism of anteroposterior axis specification in vertebrates. Lessons from the amphibians. Development. 1992;114:285–302. [PubMed: 1350531]
- 30.
- Nieuwkoop P. Activation and organization of the central nervous system in amphibians. J Exp Zool. 1952;120:1–108.
- 31.
- Nieuwkoop P. Inductive interactions in early amphibian development and their general nature. J Embryol Exp Morph. 1985;89(suppl):333–347. [PubMed: 3831218]
- 32.
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. [PubMed: 8895453]
- 33.
- Jessell TM. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat Rev Genet. 2000;1(1):20–29. [PubMed: 11262869]
- 34.
- Le Douarin N, Kalcheim C. The Neural Crest. 2nd ed. Cambridge Univesity Press. 1999
- 35.
- Keynes R, Krumlauf R. Hox genes and regionalization of the nervous system. Ann Rev Neurosci. 1994;17:109–132. [PubMed: 7911650]
- 36.
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–428. [PubMed: 2644541]
- 37.
- Fraser S, Keynes R, Lumsden A. Segmentation in the chick embryo hindbrain is defined by cell lineage restrictions. Nature. 1990;344:431–435. [PubMed: 2320110]
- 38.
- Akam M, Dawson I, Tear G. Homeotic genes and the control of segment diversity. Development. 1988;104(Supplement Mechanisms of Segmentation):123–133.
- 39.
- Chisaka O, Capecchi MR. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox1.5. Nature. 1991;350:473–479. [PubMed: 1673020]
- 40.
- Condie BG, Capecchi MR. Mice homozygous for a targeted disruption of Hoxd-3(Hox-4.1) exhibit anterior transformations of the first and second cervical vertebrae, the atlas and axis. Development. 1993;119:579–595. [PubMed: 7910549]
- 41.
- Gendron-Maguire M, Mallo M, Zhang M. et al. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. [PubMed: 7903600]
- 42.
- Jeannottee L, Lemieux M, Charron J. et al. Specification of axial identity in the mouse: Role of the Hoxa-5(Hox1.3) gene. Genes Dev. 1993;7:2085–2096. [PubMed: 7901120]
- 43.
- Rijli FM, Mark M, Lakkaraju S. et al. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell. 1993;75:1333–1349. [PubMed: 7903601]
- 44.
- Ramirez-Solis R, Zheng H, Whiting J. et al. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell. 1993;73:279–294. [PubMed: 8097432]
- 45.
- Small KS, Potter S. Homeotic transformations and limb defects in Hoxa-11 mutant mice. Genes Dev. 1993;7:2318–2328. [PubMed: 7902826]
- 46.
- Jegalian BG, De Robertis EM. Homeotic transformations in the mouse induced by over-expression of a human Hox3.3 transgene. Cell. 1992;71:901–910. [PubMed: 1360874]
- 47.
- Le Mouellic H, Lallemand Y, Brulet P. Homeosis in the mouse induced by a null mutation in the Hox-3.1 gene. Cell. 1992;69:251–264. [PubMed: 1348969]
- 48.
- Birgbauer E, Fraser SE. Violation of cell lineage restriction compartments in the chick hindbrain. Development. 1994;120:1347–1356. [PubMed: 7519542]
- 49.
- Clarke JD, Lumsden A. Segmental repetition of neuronal phenotype sets in the chick embryo hindbrain. Development. 1993;118:151–162. [PubMed: 8375332]
- 50.
- Clarke JDW, Erskine L, Lumsden A. Differential progenitor dispersal and the spatial origin of early neurons can explain the predominance of single-phenotype clones in the chick hindbrain. Dev Dyn. 1998;212:14–26. [PubMed: 9603420]
- 51.
- McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129(10):2339–2354. [PubMed: 11973267]
- 52.
- McClintock JM, Carlson R, Mann DM. et al. Consequences of Hox gene duplication in the vertebrates: An investigation of the zebrafish Hox paralogue group 1 genes. Development. 2001;128(13):2471–2484. [PubMed: 11493564]
- 53.
- Moens CB, Cordes SP, Giorgianni MW. et al. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–391. [PubMed: 9425134]
- 54.
- Moens CB, Kimmel CB. Hindbrain patterning in the zebrafish embryo. Soc Neurosci Abstr. 1995;21:118.118.
- 55.
- Moens CB, Prince VE. Constructing the hindbrain: Insights from the zebrafish. Dev Dyn. 2002;224(1):1–17. [PubMed: 11984869]
- 56.
- Zhang M, Kim HJ, Marshall H. et al. Ectopic Hoxa-1 induces rhombomere transformation in mouse hindbrain. Development. 1994;120:2431–2442. [PubMed: 7956823]
- 57.
- Prince VE, Joly L, Ekker M. et al. Zebrafish hox genes: Genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. [PubMed: 9425136]
- 58.
- Prince VE, Moens CB, Kimmel CB. et al. Zebrafish hox genes: Expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. [PubMed: 9425135]
- 59.
- Bell E, Wingate RJ, Lumsden A. Homeotic transformation of rhombomere identity after localized Hoxb1 misexpression. Science. 1999;284:21682171. [PubMed: 10381880]
- 60.
- Alexandre D, Clarke J, Oxtoby E. et al. Ectopic expression of Hoxa-1 in the zebrafish alters the fate of the mandibular arch neural crest and phenocopies a retinoic acid -induced phenotype. Development. 1996;122:735–746. [PubMed: 8631251]
- 61.
- Rossel M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126:5027–5040. [PubMed: 10529420]
- 62.
- Gaufo GO, Wu S, Capecchi MR. Contribution of Hox genes to the diversity of the hindbrain sensory system. Development. 2004;131(6):1259–1266. [PubMed: 14960494]
- 63.
- Gaufo GO, Thomas KR, Capecchi MR. Hox3 genes coordinate mechanisms of genetic suppression and activation in the generation of branchial and somatic motoneurons. Development. 2003;130:5191–5201. [PubMed: 12954718]
- 64.
- Lufkin T, Dierich A, LeMeur M. et al. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66(6):1105–1119. [PubMed: 1680563]
- 65.
- Mark M, Lufkin T, Vonesch JL. et al. Two rhombomeres are altered in Hoxa-1 mutant mice. Development. 1993;119:319–338. [PubMed: 8287791]
- 66.
- Gavalas A, Davenne M, Lumsden A. et al. Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development. 1997;124:3693–3702. [PubMed: 9367425]
- 67.
- Gavalas A, Studer M, Lumsden A. et al. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development. 1998;125:1123–1136. [PubMed: 9463359]
- 68.
- Gavalas A, Ruhrberg C, Livet J. et al. Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. Development. 2003 [PubMed: 14522873]
- 69.
- Studer M, Lumsden A, Ariza-McNaughton L. et al. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–635. [PubMed: 8967950]
- 70.
- Studer M, Gavalas A, Marshall H. et al. Genetic interaction between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. [PubMed: 9463349]
- 71.
- Lim T, Jaques K, Stern C. et al. An evaluation of myelomeres and segmentation of the chick embryo spinal cord. Development. 1991;113:227–238. [PubMed: 1722449]
- 72.
- Stern C, Jaques K, Lim T. et al. Segmental lineage restrictions in the chick embryo spinal cord depend on the adjacent somites. Development. 1991;113:239–244. [PubMed: 1764998]
- 73.
- Phelan KA, Hollyday M. Axon guidance in muscleless chick wings: The role of muscle cells in motoneuronal pathway selection and muscle nerve formation. J Neurosci. 1990;10:2699–2716. [PMC free article: PMC6570285] [PubMed: 2167355]
- 74.
- Ensini M, Tsuchida T, Belting HG. et al. The control of rostrocaudal pattern in the developing spinal cord: Specification of motor neuron subtype identity is initiated by signals from paraxial mesoderm. Development. 1998;125:969–982. [PubMed: 9463344]
- 75.
- Bronner-Fraser M. Rostrocaudal differences within the somites confer segmental pattern to trunk neural crest migration. Curr Top Dev Biol. 2000;47:279–296. [PubMed: 10595308]
- 76.
- Krumlauf R, Holland P, McVey J. et al. Developmental and spatial patterns of expression of the mouse homeobox gene, Hox 2.1. Development. 1987;99:603–617. [PubMed: 2889591]
- 77.
- Murphy P, Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991;111:61–74. [PubMed: 1673098]
- 78.
- Murphy P, Davidson DR, Hill RE. Segment-specific expression of a homeobox-containing gene in the mouse hindbrain. Nature. 1989;341:156–159. [PubMed: 2571087]
- 79.
- Prince V, Lumsden A. Hox-a2 expression in normal and transposed rhombomeres: Independent regulation in the neural tube and neural crest. Development. 1994;120:911–923. [PubMed: 7600967]
- 80.
- Frohman MA, Boyle M, Martin GR. Isolation of the mouse Hox-2.9 gene; analysis of embryonic expression suggests that positional information along the anterior-posterior axis is specified by mesoderm. Development. 1990;110:589–607. [PubMed: 1983472]
- 81.
- Geada AMC, Gaunt SJ, Azzawi M. et al. Sequence and embryonic expression of the murine Hox-3.5 gene. Development. 1992;116:497–506. [PubMed: 1363091]
- 82.
- Manzanares M, Bel-Vialer S, Ariza-McNaughton L. et al. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involves auto and cross-regulatory mechanisms. Development. 2001;128(18):3595–3607. [PubMed: 11566863]
- 83.
- Forlani S, Lawson KA, Deschamps J. Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development. 2003;130:3807–3819. [PubMed: 12835396]
- 84.
- Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. [PubMed: 15944185]
- 85.
- Carpenter EM. Hox genes and spinal cord development. Dev Neurosci. 2002;24:24–34. [PubMed: 12145408]
- 86.
- Zeltser L, Desplan C, Heintz N. Hoxb-13: Colinear expression of a new Hox gene in a distant region of the HOXB cluster. Development. 1996;122:2475–2484. [PubMed: 8756292]
- 87.
- Mahon KA, Westphal H, Gruss P. Expression of homeobox gene Hox 1.1 during mouse embryogenesis. Development. 1988;104(Suppl):187–195. [PubMed: 2908296]
- 88.
- Peterson RL, Jacobs DF, Awgulewitsch A. Hox-3.6: Isolation and characterization of a new murine homeobox gene located in the 5' region of the Hox-3 cluster. Mech Dev. 1992;37:151–166. [PubMed: 1353983]
- 89.
- Oosterveen T, Meijlink F, Deschamps J. Expression of retinaldehyde dehydrogenase II and sequential activation of 5' Hoxb genes in the mouse caudal hindbrain. Gene Expr Patterns. 2004;4:243–247. [PubMed: 15053971]
- 90.
- Hostikka SL, Capecchi MR. The mouse Hoxc11 gene: Genomic structure and expression pattern. Mech Dev. 1998;70:133–145. [PubMed: 9510030]
- 91.
- Erselius JR, Goulding MD, Gruss P. Structure and expression pattern of the murine Hox-3.2 gene. Development. 1990;110:629–642. [PubMed: 1723949]
- 92.
- Larochelle C, Tremblay M, Bernier D. et al. Multiple cis-acting regulatory regions are required for restricted spatio-temporal Hoxa5 gene expression. Dev Dyn. 1999;214:127–140. [PubMed: 10030592]
- 93.
- Sham MH, Hunt P, Nonchev S. et al. Analysis of the murine Hox-2.7 gene: Conserved alternative transcripts with differential distributions in the nervous system and the potential for shared regulatory regions. EMBO J. 1992;11:1825–1836. [PMC free article: PMC556640] [PubMed: 1582411]
- 94.
- Awgulewitsch A, Jacobs D. Differential expression of Hox 3.1 protein in subregions of the embryonic and adult spinal cord. Development. 1990;108(3):411–420. [PubMed: 1971213]
- 95.
- Liu JP, Laufer E, Jessell TM. Assigning the positional identity of spinal motor neurons. rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and Retinoids. Neuron. 2001;32(6):997–1012. [PubMed: 11754833]
- 96.
- Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425(6961):926–933. [PubMed: 14586461]
- 97.
- Carpenter EM, Goddard JM, Davis AP. et al. Targeted disruption of Hoxd-10 affects mouse hindlimb development. Development. 1997;124:4505–4514. [PubMed: 9409668]
- 98.
- Tiret L, Le Mouellic H, Maury M. et al. Increased apoptosis of motoneurons and altered somatotopic maps in the branchial spinal cord of Hoxc8-deficient mice. Development. 1998;125:279–291. [PubMed: 9486801]
- 99.
- de la Cruz CC, Der-Avakian A, Spyropoulos DD. et al. Targeted disruption of Hoxd9 and Hoxd10 alters locomotor behavior, vertebral identity, and peripheral nervous system development. Dev Biol. 1999;216:595–610. [PubMed: 10642795]
- 100.
- Lin AW, Carpenter EM. Hoxa10 and Hoxd10 coordinately regulate lumbar motor neuron patterning. J Neurobiol. 2003;56:328–337. [PubMed: 12918017]
- 101.
- Wahba GM, Hostikka SL, Carpenter EM. The paralogous Hox genes Hoxa10 and Hoxd10 interact to pattern the mouse hindlimb peripheral nervous system and skeleton. Dev Biol. 2001;231:87–102. [PubMed: 11180954]
- 102.
- Burke AC, Tabin CJ. Virally mediated misexpression of Hoxc-6 in the cervical mesoderm results in spinal nerve truncations. Dev Biol. 1996;178:192–197. [PubMed: 8812121]
- 103.
- Shah V, Drill E, Lance-Jones C. Ectopic expression of Hoxd10 in thoracic spinal segments induces motoneurons with a lumbosacral molecular profile and axon projections to the limb. Dev Dyn. 2004;231:43–56. [PubMed: 15305286]
- 104.
- Altman J, Bayer J. The development of the rat spinal cord. Adv Anat Embryol Cell Biol. 1984;85:1–166. [PubMed: 6741688]
- 105.
- Graham A, Maden M, Krumlauf R. The murine Hox-2 genes display dynamic dorsoventral patterns of expression during central nervous system development. Development. 1991;112:255–264. [PubMed: 1685115]
- 106.
- Gaunt SJ. Expression patterns of mouse Hox genes: Clues to an understanding of developmental and evolutionary strategies. Bioessays. 1991;13(10):505–513. [PubMed: 1684493]
- 107.
- Gaunt SJ, Coletta PL, Pravtcheva D. et al. Mouse Hox-3.4: Homeobox sequence and embryonic expression patterns compared with other members of the Hox gene network. Development. 1990;109(2):329–339. [PubMed: 1976088]
- 108.
- Greer JM, Capecchi MR. Hoxb8 is required for normal grooming behavior in mice. Neuron. 2002;33:23–34. [PubMed: 11779477]
- 109.
- Lufkin T. Transcriptional regulation of vertebrate Hox genes during embryogenesis. Crit Rev Eukaryot Gene Expr. 1997;7:195–213. [PubMed: 9399070]
- 110.
- Pöpperl H, Bienz M, Studer M. et al. Segmental expression of Hoxb1 is controlled by a highly conserved autoregulatory loop dependent upon exd/Pbx. Cell. 1995;81:1031–1042. [PubMed: 7600572]
- 111.
- Studer M, Pöpperl H, Marshall H. et al. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728–1732. [PubMed: 7916164]
- 112.
- Marshall H, Studer M, Pöpperl H. et al. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. [PubMed: 7914354]
- 113.
- Manzanares M, Cordes S, Kwan CT. et al. Segmental regulation of Hoxb3 by kreisler. Nature. 1997;387:191–195. [PubMed: 9144291]
- 114.
- Manzanares M, Nardelli J, Gilardi-Hebenstreit P. et al. Krox20 and kreisler cooperate in the transcriptional control of segmental expression of Hoxb3 in the developing hindbrain. EMBO J. 2002;21(3):365–376. [PMC free article: PMC125344] [PubMed: 11823429]
- 115.
- Manzanares M, Cordes S, Ariza-McNaughton L. et al. Conserved and distinct roles of kreisler in regulation of the paralogous Hoxa3 and Hoxb3 genes. Development. 1999;126:759–769. [PubMed: 9895323]
- 116.
- Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39–51. [PubMed: 9697850]
- 117.
- Whiting J, Marshall H, Cook M. et al. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 1991;5(11):2048–2059. [PubMed: 1682218]
- 118.
- Morrison A, Ariza-McNaughton L, Gould A. et al. HOXD4 and regulation of the group 4 paralog genes. Development. 1997;124:3135–3146. [PubMed: 9272954]
- 119.
- Frasch M, Chen X, Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995;121:957–974. [PubMed: 7743939]
- 120.
- Maconochie M, Krishnamurthy R, Nonchev S. et al. Regulation of Hoxa2 in cranial neural crest cells involves members of the AP-2 family. Development. 1999;126:1483–1494. [PubMed: 10068641]
- 121.
- Maconochie MK, Nonchev S, Manzanares M. et al. Differences in Krox20-dependent regulation of Hoxa2 and Hoxb2 during hindbrain development. Dev Biol. 2001;233(2):468–481. [PubMed: 11336508]
- 122.
- Maconochie MK, Nonchev S, Studer M. et al. Cross-regulation in the mouse HoxB complex: The expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev. 1997;11:1885–1896. [PubMed: 9242495]
- 123.
- Tumpel S, Cambronero F, Wiedemann LM. et al. Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes). Proc Natl Acad Sci USA. 2006;103:5419–5424. [PMC free article: PMC1459370] [PubMed: 16569696]
- 124.
- Nonchev S, Maconochie M, Vesque C. et al. The conserved role of Krox-20 in directing Hox gene expression during vertebrate hindbrain segmentation. Proc Natl Acad Sci USA. 1996;93:9339–9345. [PMC free article: PMC38429] [PubMed: 8790331]
- 125.
- Nonchev S, Vesque C, Maconochie M. et al. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development. 1996;122:543–554. [PubMed: 8625806]
- 126.
- Zhang F, Pöpperl H, Morrison A. et al. Elements both 5' and 3' to the murine Hoxd4 gene establish anterior borders of expression in mesoderm and neuroectoderm. Mech Dev. 1997;67:49–58. [PubMed: 9347914]
- 127.
- Vesque C, Maconochie M, Nonchev S. et al. Hoxb-2 transcriptional activation in rhombomeres 3 and 5 requires an evolutionarily conserved cis-acting element in addition to the Krox-20 binding site. EMBO J. 1996;15:5383–5896. [PMC free article: PMC452281] [PubMed: 8895582]
- 128.
- Sham MH, Vesque C, Nonchev S. et al. The zinc finger gene Krox-20 regulates Hoxb-2 (Hox2.8) during hindbrain segmentation. Cell. 1993;72:183–196. [PubMed: 8093858]
- 129.
- Packer AI, Crotty DA, Elwell VA. et al. Expression of the murine Hoxa4 gene requires both autoregulation and a conserved retinoic acid response element. Development. 1998;125:1991–1998. [PubMed: 9570764]
- 130.
- Sharpe J, Nonchev S, Gould A. et al. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998;17:1788–1798. [PMC free article: PMC1170526] [PubMed: 9501100]
- 131.
- Pöpperl H, Featherstone M. Identification of a retinoic acid repsonse element upstream of the murine Hox-4.2 gene. Mol Cell Biol. 1993;13(1):257–265. [PMC free article: PMC358905] [PubMed: 8093325]
- 132.
- Morrison A, Chaudhuri C, Ariza-McNaughton L. et al. Comparative analysis of chicken Hoxb-4 regulation in transgenic mice. Mech Dev. 1995;53:47–59. [PubMed: 8555111]
- 133.
- Packer AI, Mailutha KG, Ambrozewicz LA. et al. Regulation of the Hoxa4 and Hoxa5 genes in the embryonic mouse lung by retinoic acid and TGFbeta1: Implications for lung development and patterning. Dev Dyn. 2000;217(1):62–74. [PubMed: 10679930]
- 134.
- Behringer R, Crotty DA, Tennyson VM. et al. Seguences 5' of the homeobox of the Hox-1.4 gene direct tissue-specific expression of lacZ during mouse development. Development. 1993;117:823–833. [PubMed: 8100763]
- 135.
- Nowling T, Zhou W, Krieger KE. et al. Hoxa5 gene regulation: A gradient of binding activity to a brachial spinal cord element. Dev Biol. 1999;208:134–146. [PubMed: 10075847]
- 136.
- Moreau J, Jeannotte L. Sequence analysis of a Hoxa4-Hoxa5 intergenic region including shared regulatory elements. DNA Seq. 2002;13:203–209. [PubMed: 12487022]
- 137.
- Tuggle CK, Zakany J, Cianetti L. et al. Region-specific enhancers near two mammalian homeo box genes define adjacent rostrocaudal domains in the central nervous system. Genes Dev. 1990;4(2):180–189. [PubMed: 1970971]
- 138.
- Zakany J, Gérard M, Favier B. et al. Deletion of a HoxD enhancer induces transcriptional heterochrony leading to transposition of the sacrum. EMBO J. 1997;16:4393–4402. [PMC free article: PMC1170065] [PubMed: 9250683]
- 139.
- Gérard M, Chen JW, Gronemeyer H. et al. In vivo targeted mutagenesis of a regulatory element reguired for positioning the Hoxd-11 and Hoxd-10 expression boundaries. Genes Dev. 1996;10:2326–2334. [PubMed: 8824591]
- 140.
- Gould A, Morrison A, Sproat G. et al. Positive cross-regulation and enhancer sharing: Two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. [PubMed: 9106661]
- 141.
- Mansfield JH, Harfe BD, Nissen R. et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004 [PubMed: 15361871]
- 142.
- Hornstein E, Mansfield JH, Yekta S. et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. [PubMed: 16319892]
- 143.
- Williams T, Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–682. [PubMed: 2010091]
- 144.
- Mitchell PJ, Timmons PM, Hébert JM. et al. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. [PubMed: 1989904]
- 145.
- Moser M, Ruschoff J, Buettner R. Comparative analysis of AP-2α and AP-2β gene expression during mouse embryogenesis. Dev Dyn. 1997;208:115–124. [PubMed: 8989526]
- 146.
- Chazaud C, Oulad-Abdelghani M, Bouillet P. et al. AP-2.2, a novel gene related to AP-2, is expressed in the forebrain, limbs and face during mouse embryogenesis. Mech Dev. 1996;54:83–94. [PubMed: 8808408]
- 147.
- Deol MS. The abnormalities of the inner ear in kreisler mice. J Embryol Exp Morphol. 1964;12:475–490. [PubMed: 14207033]
- 148.
- McKay I, Lewis J, Lumsden A. Organization and development of facial motor neurons in the kreisler mutant mouse. Eur J Neurosci. 1997;9:1499–1506. [PubMed: 9240407]
- 149.
- McKay IJ, Muchamore I, Krumlauf R. et al. The kreisler mouse: A hindbrain segmentation mutant that lacks two rhombomeres. Development. 1994;120:2199–2211. [PubMed: 7925021]
- 150.
- Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. [PubMed: 8001130]
- 151.
- Frohman MA, Martin GR, Cordes SP. et al. Altered rhombomere-specific gene expression and hyoid bone differentiation in the mouse segmentation mutant, kreisler (kr). Development. 1993;117:925–936. [PubMed: 8100767]
- 152.
- Sadl VS, Sing A, Mar L. et al. Analysis of hindbrain patterning defects caused by the kreisler(enu) mutation reveals multiple roles of Kreisler in hindbrain segmentation. Dev Dyn. 2003;227:134–142. [PubMed: 12701106]
- 153.
- Moens CB, Yan YL, Appel B. et al. Valentino: A zebrafish gene required for normal hindbrain segmentation. Development. 1996;122:3981–3990. [PubMed: 9012518]
- 154.
- Giudicelli F, Gilardi-Hebenstreit P, Mechta-Grigoriou F. et al. Novel activities of Mafb underlie its dual role in hindbrain segmentation and regional specification. Dev Biol. 2003;253(1):150–162. [PubMed: 12490204]
- 155.
- Theil T, Ariza-McNaughton L, Manzanares M. et al. Requirement for downregulation of kreisler during late patterning of the hindbrain. Development. 2002;129(6):1477–1485. [PubMed: 11880356]
- 156.
- Nieto MA, Bradley LC, Wilkinson DG. Conserved segmental expression of Krox-20 in the vertebrate hindbrain and its relationship to lineage restriction. Development Supplement. 1991;2:59–62. [PubMed: 1688180]
- 157.
- Wilkinson DG, Bhatt S, Chavrier P. et al. Segment-specific expression of a zinc finger gene in the developing nervous system of the mouse. Nature. 1989;337:461–464. [PubMed: 2915691]
- 158.
- Schneider-Maunoury S, Topilko P, Seitandou T. et al. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993;75:1199–1214. [PubMed: 7903221]
- 159.
- Swiatek PJ, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox-20. Genes Dev. 1993;7:2071–2084. [PubMed: 8224839]
- 160.
- Voiculescu O, Taillebourg E, Pujades C. et al. Hindbrain patterning: Krox20 couples segmentation and specification of regional identity. Development. 2001;128(24):4967–4978. [PubMed: 11748134]
- 161.
- Vogel A, Tickle C. FGF-4 maintains polarizing activity of posterior limb buds cells in vivo and in vitro. Development. 1993;119(1):199–206. [PubMed: 8275856]
- 162.
- Chavrier P, Vesque C, Galliot B. et al. The segment-specific gene Krox-20 encodes a transcription factor with binding sites in the promoter of the Hox 1.4 gene. EMBO J. 1990;9:1209–1218. [PMC free article: PMC551797] [PubMed: 1969796]
- 163.
- Kim MH, Chang HH, Shin C. et al. Genomic structure and sequence analysis of human HOXA-9. DNA Cell Biol. 1998;17:407–414. [PubMed: 9628584]
- 164.
- Kim MH, Cho M, Park D. Sequence analysis of the 5'-flanking region of the gene encoding human HOXA-7. Somat Cell Mol Genet. 1998;24:371–374. [PubMed: 10763416]
- 165.
- Theil T, Frain M, Gilardi-Hebenstreit P. et al. Segmental expression of the EphA4 (Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development. 1998;125(3):443–452. [PubMed: 9425139]
- 166.
- McNabb DS, Xing Y, Guarente L. Cloning of yeast HAP5: A novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. [PubMed: 7828851]
- 167.
- Sinha S, Maity SN, Lu J. et al. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. [PMC free article: PMC42572] [PubMed: 7878029]
- 168.
- Bertagna A, Jahroudi N. The NFY transcription factor mediates induction of the von Willebrand factor promoter by irradiation. Thromb Haemost. 2001;85:837–844. [PubMed: 11372677]
- 169.
- Faniello MC, Bevilacqua MA, Condorelli G. et al. The B subunit of the CAAT-binding factor NFY binds the central segment of the Coactivator p300. J Biol Chem. 1999;274:7623–7626. [PubMed: 10075648]
- 170.
- Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene. 1999;236:197–208. [PubMed: 10452940]
- 171.
- Gilthorpe J, Vandromme M, Brend T. et al. Spatially specific expression of Hoxb4 is dependent on the ubiquitous transcription factor NFY. Development. 2002;129:3887–3899. [PubMed: 12135926]
- 172.
- Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: With partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. [PubMed: 10729834]
- 173.
- Botquin V, Hess H, Fuhrmann G. et al. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12(13):2073–2090. [PMC free article: PMC316977] [PubMed: 9649510]
- 174.
- Ambrosetti DC, Scholer HR, Dailey L. et al. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem. 2000;275(30):23387–23397. [PubMed: 10801796]
- 175.
- Boyer LA, Lee TI, Cole MF. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. [PMC free article: PMC3006442] [PubMed: 16153702]
- 176.
- Di Rocco G, Gavalas A, Popperl H. et al. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J Biol Chem. 2001;1:1. [PubMed: 11278854]
- 177.
- Mlodzik M, Fjose A, Gehring WJ. Isolation of caudal, a Drosophila homeo box-containing gene with maternal expression, whose transcripts form a concentration gradient at the preblastoderm stage. EMBO J. 1985;4:2961–2969. [PMC free article: PMC554605] [PubMed: 16453641]
- 178.
- Mlodzik M, Gehring WJ. Expression of the caudal gene in the germ line of Drosophila: Formation of an RNA and protein gradient during early embryogenesis. Cell. 1987;48:465–478. [PubMed: 2433048]
- 179.
- Duprey P, Chowdhury K, Dressler GR. et al. A mouse gene homologous to the Drosophila gene caudal is expressed in epithelial cells from the embryonic intestine. Genes Dev. 1988;2:1647–1654. [PubMed: 2905686]
- 180.
- Gamer LW, Wright CV. Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech Dev. 1993;43:71–81. [PubMed: 7902125]
- 181.
- Beck F, Erler T, Russell A. et al. Expression of Cdx-2 in the mouse embryo and placenta: Possible role in patterning of the extra-embryonic membranes. Dev Dyn. 1995;204:219–227. [PubMed: 8573715]
- 182.
- Meyer BI, Gruss P. Mouse Cdx-1 expression during gastrulation. Development. 1993;117:191–203. [PubMed: 7900985]
- 183.
- Marom K, Shapira E, Fainsod A. The chicken caudal genes establish an anterior-posterior gradient by partially overlapping temporal and spatial patterns of expression. Mech Dev. 1997;64:41–52. [PubMed: 9232595]
- 184.
- Pillemer G, Epstein M, Blumberg B. et al. Nested expression and sequential downregulation of the Xenopus caudal genes along the anterior-posterior axis. Mech Dev. 1998;71:193–196. [PubMed: 9507125]
- 185.
- Davidson AJ, Ernst P, Wang Y. et al. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–306. [PubMed: 13679919]
- 186.
- Joly JS, Maury M, Joly C. et al. Expression of a zebrafish caudal homeobox gene correlates with the establishment of posterior cell lineages at gastrulation. Differentiation. 1992;50:75–87. [PubMed: 1354191]
- 187.
- Subramanian V, Meyer BI, Gruss P. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell. 1995;83(4):641–653. [PubMed: 7585967]
- 188.
- Chawengsaksophak K, James R, Hammond VE. et al. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. [PubMed: 9052785]
- 189.
- van den Akker E, Forlani S, Chawengsaksophak K. et al. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129:2181–2193. [PubMed: 11959827]
- 190.
- Epstein M, Pillemer G, Yelin R. et al. Patterning of the embryo along the anterior-posterior axis: The role of the caudal genes. Development. 1997;124:3805–3814. [PubMed: 9367436]
- 191.
- Isaacs H, Pownall M, Slack J. Regulation of Hox gene expression and posterior development by the Xenopus caudal homolog Xcad3. EMBO J. 1998;17:3413–3427. [PMC free article: PMC1170678] [PubMed: 9628877]
- 192.
- Tabaries S, Lapointe J, Besch T. et al. Cdx protein interaction with Hoxa5 regulatory sequences contributes to Hoxa5 regional expression along the axial skeleton. Mol Cell Biol. 2005;25:1389–1401. [PMC free article: PMC548006] [PubMed: 15684390]
- 193.
- Knittel T, Kessel M, Kim MH. et al. A conserved enhancer of the human and murine Hoxa-7 gene specifies the anterior boundary of expression during embryonal development. Development. 1995;121:1077–1088. [PubMed: 7538068]
- 194.
- Min W, Cho M, Jang SI. et al. Sequence and functional analysis of an upstream regulatory region of human HOXA7 gene. Gene. 1996;182:1–6. [PubMed: 8982060]
- 195.
- Charité J, de Graaff W, Consten D. et al. Transducing positional information to the Hox henes: Critical interaction of cdx gene products with position-sensitive regulatory elements. Development. 1998;125:4349–4358. [PubMed: 9778495]
- 196.
- Shashikant CS, Ruddle FH. Combinations of closely situated cis-acting elements determine tissue-specific patterns and anterior extent of early Hoxc8 expression. Proc Natl Acad Sci USA. 1996;93:12364–12369. [PMC free article: PMC37997] [PubMed: 8901587]
- 197.
- Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: In the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129(22):5103–5115. [PubMed: 12399303]
- 198.
- Wilson JG, Warkany J. Congenital anomalies of heart and great vessels in offspring of vitamin A-deficient rats. Am J Dis Child. 1950;79:963. [PubMed: 15410706]
- 199.
- Wilson JG, Warkany J. Malformations in the genito-urinary tract induced by maternal vitamin A deficiency in the rat. Am J Anat. 1948;83:357–407. [PubMed: 18098411]
- 200.
- Wilson JG, Roth CB, Warkany J. An analysis of the syndrome of malformations induced by maternal vitamin A deficiency. Effects of restoration of vitamin A at various times during gestation. Am J Anat. 1953;92:189–217. [PubMed: 13030424]
- 201.
- Morriss GM. Morphogenesis of the malformations induced in rat embryos by maternal hypervitaminosis A. J Anat. 1972;113:241–250. [PMC free article: PMC1271684] [PubMed: 4664235]
- 202.
- Morriss-Kay GM, Murphy P, Hill RE. et al. Effects of retinoic acid excess on expression of Hox-2.9 and Krox-20 and on morphological segmentation in the hindbrain of mouse embryos. EMBO J. 1991;10:2985–2995. [PMC free article: PMC453013] [PubMed: 1915273]
- 203.
- Simeone A, Avantaggiato V, Moroni MC. et al. Retinoic acid induces stage-specific antero-posterior transformation of rostral central nervous system. Mech Dev. 1995;51(1):83–98. [PubMed: 7669695]
- 204.
- Papalopulu N, Clarke J, Bradley L. et al. Retinoic acid causes abnormal development and segmental patterning of the anterior hindbrain in Xenopus embryos. Development. 1991;113:1145–1159. [PubMed: 1811933]
- 205.
- Leonard L, Horton C, Maden M. et al. Anteriorization of CRABP-I expression by retinoic acid in the developing mouse central nervous system and its relationship to teratogenesis. Dev Biol. 1995;168:514–528. [PubMed: 7729586]
- 206.
- Hill J, Clarke JDW, Vargesson N. et al. Exogenous retinoic acid causes specific alterations in the development of the midbrain and hindbrain of the zebrafish embryo including positional respecification of the Mauthner neuron. Mech Dev. 1995;50:3–16. [PubMed: 7605750]
- 207.
- Holder N, Hill J. Effects of retinoic acid on zebrafish embryos: Modulation of engrailed protein expression at the midbrain-hindbrain border and development of cranial ganglia. Development. 1991;113:1159–1170. [PubMed: 1811934]
- 208.
- Durston AJ, Timmermans JP, Hage WJ. et al. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature. 1989;340:140–144. [PubMed: 2739735]
- 209.
- Sive H, Draper B, Harland R. et al. Identification of a retinoic acid-sensitive period during primary axis formation in Xenopus laevis. Genes Dev. 1990;4:932–942. [PubMed: 2384214]
- 210.
- Niederreither K, Vermot J, Schuhbaur B. et al. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127(1):75–85. [PubMed: 10654602]
- 211.
- White JC, Highland M, Kaiser M. et al. Vitamin A-deficiency in the rat embryo results in anteriorization of the posterior hindbrain which is prevented by maternal consumption of retinoic acid or retinol. Dev Biol. 2000;220:263–284. [PubMed: 10753515]
- 212.
- Maden M, Gale E, Kostetskii I. et al. Vitamin A deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6:417–426. [PubMed: 8723346]
- 213.
- Dupe V, Lumsden A. Hindbrain patterning involves graded responses to retinoic acid signalling. Development. 2001;128(12):2199–2208. [PubMed: 11493540]
- 214.
- Dupé V, Ghyselinck NB, Wendling O. et al. Key roles of retinoic acid receptors alpha and beta in the patterning of the caudal hindbrain, pharyngeal arches and otocyst in the mouse. Development. 1999;126:5051–5059. [PubMed: 10529422]
- 215.
- Blumberg B, Bolado J, Moreno T. et al. An essential role for retinoid signaling in anteroposterior neural patterning. Development. 1997;124:373–379. [PubMed: 9053313]
- 216.
- Conlon RA, Rossant J. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development. 1992;116(2):357–368. [PubMed: 1363087]
- 217.
- Marshall H, Nonchev S, Sham MH. et al. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature. 1992;360:737–741. [PubMed: 1361214]
- 218.
- Simeone A, Acampora D, Arcioni L. et al. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature. 1990;346:763–766. [PubMed: 1975088]
- 219.
- Papalopulu N, Lovell-Badge R, Krumlauf R. The expression of murine Hox-2 genes is dependent on the differentiation pathway and displays a collinear sensitivity to retinoic acid in F9 cells and Xenopus embryos. Nuc Acids Res. 1991;19(20):5497–5506. [PMC free article: PMC328948] [PubMed: 1682879]
- 220.
- Simeone A, Acampora D, Nigro V. et al. Differential regulation by retinoic acid of the homeobox genes of the four HOX loci in human embryonal carcinoma cells. Mech Dev. 1991;33:215–227. [PubMed: 1677812]
- 221.
- Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3' of the murine homeobox gene Hox-1.6. Mech Dev. 1992;38(3):217–228. [PubMed: 1360810]
- 222.
- Langston A, Thompson J, Gudas L. Retinoic acid-responsive enhancers located 3' of the HoxA and the HoxB gene clusters. J Biol Chem. 1997;272:2167–2175. [PubMed: 8999919]
- 223.
- Ogura T, Evans R. Evidence for two distinct retinoic acid response pathways for Hoxb-1 gene regulation. Proc Natl Acad Sci USA. 1995;92:392–396. [PMC free article: PMC42746] [PubMed: 7831297]
- 224.
- Morrison A, Moroni M, Ariza-McNaughton L. et al. In vitro and transgenic analysis of a human HOXD4 retinoid-responsive enhancer. Development. 1996;122:1895–1907. [PubMed: 8674428]
- 225.
- Zhang F, Nagy Kovacs E, Featherstone MS. Murine hoxd4 expression in the CNS requires multiple elements including a retinoic acid response element. Mech Dev. 2000;96(1):79–89. [PubMed: 10940626]
- 226.
- Oosterveen T, Niederreither K, Dolle P. et al. Retinoids regulate the anterior expression boundaries of 5' Hoxb genes in posterior hindbrain. EMBO J. 2003;22:262–269. [PMC free article: PMC140104] [PubMed: 12514132]
- 227.
- Dupé V, Davenne M, Brocard J. et al. In vivo functional analysis of the Hoxa1 3' retinoid response element (3' RARE). Development. 1997;124:399–410. [PubMed: 9053316]
- 228.
- Ruiz-i-Altaba A, Melton DA. Interaction between peptide growth factors and homeobox genes in the establishment of anteroposterior polarity in frog embryos. Nature. 1989;341:33–38. [PubMed: 2570357]
- 229.
- Cho K, De Robertis E. Differential activation of Xenopus homeobox genes by mesoderm-inducing growth factors and retinoic acid. Genes Dev. 1990;4:1910–1917. [PubMed: 1980476]
- 230.
- Kengaku M, Okamoto H. bFGF as a possible morphogen for the anteroposterior axis of the central nervous system in Xenopus. Development. 1995;121:3121–3130. [PubMed: 7555736]
- 231.
- Cox WG, Hemmati-Brivanlou A. Caudalization of neural fate by tissue recombination and bFGF. Development. 1995;121:4349–4358. [PubMed: 8575335]
- 232.
- Kolm P, Sive H. Regulation of the Xenopus labial homeodomain genes, HoxA1 and HoxD1: Activation by retinoids and peptide growth factors. Dev Biol. 1995;167:34–49. [PubMed: 7851655]
- 233.
- Lamb TM, Harland RM. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development. 1995;121:3627–3636. [PubMed: 8582276]
- 234.
- Pownall M, Tucker A, Slack J. et al. eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development. 1996;122:3881–3892. [PubMed: 9012508]
- 235.
- Godsave SF, Durston AJ. Neural induction and patterning in embryos deficient in FGF signaling. Int J Dev Biol. 1997;41(1):57–65. [PubMed: 9074938]
- 236.
- Ornitz DM, Itoh N. Fibroblast growth factors Genome Biol 20012 (REVIEWS3005) [PMC free article: PMC138918] [PubMed: 11276432]
- 237.
- Ornitz DM, Xu J, Colvin JS. et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. [PubMed: 8663044]
- 238.
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. [PubMed: 15475116]
- 239.
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129(16):3825–3837. [PubMed: 12135921]
- 240.
- Walshe J, Maroon H, McGonnell IM. et al. Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol. 2002;12(13):1117–1123. [PubMed: 12121619]
- 241.
- Heikinheimo M, Lawshe A, Shackleford G. et al. Fgf-8 expression in the post-gastrulation mouse suggests roles in the development of face, limbs and central nervous system. Mech Dev. 1994;48:129–138. [PubMed: 7873403]
- 242.
- Crossley PH, Martin GR. The mouse FGF-8 gene encodes a family of polypeptides expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. [PubMed: 7768185]
- 243.
- Marin F, Charnay P. Hindbrain patterning: FGFs regulate Krox20 and mafB/kr expression in the otic/preotic region. Development. 2000;127(22):4925–4935. [PubMed: 11044406]
- 244.
- Mahmood R, Kiefer P, Guthrie S. et al. Multiple roles for FGF-3 during cranial neural development in the chicken. Development. 1995;121(5):1399–1410. [PubMed: 7789270]
- 245.
- Mahmood R, Mason IJ, Morriss-Kay GM. Expression of Fgf-3 in relation to hindbrain segmentation, otic pit position and pharyngeal arch morphology in normal and retinoic acid-exposed mouse embryos. Anat Embryol (Berl). 1996;194:13–22. [PubMed: 8800419]
- 246.
- Lombardo A, Isaacs HV, Slack JM. Expression and functions of FGF-3 in Xenopus development. Int J Dev Biol. 1998;42:1101–1107. [PubMed: 9879707]
- 247.
- Irving C, Mason I. Signalling by FGF8 from the isthmus patterns anterior hindbrain and establishes the anterior limit of Hox gene expression. Development. 2000;127:177–186. [PubMed: 10654611]
- 248.
- Diez del Corral R, Breitkreuz DN, Storey KG. Onset of neuronal differentiation is regulated by paraxial mesoderm and requires attenuation of FGF signalling. Development. 2002;129:1681–1691. [PubMed: 11923204]
- 249.
- Diez del Corral R, Olivera-Martinez I, Goriely A. et al. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40(1):65–79. [PubMed: 14527434]
- 250.
- Houle M, Prinos P, Iulianella A. et al. Retinoic acid regulation of Cdx1: An indirect mechanism for retinoids and vertebral specification. Mol Cell Biol. 2000;20(17):6579–6586. [PMC free article: PMC86138] [PubMed: 10938132]
- 251.
- Lohnes D. The Cdx1 homeodomain protein: An integrator of posterior signaling in the mouse. Bioessays. 2003;25(10):971–980. [PubMed: 14505364]
- 252.
- Gaunt SJ, Drage D, Trubshaw RC. cdx4/lacZ and cdx2/lacZ protein gradients formed by decay during gastrulation in the mouse. Int J Dev Biol. 2005;49:901–908. [PubMed: 16281167]
- 253.
- Gaunt SJ, Cockley A, Drage D. Additional enhancer copies, with intact cdx binding sites, anteriorize Hoxa-7/lacZ expression in mouse embryos: Evidence in keeping with an instructional cdx gradient. Int J Dev Biol. 2004;48:613–622. [PubMed: 15470633]
- 254.
- Gaunt SJ, Drage D, Cockley A. Vertebrate caudal gene expression gradients investigated by use of chick cdx-A/lacZ and mouse cdx-1/lacZ reporters in transgenic mouse embryos: Evidence for an intron enhancer. Mech Dev. 2003;120:573–586. [PubMed: 12782274]
- 255.
- Gaunt SJ. Gradients and forward spreading of vertebrate Hox gene expression detected by using a Hox/lacZ transgene. Dev Dyn. 2001;221:26–36. [PubMed: 11357191]
- 256.
- Carpenter EM, Goddard JM, Chisaka O. et al. Loss of Hoxa-1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development. 1993;118:1063–1075. [PubMed: 7903632]
- 257.
- Goddard J, Rossel M, Manley N. et al. Mice with targeted disruption of Hoxb1 fail to form the motor nucleus of the VIIth nerve. Development. 1996;122:3217–3228. [PubMed: 8898234]
- 258.
- Jungbluth S, Bell E, Lumsden A. Specification of distinct motor neuron identities by the singular activities of individual Hox genes. Development. 1999;126(12):2751–2758. [PubMed: 10331985]
- 259.
- Arenkiel BR, Tvrdik P, Gaufo GO. et al. Hoxb1 functions in both motoneurons and in tissues of the periphery to establish and maintain the proper neuronal circuitry. Genes Dev. 2004;18(13):1539–1552. [PMC free article: PMC443517] [PubMed: 15198977]
- 260.
- Davenne M, Maconochie MK, Neun R. et al. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron. 1999;22:677–691. [PubMed: 10230789]
- 261.
- Guidato S, Prin F, Guthrie S. Somatic motoneurone specification in the hindbrain: The influence of somite-derived signals, retinoic acid and Hoxa3. Development. 2003;130:2981–2996. [PubMed: 12756180]
- 262.
- Wingate RJ, Hatten ME. The role of the rhombic lip in avian cerebellum development. Development. 1999;126:4395–4404. [PubMed: 10498676]
- 263.
- Wahba GM, Hostikka SL, Carpenter EM. The paralogous Hox genes Hoxa10 and Hoxd10 interact to pattern the mouse hindlimb peripheral nervous system and skeleton. Dev Biol. 2001;231(1):87–102. [PubMed: 11180954]
- 264.
- Economides KD, Zeltser L, Capecchi MR. Hoxb13 mutations cause overgrowth of caudal spinal cord and tail vertebrae. Dev Biol. 2003;256:317–330. [PubMed: 12679105]
- 265.
- Gould A. Functions of mammalian Polycomb-group and trithorax-group related genes. Curr Opin Gen Dev. 1997;7:488–494. [PubMed: 9309179]
- 266.
- Tümpel S, Maconochie M, Wiedemann LM. et al. Conservation and diversity in the cis-regulatory networks that integrate information controlling expression of Hoxa2 in hindbrain and cranial neural crest cells in vertebrates. Dev Biol. 2002;246(1):45–56. [PubMed: 12027433]
- 267.
- Pöpperl H, Featherstone M. An autoregulatory element of the murine Hox-4.2 gene. EMBO J. 1992;11(10):3673–3680. [PMC free article: PMC556827] [PubMed: 1356763]
- 268.
- Gavalas A, Trainor P, Ariza-McNaughton L. et al. Synergy between Hoxa1 and Hoxb1: The relationship between arch patterning and the generation of cranial neural crest. Development. 2001;128(15):3017–3027. [PubMed: 11532923]
- 269.
- Arcioni L, Simeone A, Guazzi S. et al. The upstream region of the human homeobox gene HOX3D is a target for regulation by retinoic acid and HOX homeoproteins. EMBO J. 1992;11:265–277. [PMC free article: PMC556447] [PubMed: 1346761]
- 270.
- Nonchev S, Maconochie M, Gould A. et al. Cross-regulatory interactions between Hox genes and the control of segmental expression in the vertebrate central nervous system. Cold Spring Harb Symp Quant Biol. 1997;62:313–323. [PubMed: 9598365]
- 271.
- Yau TO, Kwan CT, Jakt LM. et al. Auto/cross-regulation of Hoxb3 expression in posterior hindbrain and spinal cord. Dev Biol. 2002;252(2):287–300. [PubMed: 12482716]
- 272.
- Ferretti E, Cambronero F, Tümpel S. et al. The Hoxb1 enhancer and control of rhombomere 4 expression: Complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol Cell Biol. 2005;25:8541–8552. [PMC free article: PMC1265741] [PubMed: 16166636]
- 273.
- Ferretti E, Marshall H, Pöpperl H. et al. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127:155–166. [PubMed: 10654609]
- 274.
- Ferretti E, Schulz H, Talarico D. et al. The PBX-regulating protein PREP1 is present in different PBX-complexed forms in mouse. Mech Dev. 1999;83(1-2):53–64. [PubMed: 10381567]
- 275.
- Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. [PMC free article: PMC84356] [PubMed: 10373562]
- 276.
- Chang CP, Shen WF, Rozenfeld S. et al. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. [PubMed: 7729685]
- 277.
- Piper DE, Batchelor AH, Chang CP. et al. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: Role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96(4):587–597. [PubMed: 10052460]
- 278.
- Berthelsen J, Zappavigna V, Ferretti E. et al. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17(5):1434–1445. [PMC free article: PMC1170491] [PubMed: 9482740]
- 279.
- Fibi M, Zink B, Kessel M. et al. Coding sequence and expression of the homeobox gene Hox 1.3. Development. 1988;102(2):349–359. [PubMed: 2901335]
- 280.
- Deflorian G, Tiso N, Ferretti E. et al. Prep1.1 has essential genetic functions in hindbrain development and cranial neural crest cell differentiation. Development. 2004;131(3):613–627. [PubMed: 14711874]
- 281.
- Popperl H, Rikhof H, Chang H. et al. Lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell. 2000;6(2):255–267. [PubMed: 10983974]
- 282.
- Waskiewicz AJ, Rikhof HA, Moens CB. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev Cell. 2002;3(5):723–733. [PubMed: 12431378]
- 283.
- Kimmel CB, Miller CT, Moens CB. Specification and morphogenesis of the zebrafish larval head skeleton. Dev Biol. 2001;233(2):239–257. [PubMed: 11336493]
Footnotes
- a
In the chick the autonomic MNs are called the Column of Terni neurons.
- b
These borders differences are highlighted in Figure 4 by the presence of a red rectangle.
- c
In the mouse, these are Cdx1, Cdx2, and Cdx4. The chicken and Xenopus orthologs are CdxA, CdxB, and CdxC; and Xcad1, Xcad2, and Xcad3, respectively.
- Expression of Hox Genes in the Nervous System of Vertebrates - Madame Curie Bios...Expression of Hox Genes in the Nervous System of Vertebrates - Madame Curie Bioscience Database
- The Function of Toll-Like Receptors - Madame Curie Bioscience DatabaseThe Function of Toll-Like Receptors - Madame Curie Bioscience Database
- Gastroenterologic and Hepatic Diseases - Madame Curie Bioscience DatabaseGastroenterologic and Hepatic Diseases - Madame Curie Bioscience Database
- Human, Mouse and Yeast Telomerase - Madame Curie Bioscience DatabaseHuman, Mouse and Yeast Telomerase - Madame Curie Bioscience Database
- In Situ Activation of Caspases Revealed by Affinity Labeling Their Enzymatic Sit...In Situ Activation of Caspases Revealed by Affinity Labeling Their Enzymatic Sites - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...