NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Telomeres are crucial components required for genomic stability. Telomere dysfunction can result in enormously elevated rates of chromosomal alterations, particularly in subtelomeric regions. Interestingly, the chromosomal regions in the vicinity of telomeres are often among the most rapidly evolving in the genome. These facts could suggest that the protective capping function of telomeres has not evolved to be fail-safe but instead to permit a certain rate of failure that can foster evolution through subtelomeric rearrangements.
Introduction
Chromosome Ends: The Wild West of the Genome?
Just as the geography of the earth has influenced the nature and evolution of human communities, so too has the geography of chromosomes influenced the nature and evolution of genes. A prominent example of this is subtelomeric DNA. Subtelomeric regions are the frontier outposts of the genomes and existence for genes there can be precarious and not always governed by the laws that regulate genes in other parts of the chromosome. Like the inhabitants of frontier towns, the genes in subtelomeric regions are often not representative of those found elsewhere. Instead, subtelomeric regions are often enriched for genes that allow rapid adaptation to new environments. In this chapter, I examine the special consequences that can result from a location near a chromosome end and how this may relate to the structure, function and evolution of subtelomeric sequences.
Telomeric and Broken DNA Ends and the Processes that Act on Them
The long linear DNA molecules that constitute eukaryotic chromosomes can have two types of DNA ends, telomeric ends and broken ends.1 The fundamental difference between a telomere and a broken DNA end is that the former is the natural stable end of the chromosome and the latter is something that typically occurs from damage and that the cell will usually rapidly repair. Telomeres and broken ends are therefore usually treated by the cell in opposite ways.
Telomeric ends are the normal targets for sequence addition by telomerase, which counteracts the gradual sequence loss from ends that occurs as a consequence of the ‘end replication problem’.2 Telomeric ends also function to prevent chromosome ends from acting like broken DNA ends. This protective role is commonly known as capping. In the great majority of eukaryotes, the distinctive features that distinguish telomeres from broken ends result from the specific DNA sequences that make up telomeres as well as the specific proteins that bind them. In the large majority of eukaryotes, telomeric DNA is composed of tandem arrays of a short repeat (5-26 bp) the sequence of which is specified by the template region of the telomerase RNA.3,4 Telomere capping generally involves telomere-specific DNA binding proteins. These include proteins that bind double stranded telomeric repeats as well as those that bind the single stranded 3' overhangs at telomeric termini.5 Paradoxically, some proteins that bind to broken DNA ends also make critical contributions to telomere capping. Additionally, some organisms appear to use t-loops, structures where the 3' overhang is thought to be strand invaded into more internal telomeric repeats, as a means to help cap telomeres.6
Telomeric repeat sequences are not always limited to being at chromosome ends. Many species have telomere-like repeat arrays present at interstitial locations in chromosomes. These likely arise from a number of mechanisms including telomere-telomere fusions and aberrant repair of DNA double strand breaks (DSBs).7 In at least some cases, these interstitial telomeric repeats can influence chromosome structure and stability by being hotspots for chromosome breakage or seed sequences for formation of a new telomere. The incidence and effects of interstitial telomeric repeats are reviewed elsewhere.8
In contrast to telomeric ends, broken DNA ends (such as produced by DSBs) are severe forms of DNA damage and are precursors of many if not most chromosomal rearrangements. DSBs are typically repaired by either of two repair pathways, nonhomologous end joining (NHEJ) or homologous recombination (HR) and are not normally substrates for addition by telomerase.9 NHEJ is a ligation reaction requiring the specialized ligase IV enzyme plus certain additional proteins.10,11 Mitotic DSB repair by HR is thought to commonly proceed through a synthesis-dependent strand annealing mechanism that results in localized gene conversion but not in cross-overs. It requires an intact homologous sequence as a template and this is thought to be most commonly supplied by a sister chromatid. DSB repair through either NHEJ or HR will generally result in the two broken arms of a chromosome being rejoined. HR will normally bring about precise repair (albeit sometimes with gene conversions) while NHEJ is imprecise and will often incorporate small insertions or deletions at the junction point.
Telomeres therefore serve as key guardians of chromosomal integrity. By blocking chromosome ends from being subjected to NHEJ or HR events, telomeres act to preserve the integrity of the genome, particularly in and around subtelomeric regions. As will be discussed below, DNA repair events triggered by telomere failure or by DSBs near telomeres can be crucial factors in shaping the sequences that are present in subtelomeric regions.
Immediate Subtelomeric Regions and Their Possible Functions
Subtelomeric regions are the DNA sequences in the vicinity of chromosome ends. An exact definition is not possible but an approximate definition would be those sequences adjacent to the telomeres that have features that differentiate them from other regions of chromosomes. These regions are known from many organisms for unusual characteristics that include a complex repetitive structure, rapid evolution and a frequently heterochromatic nature.12-14
Subtelomeric sequences can loosely be grouped into two categories; regions immediately adjacent to the telomeric repeats that often lack genes and are present at a large percentage of the chromosome ends and more internal gene-containing regions that are present at smaller subsets of chromosome ends. When subtelomeric sequences are present at more than one chromosome end, they strongly tend to have the same orientation with respect to the telomeres where they are present. The functions of the generally gene-free, immediate subtelomeric sequences are not fully clear and may vary between species. Although they do not appear to be vital to the basic protective capping function of telomeres, there is evidence that they can contribute to telomere length regulation.15-19 Subtelomeric sequences can also contribute to telomere position effect (TPE), the transcriptional repression that occurs next to telomeres.20-22
Another function of immediate subtelomeric elements can sometimes be to serve as templates for telomere repair through homologous recombination. In S. cerevisiae, mutants lacking telomerase frequently amplify subtelomeric Y' sequences and spread them to chromosome ends where they were not originally present.23 Y' elements are DNA sequences of several kilobases in size that are present in one or two copies next to many but not all telomeres in wild type cells. In the related yeast, Kluyveromyces lactis, subtelomeric sequences can also be spread to other telomeres when telomeres become short and prone to recombination.24 These recombination events are thought to represent break-induced replication (BIR), a nonreciprocal HR event copying a sequence from one chromosome arm to another that acts to restore telomeric sequences to chromosome ends that have lost most or all of their telomeric repeats.25
It is interesting to speculate that immediate subtelomeric repeats could sometimes have characteristics of selfish DNA elements. Replacement of part or all of such an element at one telomere by a BIR event would clearly involve competition between other such subtelomeric elements in the cell. If subtelomeric BIR events are not strictly limited to mutants with compromised telomere function, this would have the potential to select for elements particularly able to promote their own spread by BIR events. The common presence of families of small direct repeats in immediate subtelomeric sequences conceivably might act to facilitate the homology search of a Rad51-coated DNA filament that would be expected to initiate most BIR events.
Telomeres have been shown to be involved in the early stages of homologous chromosome pairing during meiosis.26,27 However, yeast subtelomeric sequences appear to be relatively resistant to being sites of meiotic cross over events. Work in S. cerevisiae has shown that subtelomeric sequences, even if moved away from a telomere, are relatively resistant to crossover formation.28 Those crossovers that do occur in subtelomeric DNA appear poorly able to bring about proper chromosome segregation at meiosis I.29 Consistent with these data, cleavage by Spo11, the nuclease that makes the DNA double strand breaks that initiate meiotic recombination, is infrequent in subtelomeric DNA.30,31 These data suggest that one function of subtelomeric DNA in yeast is to prevent Spo11-induced meiotic crossovers from occurring in regions of the chromosome where they would not be able to properly function. Meiotic recombination maps in humans, on the other hand, show an increase in recombination at the most distal markers.32,33
Subtelomeric Regions Are Often Enriched in Contingency Genes
The genes in subtelomeric regions often include rapidly evolving gene families and “contingency genes”34 involved in adaptation to changing environments. The most notable examples of this come from a number of eukaryotic pathogens where variant surface antigens or other gene families associated with rapid adaptation to the host's immune system are typically found at subtelomeric locations.35 Organisms where this occurs include Typanosoma brucei,36,37 Plasmodium falciparum,38 Pneumocystis carinii,39 and Candida glabrata.40 In T. brucei, for example, there are hundreds of silent variant surface glycoprotein (VSG) genes but only one of these is transcriptionally expressed in a given cell at a time. Switching between different expressed copies of the gene occurs at high frequency in the mammalian host through a mechanism that generally involves being duplicated into one of 20-30 bloodstream expression sites residing next to a telomere.36,37
Multiple lines of evidence indicate that human subtelomeric regions evolve rapidly. An estimated half of known human subtelomeric DNA, which exists as patchworks of interchromosomal segmental duplications, is estimated to have arisen since the human-chimp split.14,41,42 Among individuals, particular subtelomeric regions are often highly polymorphic. Subtelomeric mutations are thought to underlie ˜5% of idiopathic mental retardation.43 Human subtelomeric regions are also observed to undergo high rates of sister chromatid exchange.44,45
Rapid subtelomeric evolution is also apparent in Saccharomyces. A number of gene families relevant to adaptation to novel environments are present at subtelomeric locations in S. cerevisiae. The subtelomeric SUC and RTM gene families (encoding invertase and a resistance to molasses determinant, respectively) vary widely in copy number between yeast strains.46-49 A comparative study of the genomes of four closely related Saccharomyces species found that the majority of genes without clear corresponding genes in other species were subtelomeric.50 Yeast subtelomeric regions are also enriched for mutationally-inactivated genes.50-52
Although S. cerevisiae is normally free living, its subtelomeric gene families might be subject to changing selective pressures similar to those experienced by the cell surface protein gene families of parasites such as T. brucei. A number of the subtelomeric gene families in S. cerevisiae encode proteins that are secreted. These provide the potential for “cheaters”, that produce little or none of the communal resource (the secreted enzyme), to be favored by natural selection at high cell concentrations. Recent work has demonstrated the plausibility of this idea for the subtelomeric SUC family in yeast.53 The SUC2 genes encode invertase, a secreted enzyme required for utilization of sucrose. Isogenic strains either lacking SUC2 or containing two SUC2 genes near different telomeres were passaged together at different cell densities on medium containing sucrose as a carbon source. While the cells lacking SUC2 were less fit at low cell densities, they were more fit at high cell densities than the strain with SUC2 genes. These fitness differences were specific to growth on sucrose. Thus, S. cerevisiae cells are likely to sometimes experience fluctuating environmental conditions that select for changing expression levels or copy number of SUC2 genes.
Subtelomeric DNA is Intrinsically Tolerant of Rearrangement
What forces help govern the often repetitive structure and rapid evolution of subtelomeric sequences? A key factor is simply their location near chromosome ends. To understand why this is true, it is useful to first discuss the kinds of aberrant repair events that can occur at a DSB. I will refer to any DSB repair that rejoins the original two chromosome arms, regardless of whether mutations arise at the junction point, as ‘conservative’ repair and any other outcomes as ‘nonconservative’ repair. To restore the integrity of a chromosome suffering a DSB it is necessary for broken ends to be eliminated and for the chromosome to terminate with telomeres. There are only two general routes to eliminate a broken end. Either it can be joined to a second end, thereby eliminating both, or it can be directly or indirectly extended to acquire a telomere. End joinings that create dicentric chromosomes (such as fusions between sister chromatids lacking a telomere) will be unstable and will likely lead to new DSBs through breakage-fusion-bridge cycles.54-56 End joinings circularizing a chromosome fragment (which would require loss or failure of the other telomere) can function in mitotic cells but will lead to dicentric chromosomes from crossover formation in meiosis. Hence they are not likely to be maintained in populations. The type of nonconservative NHEJ event most likely to be tolerated would probably be mis-joining two simultaneous DSBs to produce a reciprocal translocation.
The second route to eliminate a broken end, acquiring a telomere, could occur in at least two ways. De novo telomere formation could occur by direct addition of telomeric repeats to a broken end by telomerase. There is evidence that this occasionally occur at broken ends, particularly at ends with coincidental similarity to telomeric sequence.57-60 The most likely result of de novo telomere addition would be a terminal truncation of the affected chromosome and loss of the acentric fragment. A second way for a broken end to acquire a telomere would be for a BIR event to extend the sequence at a broken end by copying another chromosome arm all the way to the telomere. This would not only add telomeric repeats but also some amount, perhaps very considerable, of additional DNA and result in a duplication (nonreciprocal translocation) of the copied chromosome arm.
In addition to restoring the presence of a telomere, the nonconservative DSB repair events discussed above tend to have other features in common. For one, in moving or duplicating DNA regions, they typically maintain sequence orientation relative to the telomere. This is significant because, as mentioned above, sequences present at multiple subtelomeric regions within an organism commonly have the same orientation relative to the telomere. Second, there is less selective pressure for conservative repair of DSBs close to telomeres than there is for DSBs far from telomeres. At most positions within a chromosome, there will be strong selection for conservative DSB repair. This is because the aneuploidies that would result from the nonconservative repair would be smaller the closer the DSB had been to a telomere. Simply put, small terminal deletions or duplications would be less detrimental than large ones (Fig. 1). The important implication of this is that nonconservative DSB repair is more likely to be survivable the closer it occurs to telomeres. The corollary of this is that subtelomeric DNA will intrinsically be more tolerant of accumulating DNA rearrangements formed by nonconservative DSB repair than will other parts of the chromosome.
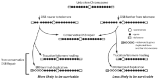
Figure 1
Subtelomeric regions are more tolerant of nonconservative DSB repair. See text for details.
Direct evidence for the tolerance of subtelomeric regions to nonconservative DSB repair has come from work in S. cerevisiae. Yeast strains engineered to have a DSB at any of multiple places along a single chromosome were examined for their rate of survival and mode of DNA repair.61 As the strains were haploid, no HR-mediated gene conversion using a homologue as template was possible. Also, because the chromosome break was actually a pair of closely spaced DSBs with noncompatible overhangs, repair by simple NHEJ was inefficient. Results from this study found that DSBs in subtelomeric regions were more frequently able to produce surviving cells. The reason for this was an increase in the number of recovered cells that had undergone nonconservative DSB repair. Most prevalent were BIR events that copied terminal regions of other chromosomal arms. However, instances of de novo telomere formation were also found.61 Interestingly, repair of subtelomeric DSBs (but not DSBs at other locations), by both conservative NHEJ and other mechanisms, was found to mostly depend on telomeric tethering to the nuclear periphery.62 This suggests the possibility that repair of subtelomeric DNA could be subject to regulation in a manner non-identical to that of DSBs elsewhere in the genome.
There is expected to be a synergistic evolutionary relationship between the tolerance of subtelomeric terminal deletions/duplications and the occurrence of nonconservative DSB repair events. Once a chromosomal terminus has been duplicated onto another chromosome end, this could greatly increase the likelihood that future repair of DSBs in or near the duplicated region will involve homologous recombination between those different chromosomes.
The Differences between Uncapped Telomeres and Broken DNA Ends
As described above, broken DNA ends formed occasionally near telomeres have clear potential to influence subtelomeric evolution. Another factor of potentially even greater importance in subtelomere structure and evolution is the influence of telomeres. Although the prime function of telomeres is to protect chromosome ends from acting like broken ends, this function can fail for a variety of reasons and such telomere uncapping can be a powerful engine for generating chromosomal rearrangements, particularly those affecting subtelomeric regions. Before discussing how this could happen it is useful to review certain features of uncapped telomeres and how these differ from broken ends.
Disruption of telomere capping can subject telomeres to NHEJ and HR, the same repair pathways that act on broken ends. However, uncapped telomeres differ from broken ends in a number of important ways. An obvious difference is that uncapped telomeres are limited to occurring at chromosome ends while broken ends can occur anywhere in the genome. This can strongly bias DNA rearrangements derived from aberrant repair of uncapped telomeres to occur in subtelomeric regions.
The causes of broken ends and uncapped telomeres are also typically different. Uncapped telomeres are not after all, DNA breaks, but rather malfunctioning natural DNA ends. Telomere uncapping can occur from either of two basic causes. Either telomeric DNA repeat arrays become too short to properly function or the function of one or more telomeric capping proteins are disrupted in some way. The former is observed when cells divide repeatedly in the absence of telomerase such as is the case in human somatic cells naturally lacking telomerase and in yeast mutants deleted for telomerase.25,63 Although multiple telomeres in such cells can acquire capping defects more or less simultaneously, this type of capping defect appears to act at individual telomeres independently. Thus, the shortest telomeres in a cell may become uncapped at a time when other, longer telomeres do not. In contrast, capping defects occurring through mutation or imbalance of a telomeric protein will commonly affect all telomeres in the cell simultaneously. An additional important point is that, because of the varied roles of telomeric proteins, capping defects resulting from protein defects tend to be more varied in their phenotype than capping defects caused by telomere shortening.
Unlike broken DNA ends, which are an essentially all or nothing phenomenon, telomere uncapping is more variable. In some cases it might be total, where a telomere behaves indistinguishably from a broken end. However, in most cases it is more likely to be partial, where a telomere might have properties intermediate between a broken end and a normally functioning telomere. Different kinds of partial capping defects can easily be imagined. For example, telomere failure might occur stochastically, rendering a chromosome end subject to DNA repair pathways is some, but not all, cell divisions. Alternatively, a capping defect might cause a telomere to be prone to one type of DSB repair but not to the other. NHEJ favors ends that are blunt or with minimal overhangs while HR is favored by the long 3' single stranded tails that can be produced by exonucleolytic processing. Protection against these two processes is thus likely to require at least partially different mechanisms that might be differentially affected by a particular capping defect. To summarize, the effects of telomere capping defects are potentially more subtle and variable than those of broken ends. This gives them appreciable flexibility for influencing subtelomeric evolution.
Other important features of broken ends and uncapped telomeres concern both their structure and their manner of repair. Broken ends occur in two basic types. Those that occur in pairs from a DSB and those that occur singly from collapsed or stalled replication forks. The former are normally repaired conservatively by HR or NHEJ resulting in rejoining of the two ends. The latter are repaired through a HR-dependent process that leads to restarting the replication fork.64 Uncapped telomeres, in contrast, can occur singly or in multiple and cannot be conservatively repaired by being joined to another end. Uncapped telomeres also typically contain some residual number of telomeric repeats. These have the potential to greatly bias DNA repair activity. They would, for example, certainly be expected to enhance the likelihood that telomerase (if present in the cell) would act on an uncapped telomere but they might also affect the ability of HR or NHEJ to act as well.
How uncapped telomeres are repaired varies depending upon circumstances and organism. The most simple repair would be to restore a functional telomere. If uncapping is due to telomere shortening, this can be straightforward. If telomerase is present, it should be able to directly elongate the telomere to a size that will permit capping. If telomerase is not present, an uncapped telomere may undergo HR reactions that can lengthen or replace it.23,65 The simplest mechanism for this would be for the short telomere to strand invade a longer telomere and copy its sequence in a BIR event. Such events have been shown to be very efficient in yeast when a long telomere is available as a substrate.66 However, this mechanism may not be feasible when all telomeres are becoming critically short together, such as in a typical senescing yeast telomerase deletion mutant. In such cases though, HR processes can still occasionally generate elongated telomeres, apparently by first producing a small telomeric circle and then using it as a template for a BIR event to make an elongated telomere.67,68 Other cells, such as most senescing human somatic cells, do not appear to be able to lengthen telomeres using HR. As the replicative senescence caused by telomere shortening in human cells is thought to help prevent carcinogenesis, this failure of HR to act at short telomeres might be an adaptive trait. In the event that telomere uncapping is due to a defect in a telomeric protein, lengthening the telomere by telomerase or HR may not be able to restore a capped state. In such cases, telomeres might persist in a chronically uncapped state.
NHEJ has been shown to act on uncapped telomeres in a number of circumstances.69-75 The most common outcome of this is for fusion to occur between two uncapped telomeres. This can occur between two telomeres of different chromosome arms or between identical replicated sister chromosome arms, the latter favored when only one uncapped telomere is present. In some cases fusions may occur between chromosome ends that retain telomeric repeats and in others it may occur only after all telomeric repeats have been lost. In extreme cases, all chromosome ends in the cell may undergo fusions.76-78 Through eliminating chromosome ends altogether, telomere fusions can at least transiently eliminate any type of telomere capping defect. However, the dicentric chromosomes created by such fusions promote recurrent breakage-fusion-bridge cycles that tear apart and reform chromosomes.
Disruption of Telomere Capping Can Trigger High Rates of Subtelomeric Change
The consequences of telomere dysfunction have been documented in numerous studies done in multiple organisms. In some of these, dramatic effects on subtelomeric sequences have been observed. In an early example, a temperature-sensitive defect in S. cerevisiae Cdc13, (a protein that protects 3' overhangs and helps recruit telomerase) was found to lead to a large increase in mitotic recombination in subtelomeric regions after a period at the restrictive temperature.79 The increase was greatest near the telomere but extended ˜50 kb distance from it. Amplification of subtelomeric Y' elements were found to characterize the major type of post-senescence survivors that occur through HR in S. cerevisiae mutants lacking telomerase.23,80 Y' element amplification in ‘Type I’ survivors generates large tandem arrays of the elements and spreads them to most or all chromosome ends. The amplified DNA can be up to several percent of total genomic DNA in these survivors. This subtelomeric amplification likely depends on the presence of short blocks of telomeric repeats that are typically present just internal to the Y' elements in both wild type cells and the Type I survivors. The shortening terminal telomeric repeat arrays likely become prone to HR and strand invade into the subtelomeric blocks of telomere repeats and thereby initiate the events leading to the large scale amplification. In K. lactis, where wild type cells have telomeric repeats only at chromosome ends, telomerase deletion postsenescence survivors do not exhibit subtelomeric amplification.65 However, if even a single block of telomeric repeats is engineered to be internal to a subtelomeric marker gene, postsenescence survivors can readily form long tandem arrays of the alternating telomeric and marker gene sequences.81
Other types of subtelomeric rearrangements have also been observed at highly elevated frequencies in yeast telomerase deletion mutants during or after the period when telomeres were very short and malfunctioning.82-84 These included terminal deletions, BIR events that replaced one subtelomeric region with duplicated subtelomeric sequence from another chromosome arm and fusions between chromosome ends that had lost telomeric and some subtelomeric sequence. Very recently, base substitution and frame shift mutations in a subtelomeric gene (but not an internal gene) were also found to be significantly elevated in senescing S. cerevisiae cells without telomerase.85 Both the base mutations and the gross chromosomal rearrangements were found to be largely dependent on the Rev1 and Pol ζ DNA polymerases that are involved in the error-prone bypass of DNA damage. Other examples of large increases in the rate of subtelomeric BIR events have been observed in K. lactis. Telomerase deletion mutants with short dysfunctional telomeres can readily spread a subtelomeric marker gene to most or all other telomeres in the cell via shared subtelomeric homology.24 In this manner, a “gene family” with copies of the marker gene spread to most or all subtelomeres can be generated very rapidly in the laboratory. Elevated subtelomeric BIR has also been seen in other situations when telomere function has been compromised in K. lactis. Most notably, cells with telomeres stable at appreciably shorter than normal lengths and that do not undergo growth senescence can experience subtelomeric BIR rates that can be 40-200 fold elevated relative to wild type cells.24 This elevated BIR rate likely stems from the telomeres in these cells frequently dropping below a critical length that is able to prevent the telomere from engaging in HR. Work using tagged telomeric sequence in telomerase deletion mutants has shown that K. lactis telomeres can undergo recombination once they drop below about 100 bp in size.66 How a chromosome end retaining telomeric repeats could lead to subtelomeric gene conversions is not certain. One possibility is that the uncapped chromosome end is processed to have a 3' single strand tail that becomes bound by the Rad51 protein and the resulting strand invasion into another telomere could be followed by branch migration into subtelomeric regions. Certain resolutions of such structures might then be easily imagined to cause subtelomeric gene conversions.
Data showing that telomere dysfunction can promote subtelomeric recombination is not limited to yeast. In Trypanosoma brucei, telomere shortening caused by loss of telomerase has been shown to promote a duplicative gene conversion (thought to be BIR) that replaced the expressed variant surface gene at the expression site with another VSG gene that was previously unexpressed.86 This had led to the suggestion that short telomere lengths in trypanosomes may be associated with a higher rate of antigenic switching.87
Adaptive Telomere Failure: A Fast Track for Subtelomeric Evolution?
Telomeres are clearly of huge importance in guarding the stability of subtelomeric regions. However, this very property makes them potentially ideal agents for regulating and promoting subtelomeric evolution. I suggest that regulated failure of the telomere capping function could be an adaptive trait that can facilitate evolution of subtelomeric sequences. In essence, this hypothesis states that telomere capping in some, perhaps many, organisms has evolved to be less than 100% effective at protecting telomeric ends from engaging in DNA repair processes. It is reasonable to further predict that the repair process that would be primarily favored by adaptive telomere failure would be homologous recombination. HR would have the ability to recombine or amplify subtelomeric gene families with less risk of detrimental formation of dicentric chromosomes.
Several variants of this basic adaptive telomere failure model are conceivable. A simple possibility is that telomere capping function in some species might have evolved to have a finite and perhaps appreciable failure rate under all circumstances. This would make the testable prediction that telomere capping in such a species could be improved by mutation or overexpression of capping components in ways that would reduce the background rates of subtelomeric rearrangements.
An especially intriguing possibility is that an increase in the rate of telomere failure might be inducible. In this scenario, telomeres might function with a very low failure rate in most situations but with an appreciably higher failure rate under certain other circumstances. Unicellular eukaryotes subjected to starvation or other stress conditions could represent an example of a situation where increased rates of subtelomeric rearrangements might be evolutionarily favorable. Inducing an increased telomere failure rate would be trivial mechanistically. The decreased expression, or modification of a single telomere capping component might easily be sufficient and could affect all telomeres equally. It is also possible that an inducible telomere capping defect could evolve that was specific to a single telomere, perhaps through affecting transcription or a DNA binding protein located next to a particular telomere.
Increased telomere failure rates could obviously also arise through mutation. Sufficiently large populations, such as occur in many microbes, that are subject to strong selective pressures can favor the emergence of mutator phenotypes that produce elevated mutations rates.88 A mutation arising in a telomere capping component that elevated the rate of formation subtelomeric rearrangements would serve as a very effective region-specific mutator. Once the original selective pressure was relaxed, reversion or suppression of the telomeric capping mutation would likely occur. In higher eukaryotes, it is interesting to speculate that alleles that cause increased rates of telomere failure might be present in a subset of individuals in populations. These alleles might boost the rates of subtelomeric rearrangements in the individuals where they occurred. Such alleles might affect all cells within individuals carrying them or they could be imagined to act in a germline-specific manner.
An obvious potential example of adaptive telomere failure identified to date is the telomere shortening that occurs in most human somatic cells because of the transcriptional repression of telomerase in embryonic development. Human cells containing some small number of telomeres shortened to a critical degree are subject to a permanent growth arrest called replicative senescence.89 This phenomenon is widely believed to be an adaptation that blocks the unlimited replication of cancerous or precancerous cells. In this case, the adaptive role of the telomere dysfunction is limited to somatic cells and is thought to involve fostering genetic stability rather than genetic change. Interestingly, human cells lacking the pathway that arrests growth in response to short telomeres eventually experience massive cell death, apparently from the deleterious consequences of high rates of telomere fusions. The rare cells that survive beyond this crisis emerge with an active telomere maintenance pathway (either telomerase or HR-mediated ALT) and often exhibit considerable karyotypic abnormality.56,89 In such cases, the telomere dysfunction is thought to potentially serve as a mutator mechanism that can help promote cancer cell formation.
Telomere Position Effect Furthers the Adaptive Plasticity of Subtelomeric DNA
In addition to enhanced genetic plasticity, subtelomeric regions of chromosomes also often have a well-documented epigenetic plasticity. Subtelomeric regions in many organisms are heterochromatic. In several species, including Drosophila,90,91 S. cerevisiae,92,93 S. pombe,94 Candida glabrata,95 Plasmodium falciparum,96 Trypanosoma brucei,97 plants98 and vertebrates,99-101 genes inserted near telomeres are subject to telomere position effect (TPE) where they are able to switch at low but appreciable rates between transcriptionally silenced and transcriptionally active states. Where examined, this TPE is mediated in part through the presence or absence of specific modifications to histones present at subtelomeric regions.
Telomere function and TPE have important links connecting them. Proper telomere function is critical for TPE. For example, proteins interacting directly with telomeres, such as Rap1 and Ku in yeast, are required for TPE.102,103 Also, telomere length can influence TPE. In both yeast and humans, short telomeres are associated with diminished TPE.99,102 Similarly, the chromatin modifications associated with TPE can influence telomere function. In mammalian cells lacking histone methyltransfereases thought to modify subtelomeric chromatin, telomeres become abnormally elongated.104 Additionally, in S. cerevisiae, Rif proteins involved in telomere length regulation compete with Sir proteins involved in TPE for the ability to bind to the telomeric Rap1 protein.102
TPE can readily be imagined to be able to act synergistically with the genetic plasticity of subtelomeric regions to allow populations of organisms to have considerable adaptive headroom to respond to environmental challenges. TPE might, for example, act as a buffering force to mitigate any reduced fitness from increased gene dosage caused by a nonreciprocal translocation of a subtelomeric region to another chromosome end. This could allow adequate time for mutational mechanisms to suppress the reduced fitness and perhaps also permit further advantageous divergence of the duplicated sequence.
The Relationship between Chromosome Ends and Centromeres
A final area of discussion about the influence of telomeres on chromosome evolution concerns centromeres. Telomeres and their adjoining subtelomeric regions often share functional, structural and/or evolutionary ties with centromeres. Not only are telomeres and centromeres essential components of chromosomes but they also must evolve together to remain in a fixed two-to-one ratio. Any splitting or fusion of chromosomes must ultimately lead to a corresponding change in the number of both telomeres and centromeres. Subtelomeric and pericentromeric sequences are also often physically nearby on a chromosome and may even effectively overlap in acrocentric and telocentric chromosomes. For example, the short p arms of mouse chromosomes, have only 1.8-11 kb of DNA separating telomeres from pericentric repeats.105 The very fact that some organisms, such as mice, have only telocentric or acrocentric chromosomes clearly indicates that mutational or repair processes that can alter chromosome structure can favor outcomes where centromeres are adjacent to a telomere.
An additional tie between subtelomeric and centromeric regions is that both tend to be heterochromatic and composed of complex repeat families, some of which may even be shared in some organisms. Shared sequences would likely permit some level of ectopic homologous recombination to occur between subtelomeric and centromeric sequences. Indeed, the centromeric regions of Schizosaccharomyces pombe have been suggested to have evolved from head to head fusion of two sets of subtelomeric sequences.106 Even the key function of centromeres, serving as attachment points for permitting microtubule-driven chromosome movement, can in some contexts be done by chromosome ends. The telomeres can lead chromosome movements in a microtubule-dependent fashion in the prophase of meiosis I.26,107,108 Considerable more work will be needed to fully understand the multiple ways that telomeric and centromeric regions are interconnected.
Conclusion
Subtelomeric regions of chromosomes characteristically show a high level of genetic and epigenetic plasticity. A variety of reasons likely contribute to this. Perhaps the most fundamental is that, relative to more internal parts of chromosomes, subtelomeric regions are inherently more tolerant of nonconservative repair of DNA double strand breaks that produce terminal duplications and truncations. This in turn produces a genomic environment favorable for genes and sequences that are tolerant of and may benefit from, frequent recombination, changing copy number and the transcriptional irregularity of telomere position effect. Of huge importance to the genetic stability of subtelomeric sequences is the protective capping function of telomeres. Clearly the predominant role of telomeres in cells is to protect chromosome ends from eliciting DNA repair processes. However, it is hypothesized that a regulated relaxation of the protective capping function, termed adaptive telomere failure, could be an important mechanism helping to shape the genetic plasticity of subtelomeric regions in at least some organisms. Experiments are needed to test predictions made by this hypothesis.
References
- 1.
- de Lange T, Lundblad V, Blackburn EH.eds.Telomeres, second edition Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press 2006 .
- 2.
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337(6205):331–337. [PubMed: 2463488]
- 3.
- Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116(2):273–279. [PubMed: 14744437]
- 4.
- Autexier C, Lue NF. The Structure and Function of Telomerase Reverse Transcriptase. Annu Rev Biochem. 2006;75:493–517. [PubMed: 16756500]
- 5.
- Bertuch AA, Lundblad V. The maintenance and masking of chromosome termini. Curr Opin Cell Biol. 2006;18(3):247–253. [PubMed: 16682180]
- 6.
- Griffith JD, Comeau L, Rosenfield S. Mammalian telomeres end in a large duplex loop Cell. 1999. pp. 503–514. [PubMed: 10338214]
- 7.
- Azzalin CM, Nergadze SG, Giulotto E. Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma. 2001;110(2):75–82. [PubMed: 11453557]
- 8.
- Bolzan AD, Bianchi MS. Telomeres, interstitial telomeric repeat sequences and chromosomal aberrations. Mutat Res. 2006;612(3):189–214. [PubMed: 16490380]
- 9.
- Aylon Y, Kupiec M. DSB repair: the yeast paradigm. DNA Repair (Amst). 2004;3(8-9):797–815. [PubMed: 15279765]
- 10.
- Daley JM, Palmbos PL, Wu D. et al. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. [PubMed: 16285867]
- 11.
- Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by nonhomologous end joining. DNA Repair (Amst). 2005;4(6):639–648. [PubMed: 15907771]
- 12.
- Pryde FE, Gorham HC, Louis EJ. Chromosome ends: all the same under their caps. Curr Opin Genet Dev. 1997;7(6):822–828. [PubMed: 9468793]
- 13.
- Mewborn SK, Lese Martin C, Ledbetter DH. The dynamic nature and evolutionary history of subtelomeric and pericentromeric regions. Cytogenet Genome Res. 2005;108(1-3):22–25. [PubMed: 15545712]
- 14.
- Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet. 2002;3(2):91–102. [PubMed: 11836503]
- 15.
- Craven RJ, Petes TD. Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics. 1999;152(4):1531–1541. [PMC free article: PMC1460705] [PubMed: 10430581]
- 16.
- Ray A, Runge KW. The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol Cell Biol. 1999;19(1):31–45. [PMC free article: PMC83863] [PubMed: 9858529]
- 17.
- Figueiredo LM, Freitas-Junior LH, Bottius E. et al. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21(4):815–824. [PMC free article: PMC125872] [PubMed: 11847128]
- 18.
- Jacob NK, Stout AR, Price CM. Modulation of telomere length dynamics by the subtelomeric region of tetrahymena telomeres. Mol Biol Cell. 2004;15(8):3719–3728. [PMC free article: PMC491831] [PubMed: 15169872]
- 19.
- Berthiau AS, Yankulov K, Bah A. et al. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J. 2006;25(4):846–856. [PMC free article: PMC1383556] [PubMed: 16467854]
- 20.
- Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18(9):2538–2550. [PMC free article: PMC1171335] [PubMed: 10228167]
- 21.
- Fourel G, Revardel E, Koering CE. et al. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18(9):2522–2537. [PMC free article: PMC1171334] [PubMed: 10228166]
- 22.
- Cryderman DE, Morris EJ, Biessmann H. et al. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. EMBO J. 1999;18(13):3724–3735. [PMC free article: PMC1171449] [PubMed: 10393187]
- 23.
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73(2):347–360. [PubMed: 8477448]
- 24.
- McEachern MJ, Iyer S. Short telomeres in yeast are highly recombinogenic. Mol Cell. 2001;7(4):695–704. [PubMed: 11336694]
- 25.
- McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. [PubMed: 16756487]
- 26.
- Chikashige Y, Ding DQ, Funabiki H. et al. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264(5156):270–273. [PubMed: 8146661]
- 27.
- Bass HW. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell Mol Life Sci. 2003;60(11):2319–2324. [PubMed: 14625678]
- 28.
- Barton AB, Su Y, Lamb J. et al. A function for subtelomeric DNA in Saccharomyces cerevisiae. Genetics. 2003;165(2):929–934. [PMC free article: PMC1462788] [PubMed: 14573499]
- 29.
- Ross LO, Maxfield R, Dawson D. Exchanges are not equally able to enhance meiotic chromosome segregation in yeast. Proc Natl Acad Sci USA. 1996;93(10):4979–4983. [PMC free article: PMC39391] [PubMed: 8643515]
- 30.
- Klein S, Zenvirth D, Dror V. et al. Patterns of meiotic double-strand breakage on native and artificial yeast chromosomes. Chromosoma. 1996;105(5):276–284. [PubMed: 8939820]
- 31.
- Gerton JL, DeRisi J, Shroff R. et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97(21):11383–11390. [PMC free article: PMC17209] [PubMed: 11027339]
- 32.
- Kong A, Gudbjartsson DF, Sainz J. et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31(3):241–247. [PubMed: 12053178]
- 33.
- Matise TC, Sachidanandam R, Clark AG. et al. A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set. Am J Hum Genet. 2003;73(2):271–284. [PMC free article: PMC1180367] [PubMed: 12844283]
- 34.
- Moxon ER, Rainey PB, Nowak MA. et al. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4(1):24–33. [PubMed: 7922307]
- 35.
- Barry JD, Ginger ML, Burton P. et al. Why are parasite contingency genes often associated with telomeres? Int J Parasitol. 2003;33(1):29–45. [PubMed: 12547344]
- 36.
- Horn D, Barry JD. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 2005;13(5):525–533. [PubMed: 16132817]
- 37.
- Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109(1):5–8. [PubMed: 11955440]
- 38.
- Figueiredo L, Scherf A. Plasmodium telomeres and telomerase: the usual actors in an unusual scenario. Chromosome Res. 2005;13(5):517–524. [PubMed: 16132816]
- 39.
- Keely SP, Renauld H, Wakefield AE. et al. Gene arrays at Pneumocystis carinii telomeres. Genetics. 2005;170(4):1589–1600. [PMC free article: PMC1449779] [PubMed: 15965256]
- 40.
- De Las Penas A, Pan SJ, Castano I. et al. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003;17(18):2245–2258. [PMC free article: PMC196462] [PubMed: 12952896]
- 41.
- Riethman H, Ambrosini A, Paul S. Human subtelomere structure and variation. Chromosome Res. 2005;13(5):505–515. [PubMed: 16132815]
- 42.
- Linardopoulou EV, Williams EM, Fan Y. et al. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437(7055):94–100. [PMC free article: PMC1368961] [PubMed: 16136133]
- 43.
- Knight SJ, Flint J. The use of subtelomeric probes to study mental retardation. Methods Cell Biol. 2004;75:799–831. [PubMed: 15603454]
- 44.
- Cornforth MN, Eberle RL. Termini of human chromosomes display elevated rates of mitotic recombination. Mutagenesis. 2001;16(1):85–89. [PubMed: 11139603]
- 45.
- Rudd MK, Friedman C, Parghi SS. et al. Elevated rates of sister chromatid exchange at chromosome ends. PLoS Genet. 2007;3(2):e32. [PMC free article: PMC1802831] [PubMed: 17319749]
- 46.
- Carlson M, Celenza JL, Eng FJ. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol Cell Biol. 1985;5(11):2894–2902. [PMC free article: PMC369100] [PubMed: 3018485]
- 47.
- Teunissen AW, Steensma HY. Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast. 1995;11(11):1001–1013. [PubMed: 7502576]
- 48.
- Denayrolles M, de Villechenon EP, Lonvaud-Funel A. et al. Incidence of SUC-RTM telomeric repeated genes in brewing and wild wine strains of Saccharomyces. Curr Genet. 1997;31(6):457–461. [PubMed: 9211787]
- 49.
- Ness F, Aigle M. RTM1: a member of a new family of telomeric repeated genes in yeast. Genetics. 1995;140(3):945–956. [PMC free article: PMC1206678] [PubMed: 7672593]
- 50.
- Kellis M, Patterson N, Endrizzi M. et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423(6937):241–254. [PubMed: 12748633]
- 51.
- Harrison P, Kumar A, Lan N. et al. A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J Mol Biol. 2002;316(3):409–419. [PubMed: 11866506]
- 52.
- Lafontaine I, Fischer G, Talla E. et al. Gene relics in the genome of the yeast Saccharomyces cerevisiae. Gene. 2004;335:1–17. [PubMed: 15194185]
- 53.
- Greig D, Travisano M. The Prisoner's Dilemma and polymorphism in yeast SUC genes. Proc Biol Sci. 2004;271(Suppl 3):S25–26. [PMC free article: PMC1810003] [PubMed: 15101409]
- 54.
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. [PMC free article: PMC1209127] [PubMed: 17247004]
- 55.
- McClintock B. The fusion of broken ends of chromosomes following nuclear fusion. Proc Natl Acad Sci USA. 1942;28:458–463. [PMC free article: PMC1078518] [PubMed: 16578057]
- 56.
- Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst). 2006;5(9-10):1082–1092. [PubMed: 16784900]
- 57.
- Kramer KM, Haber JE. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 1993;7(12A):2345–2356. [PubMed: 8253381]
- 58.
- Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76(1):145–155. [PubMed: 8287473]
- 59.
- Mangahas JL, Alexander MK, Sandell LL. et al. Repair of chromosome ends after telomere loss in Saccharomyces. Mol Biol Cell. 2001;12(12):4078–4089. [PMC free article: PMC60777] [PubMed: 11739802]
- 60.
- Pennaneach V, Putnam CD, Kolodner RD. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol. 2006;59(5):1357–1368. [PubMed: 16468981]
- 61.
- Ricchetti M, Dujon B, Fairhead C. Distance from the chromosome end determines the efficiency of double strand break repair in subtelomeres of haploid yeast. J Mol Biol. 2003;328(4):847–862. [PubMed: 12729759]
- 62.
- Therizols P, Fairhead C, Cabal GG. et al. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol. 2006;172(2):189–199. [PMC free article: PMC2063549] [PubMed: 16418532]
- 63.
- Stewart SA, Weinberg RA. Telomeres: cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–557. [PubMed: 16824017]
- 64.
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7(12):932–943. [PubMed: 17139333]
- 65.
- McEachern MJ, Blackburn EH. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10(14):1822–1834. [PubMed: 8698241]
- 66.
- Topcu Z, Nickles K, Davis C. et al. Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc Natl Acad Sci USA. 2005;102:3348–3353. [PMC free article: PMC552925] [PubMed: 15713803]
- 67.
- Natarajan S, McEachern MJ. Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol. 2002;22(13):4512–4521. [PMC free article: PMC133910] [PubMed: 12052861]
- 68.
- Lin CY, Chang HH, Wu KJ. et al. Extrachromosomal telomeric circles contribute to Rad52-, Rad50- and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4(2):327–336. [PMC free article: PMC549320] [PubMed: 15701795]
- 69.
- Smogorzewska A, Karlseder J, Holtgreve-Grez H. et al. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol. 2002;12(19):1635–1644. [PubMed: 12361565]
- 70.
- Mieczkowski PA, Mieczkowska JO, Dominska M. et al. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc Natl Acad Sci USA. 2003;100(19):10854–10859. [PMC free article: PMC196892] [PubMed: 12963812]
- 71.
- Ferreira MG, Cooper JP. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 2004;18(18):2249–2254. [PMC free article: PMC517518] [PubMed: 15371339]
- 72.
- Heacock M, Spangler E, Riha K. et al. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004;23(11):2304–2313. [PMC free article: PMC419913] [PubMed: 15141167]
- 73.
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7(7):712–718. [PubMed: 15968270]
- 74.
- Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8(8):885–890. [PubMed: 16845382]
- 75.
- Carter SD, Iyer S, Xu J. et al. The Role Of Nonhomologous End-Joining Components in Telomere Metabolism in Kluyveromyces lactis. Genetics. 2007;175(3):1035–1045. [PMC free article: PMC1840097] [PubMed: 17237517]
- 76.
- Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues Nat Genet 1998; 20(2):203-206. [PubMed: 9771717]
- 77.
- Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase Science. 1998. pp. 493–496. [PubMed: 9774280]
- 78.
- McEachern MJ, Iyer S, Fulton TB. et al. Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc Natl Acad Sci USA. 2000;97:11409–11414. [PMC free article: PMC17213] [PubMed: 11016977]
- 79.
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15(11):6128–6138. [PMC free article: PMC230864] [PubMed: 7565765]
- 80.
- Teng SC, Zakian VA. Telomere-Telomere Recombination Is an Efficient Bypass Pathway for Telomere Maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(12):8083–8093. [PMC free article: PMC84893] [PubMed: 10567534]
- 81.
- Natarajan S, Groff-Vindman C, McEachern MJ. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot Cell. 2003;2(5):1115–1127. [PMC free article: PMC219379] [PubMed: 14555494]
- 82.
- Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106(3):275–286. [PubMed: 11509177]
- 83.
- Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411(6841):1073–1076. [PubMed: 11429610]
- 84.
- Liti G, Louis EJ. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol Cell. 2003;11(5):1373–1378. [PubMed: 12769859]
- 85.
- Meyer DH, Bailis AM. Telomere Dysfunction Drives Increased Mutation by Error-Prone Polymerases Rev1 and {zeta} in Saccharomyces cerevisiae. Genetics. 2007;175(3):1533–1537. [PMC free article: PMC1840081] [PubMed: 17151233]
- 86.
- Dreesen O, Cross GA. Consequences of telomere shortening at an active VSG expression site in telomerase-deficient Trypanosoma brucei. Eukaryot Cell. 2006;5(12):2114–2119. [PMC free article: PMC1694812] [PubMed: 17071826]
- 87.
- Dreesen O, Li B, Cross GA. Telomere structure and function in trypanosomes: a proposal. Nat Rev Microbiol. 2007;5(1):70–75. [PubMed: 17160000]
- 88.
- Denamur E, Matic I. Evolution of mutation rates in bacteria. Mol Microbiol. 2006;60(4):820–827. [PubMed: 16677295]
- 89.
- Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26(5):867–874. [PubMed: 15471900]
- 90.
- Hazelrigg T, Levis R, Rubin GM. Transformation of white locus DNA in Drosophila: dosage compensation, zeste interaction and position effects. Cell. 1984;36(2):469–481. [PubMed: 6319027]
- 91.
- Levis R, Hazelrigg T, Rubin GM. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1985;229(4713):558–561. [PubMed: 2992080]
- 92.
- Gottschling DE, Aparicio OM, Billington BL. et al. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63(4):751–762. [PubMed: 2225075]
- 93.
- Tham WH, Zakian VA. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene. 2002;21(4):512–521. [PubMed: 11850776]
- 94.
- Nimmo ER, Cranston G, Allshire RC. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 1994;13(16):3801–3811. [PMC free article: PMC395293] [PubMed: 8070408]
- 95.
- Castano I, Pan SJ, Zupancic M. et al. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol Microbiol. 2005;55(4):1246–1258. [PubMed: 15686568]
- 96.
- Scherf A, Hernandez-Rivas R, Buffet P. et al. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17(18):5418–5426. [PMC free article: PMC1170867] [PubMed: 9736619]
- 97.
- Horn D, Cross GA. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995;83(4):555–561. [PubMed: 7585958]
- 98.
- Matzke MA, Moscone EA, Park YD. et al. Inheritance and expression of a transgene insert in an aneuploid tobacco line. Mol Gen Genet. 1994;245(4):471–485. [PubMed: 7808397]
- 99.
- Baur JA, Zou Y, Shay JW. et al. Telomere position effect in human cells. Science. 2001;292(5524):2075–2077. [PubMed: 11408657]
- 100.
- Koering CE, Pollice A, Zibella MP. et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3(11):1055–1061. [PMC free article: PMC1307600] [PubMed: 12393752]
- 101.
- Pedram M, Sprung CN, Gao Q. et al. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol. 2006;26(5):1865–1878. [PMC free article: PMC1430234] [PubMed: 16479005]
- 102.
- Kyrion G, Liu K, Liu C. et al. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7(7A):1146–1159. [PubMed: 8319907]
- 103.
- Boulton SJ, Jackson SP. Components of the Ku-dependent nonhomologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17(6):1819–1828. [PMC free article: PMC1170529] [PubMed: 9501103]
- 104.
- Gonzalo S, Jaco I, Fraga MF. et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. Apr. 2006;8(4):416–424. [PubMed: 16565708]
- 105.
- Kalitsis P, Griffiths B, Choo KH. Mouse telocentric sequences reveal a high rate of homogenization and possible role in Robertsonian translocation. Proc Natl Acad Sci USA. 2006;103(23):8786–8791. [PMC free article: PMC1482656] [PubMed: 16731628]
- 106.
- Hansen KR, Ibarra PT, Thon G. Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res. 2006;34(1):78–88. [PMC free article: PMC1326240] [PubMed: 16407326]
- 107.
- Chikashige Y, Ding DQ, Imai Y. et al. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 1997;16(1):193–202. [PMC free article: PMC1169626] [PubMed: 9009280]
- 108.
- Scherthan H. Telomere attachment and clustering during meiosis. Cell Mol Life Sci. 2007;64(2):117–124. [PubMed: 17219025]
- Introduction
- Chromosome Ends: The Wild West of the Genome?
- Telomeric and Broken DNA Ends and the Processes that Act on Them
- Immediate Subtelomeric Regions and Their Possible Functions
- Subtelomeric Regions Are Often Enriched in Contingency Genes
- Subtelomeric DNA is Intrinsically Tolerant of Rearrangement
- The Differences between Uncapped Telomeres and Broken DNA Ends
- Disruption of Telomere Capping Can Trigger High Rates of Subtelomeric Change
- Adaptive Telomere Failure: A Fast Track for Subtelomeric Evolution?
- Telomere Position Effect Furthers the Adaptive Plasticity of Subtelomeric DNA
- The Relationship between Chromosome Ends and Centromeres
- Conclusion
- References
- Telomeres: Guardians of Genomic Integrity or Double Agents of Evolution? - Madam...Telomeres: Guardians of Genomic Integrity or Double Agents of Evolution? - Madame Curie Bioscience Database
- Preprotein Translocation through the Sec Translocon in Bacteria - Madame Curie B...Preprotein Translocation through the Sec Translocon in Bacteria - Madame Curie Bioscience Database
- Functional Evidence of Pain Control by the Immune System - Madame Curie Bioscien...Functional Evidence of Pain Control by the Immune System - Madame Curie Bioscience Database
- Future Perspectives of Interstitial and Perfusional Hyperthermia - Madame Curie ...Future Perspectives of Interstitial and Perfusional Hyperthermia - Madame Curie Bioscience Database
- α1-Microglobulin - Madame Curie Bioscience Databaseα1-Microglobulin - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...
