NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The ANGUSTIFOLIA (AN) gene in Arabidopsis thaliana (L.) Heynh. is the first homolog of the CtBP/BARS gene family identified in plants and is responsible for the polarity-dependent control of leaf cell expansion. This review compares the sequence homology and functional similarity of the AN protein with authentic animal CtBP/BARS family proteins. AN homologs have been found in both angiosperms and mosses, suggesting AN is conserved in terrestrial plant genomes. The AN subfamily is unique in having not only the D-isomer-specific 2-hydroxy acid dehydrogenase (D2-HDH) motif that is conserved among the CtBP/BARS family but also putative LxCxE/D and nuclear localization signal (NLS) motifs and a long C-terminal region. The absence of the catalytic triad, which is conserved in all D2-HDH sequences and is believed to be essential for the corepression activity of CtBP, suggests that AN might differ, at least in part, from CtBPs in molecular function. In addition, the distribution and density of the Golgi apparatus is normal in a null allele of the an mutant, suggesting that AN might not have a BARS function. An analysis of cytoskeletons in an mutant leaf cells suggests that AN might play an important role in controlling the arrangement of cortical microtubules that is plant-specific cytoskeletons. With all these attributes, AN appears to be the third member of an enigmatic family, CBA = CtBP/BARS/AN, which regulates aspects of developmental and organelle control in animals and plants.
Angustifolia—A Polarity-Dependent Regulator of Leaf Cell Expansion
Focusing on mechanisms that govern the polarized growth of leaves in the model plant Arabidopsis thaliana (L.) Heynh. (arabidopsis), we used mutational studies to identify two genes that act independently of each other to regulate polar cell elongation in leaves: AN regulates the width of leaves, and ROT3 regulates the length.1-4 The angustifolia (an) mutant of arabidopsis (Fig. 1) was originally isolated by Rédei.5 The mutation in the leaf-specific an phenotype is caused by a specific defect in the elongation of leaf cells in the transverse (width) direction1,2 (Fig. 1A,C). This polar defect was observed in all the leaf cells examined, including epidermal cells, trichomes, and parenchymatous cells (Fig. 1B-D). The altered direction of cell elongation was particularly evident in palisade cells, where expansion in the leaf-width direction was decreased, while elongation in the leaf-thickness direction was increased (Fig. 1C). Coincident with the defect in the palisade cells, the number of protrusions in epidermal cells was decreased, particularly in the leaf-width direction (Fig. 1D). A decreased number of branchings in trichome cells is also attributable to the same defect (Fig. 1B). Thus, the AN gene is thought to be the key gene to regulating the polar elongation of leaf cells in the leaf-width direction.2 Cytological analysis showed that the an mutant has abnormally arranged cortical microtubules in leaf cells (Fig. 1D),6,7 suggesting that AN might regulate polarity-dependent elongation of leaf cells via control of the arrangement of cortical microtubules.6
Recently we cloned the ANGUSTIFOLIA (AN) gene, a new member of the CtBP/BARS family from arabidopsis.6,7 Microarray analysis suggested that the AN gene might function as a transcriptional corepressor, like the CtBPs.6 Despite such speculation, the molecular function of AN is still unclear. Does AN regulate the arrangement of cortical microtubules by functioning as a plant CtBP? AN sequence data shows that the AN gene contains not only the D-isomer-specific 2-hydroxy acid dehydrogenase (D2-HDH) motif that is conserved among CtBP/BARS but also a putative LxCxE/D motif, which may be responsible for binding to retinoblastoma (Rb) protein,8-10 the PEST motifs that are thought to be responsible for degradation, 11 a putative phosphorylation site for casein kinase II, and a nuclear localization signal (NLS; Fig. 2). A long C-terminal region is also unique to AN (Fig. 2). We previously reported that the an-1 mutant allele has a nucleotide insertion in the C-terminus and that the an-2 mutant allele has a nucleotide transition resulting in the introduction of a termination codon in the conserved D2-HDH motif.6 Moreover, all known members of the AN subfamily have long C-terminal domains (Fig. 2). These data suggest that the C-terminal region might have an important role in the function of AN. More detailed comparative studies are required to determine whether the C-terminal region really superfluous to AN functions or not. Given that CtBP and BARS have similar amino acid sequences yet are functionally different, it is plausible that AN, although very closely related to CtBP/BARS, might differ from both in molecular function. This review compares AN protein with authentic CtBP family proteins.
Is AN a CtBP-Like Corepressor?
If AN is a corepressor like CtBPs, then transcription from certain genes should be up-regulated in the an mutant. We carried out microarray analysis between wild-type and an-1 mutant plants under the auspices of the Monsanto Arabidopsis Microarray Program6 and found that some genes were up-regulated in the an mutant but down-regulated in wild-type plants. RT-PCR analysis among wild type, an-1 mutant, and transgenic an plants that expressed the wild-type AN gene confirmed the results. In particular, the expression of genes in the xyloglucan endotransglucosylase/hydrolase (XTH) family showed characteristic patterns among the plants. Quantitative, real-time RT-PCR showed that one of the XTH genes, MERI5, was expressed at a three-fold higher level in rosette leaves of the an-1 mutant than in the wild type, while two other XTH genes, EXGT-A1 and EXGT-A2M, showed no differences in expression.6 These data appear to support the idea that AN can act as a corepressor of a particular set of genes in arabidopsis. However, we should note that the double mutant an-1/meri5 does not differ in morphology from the an-1 single mutant (Yokoyama, R., personal commun). This suggests that the down-regulation of MERI5 mRNA is not essential for the function of AN.
Does AN Act as a Molecular Bridge as CtBPs?
The CtBP of drosophila (dCtBP) and mouse (mCtBP2) self-associate in a yeast two-hybrid system,12,13 and it has been suggested that the dimerization of CtBP is important for its molecular function.14 Dimerized CtBPs are believed to function as a molecular bridge between a DNA-binding protein and a transcriptional repressor. We previously showed that AN self-associates as do CtBPs.6 Does AN act as a molecular bridge as does authentic CtBP? The CtBPs have an intrinsic dehydrogenase activity,15,16 and the NAD+-dependent conformational change is thought to be essential to the corepression activity of CtBP.15 In animal CtBPs, NAD+-dependent dehydrogenase activity is also thought to be linked to the regulation of protein-protein interactions via the PxDLS recognition motif.15 On the other hand, the AN gene subfamily is distinguishable from the CtBPs by its lack of the catalytic triad (His315/ Glul295/Arg266) conserved in all D2-HDH15 (Fig. 2, shown by asterisks). A mutant CtBP in which His315 was changed to a valine lacks detectable dehydrogenase activity.16 Thus, AN is expected also to lack dehydrogenase activity. In light of the above observation, AN appears to be unable to associate with the PxDLS motif. Moreover, AN does not have a GxGxxG(17x)D motif (Fig. 2), and a mutation of the GxGxxG(17x)D motif of CtBP has been shown to strongly inhibit the ability of CtBP to bind the PxDLS motif.15 To determine whether AN interacts with E1A, we carried out a yeast two-hybrid analysis using the C-terminal region of E1A as bait and the full-length AN as prey. The results indicated that AN does not interact with the E1A protein, which contains the PxDLS consensus recognition motif for animal CtBPs. (K.-H. Cho, G.-T. Kim, and H. Tsukaya, unpublished result), Thus, we conclude that AN cannot associate with proteins harboring the PxDLS motif.
Is AN a Homolog of BARS?
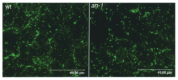
If AN is a functional homolog of CtBP, it should be localized in cell nuclei. Alternatively, if AN is a functional homolog of BARS, a member of the CtBP/BARS family that acts in the cytoplasm, an AN mutation might disrupt the Golgi apparatus, as BARS is essential for its establishment and maintenance.17 It is possible that AN functions as CtBP in the nucleus and as BARS in the cytoplasm. However, no abnormality in the shape or number of Golgi stacks was observed in the an-1 mutant leaf cells (Fig. 3). On the other hand, reports on the intracellular localization of AN6,7 are contradictory. Folkers et al7 reported that AN is localized in the cytoplasm and not in the nucleus, while our data showed that AN is detected in both nuclei and cytoplasm.6 Our recent observations on stable transgenic arabidopsis harboring ANp::AN::GUS showed that both reports might be correct; we found that the localization of AN is developmentally changed (G. Horiguchi and H. Tsukaya, unpublished observation). Further analysis on the relationship between intracellular localization and the function of AN will supply us important clues on the role of AN in plant development.
Conserved Function in Plants
As stated above, understanding the role and importance of AN-specific motifs requires detailed analyses of AN homologs from different plant species. Most AN-specific motifs are conserved in members of AN subfamily. For example, AN and IAN, an AN homolog from Japanese morning glory (Ipomoea nil), have the LxCxE motif in the N-terminal region,6,18 and MAN, an AN homolog from liverwort (Marchantia polymorpha), has a slightly different motif, LxCxD (Fig. 1). The PEST motif is conserved in IAN,18 but poorly conserved in MAN, as defined by PESTFIND (http://bioweb.pasteur.fr/seqanal/interfaces/Pestfind-simple.html). In the case of MAN, expressed sequence tag (EST) and reverse transcription-polymerase chain reaction (RT-PCR) analyses suggested that two isoforms, a long and a short type, are translated from the MAN gene by alternative splicing (H. Takano, unpublished result). Interestingly, in the case of moss (Physcomitrella patens), two AN homologs (PpAN1 and PpAN2) were found by EST analysis (M. Hasebe and H. Takano, unpublished data). While the deduced amino acid sequence of the PpAN1 gene contained the total region of plant AN protein, the PpAN2 protein excluded the C-terminus, and its size was similar to those of the short isoform of MAN and the drosophila dCtBP1 protein (H. Takano, unpublished data). If this holds true, the AN subfamily might play several roles in bryophytes. More detailed comparisons of homologs in the AN subfamily would also be informative, as some motifs found in AN are not conserved in moss homologs.
As a first step in such comparative studies, Cho et al18 analyzed IAN, the AN homolog from Ipomoea nil. The genus Ipomoea belongs to the subclass Asterids, while Arabidopsis belongs to Rosids. IAN contains not only a D2-HDH motif, which is highly conserved within the CtBP family, but also LXCXE, NLS, and PEST motifs, which are specific to the AN subfamily. The expression of IAN cDNA driven by the cauliflower mosaic virus 35S promoter restored a defect in leaf expansion in the leaf-width direction in the an-1 mutant of arabidopsis, as did the authentic AN cDNA, suggesting that IAN retains a common function with AN. By contrast, the complementation by IAN of a defect in the trichome branching pattern of the an-1 mutant was less effective than that of the defect for leaf shape. These results suggest that the mechanisms by which AN regulates leaf width and trichome branching are separable, at least, in part.18
Perspective
Because of its enigmatic functions and behaviors, the CtBP/BARS family has been one of the hot topics in biology in recent years.19,20 AN is the third subfamily of the CtBP/BARS complex found in the plant kingdom. Comparative and functional analyses of AN have just begun, but several unique features already have been discovered in the AN subfamily. Although AN is very similar to CtBP/BARS and it self-dimerizes, AN lacks the ability to associate with the PxDLS motif and possesses a unique long C-terminal domain. The mutation phenotype of the arabidopsis an mutant suggests that AN might be involved in cytoskeleton control.6 Owing to its uniqueness, AN has interested researchers of authentic CtBP/BARS.21 Considering both the unique features of AN and the commonalities between CtBP/BARS and AN, the AN subfamily might appropriately be treated as the third subfamily of the gene complex. In this vein, I propose to designate the whole gene family as the CBA family, or CtBP/BARS/AN family. Understanding of function(s) of AN subfamily is an important clue to reveal the fundamental role(s) of the CBA family in the organogenesis of multicellular organisms.
Recently, the an mutation phenotype in trichome branching was found to be rescued by the expression of drosophila CtBP driven by the cauliflower mosaic virus 35S promoter (S. Falk, M. Hülskamp, personal communication). This is an important clue to understand the common role of the CBA family. First, this data suggests that the AN-specific long C-terminal region might not be required for AN to function in trichome branching. Second, like arabidopsis, drosophila is an excellent system for analyzing gene function in organogenesis and individual development. Moreover, interacting partners of dCtBP are already well known in drosophila.22,23 Therefore, comparative and functional analyses of AN with dCtBP, for example, by interchanging experiments between AN and dCtBP in transgenic arabidopsis and transgenic drosophila, could supply some important clues on the role and evolution of the enigmatic CBA family.
Acknowledgements
The author thanks Dr. K.-H. Cho and Dr. G.-T. Kim of Dong-A University, Korea, and Dr. G. Horiguchi of NIBB, Okazaki, Japan, for their skillful analyses of ANGUSTIFOLIA. The author also thanks Dr. H. Takano (Kumamoto University, Japan) and Prof. M. Hülskamp (Köln University, Germany) for permission to cite their unpublished results. Dr. T. Ueda (University of Tokyo, Japan) kindly supplied seeds of the AtErd2::GFP strain. Dr. Richard H. Goodman (Vollum Institute for Advanced Biomedical Research, Oregon, USA) kindly supplied the C-terminal region of E1A. The present study was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, and Culture of Japan and by grants from the Bio-Design Program, Ministry of Agriculture, Forestry, and Fisheries of Japan.
References
- 1.
- Tsukaya H, Tsuge T, Uchimiya H. The cotyledon: A superior system for studies of leaf development. Planta. 1994;195:309–312.
- 2.
- Tsuge T, Tsukaya H, Uchimiya H. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development. 1996;122:1589–1600. [PubMed: 8625845]
- 3.
- Kim G-T, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998;12:2181–2191. [PMC free article: PMC317051] [PubMed: 9694802]
- 4.
- Kim G-T, Tsukaya H, Saito Y. et al. Changes in the shapes of leaves and flowers upon overexpression of the novel cytochrome P450 in Arabidopsis. Proc Natl Acad Sci, USA. 1999;96:9433–9437. [PMC free article: PMC17800] [PubMed: 10430960]
- 5.
- Rédei GP. Single locus heterosis. Z Vererbungs. 1962;93:164–170.
- 6.
- Kim GT, Shoda K, Tsuge T. et al. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 2002;21:1267–1279. [PMC free article: PMC125914] [PubMed: 11889033]
- 7.
- Folkers U, Kirik V, Schobinger U. et al. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 2002;21:1280–1288. [PMC free article: PMC125931] [PubMed: 11889034]
- 8.
- Magnaghi-Jaulin L, Groisman R, Naguibneva I. et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. [PubMed: 9468140]
- 9.
- Radulescu RT, Bellitti MR, Ruvo M. et al. Binding of the LXCXE insulin motif to a hexapeptide derived from retinoblastoma protein. Biochem Biophys Res Commun. 1995;206:97–102. [PubMed: 7818556]
- 10.
- Xie Q, Sanz-Burgos AP, Hannon GJ. et al. Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 1996;15:4900–4908. [PMC free article: PMC452227] [PubMed: 8890163]
- 11.
- Berset C, Griac P, Tempel R. et al. Transferable domain in the G(1) cyclin Cln2 sufficient to switch degradation of Sic1 from the E3 ubiquitin ligase SCFCdc4 to SCFGrr1. Mol Cell Biol. 2002;22:4463–4476. [PMC free article: PMC133886] [PubMed: 12052857]
- 12.
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: A hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. [PMC free article: PMC1170551] [PubMed: 9524128]
- 13.
- Turner J, Crossley M. Cloning and characterization of mCtBP2, a corepressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. [PMC free article: PMC1170841] [PubMed: 9724649]
- 14.
- Turner J, Crossley M. The CtBP family: Enigmatic and enzymatic transcriptional corepressors. Bioessays. 2001;23:683–690. [PubMed: 11494316]
- 15.
- Kumar V, Carlson JE, Ohgi KA. et al. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell. 2002;10:857–869. [PubMed: 12419229]
- 16.
- Balasubramanian P, Zhao LJ, Chinnadurai G. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 2003;537:157–160. [PubMed: 12606049]
- 17.
- Weigert R, Silletta MG, Spano S. et al. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402:429–433. [PubMed: 10586885]
- 18.
- Cho KH, Shindo T, Kim GT. et al. Characterization of a member of the AN subfamily, IAN, from Ipomoea nil. Plant Cell Physiol. 2005;46(1):250–255. [PubMed: 15659438]
- 19.
- Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9:213–224. [PubMed: 11864595]
- 20.
- Nardini M, Spano S, Cericola C. et al. CtBP/BARS: A dual-function protein involved in transcription corepression, and Golgi membrane fission. EMBO J. 2003;22:3122–3130. [PMC free article: PMC162135] [PubMed: 12805226]
- 21.
- de Lanerolle P, Cole AB. Cytoskeletal proteins and gene regulation: Form, function, and signal transduction in the nucleus. Sci STKE. 2002;(2002 pe30) [PubMed: 12096216]
- 22.
- Nibu Y, Zhang H, Bajor E. et al. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. [PMC free article: PMC1171049] [PubMed: 9843507]
- 23.
- Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–104. [PubMed: 9525852]
- 24.
- Thompson JD, Higgins DG, Gibson TJ. et al. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. [PMC free article: PMC308517] [PubMed: 7984417]
- 25.
- Takeuchi M, Ueda T, Sato K. et al. A dominant negative mutant of Sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 2000;23(4):517–525. [PubMed: 10972878]
- A New Member of the CtBP/BARS Family from Plants: Angustifolia - Madame Curie Bi...A New Member of the CtBP/BARS Family from Plants: Angustifolia - Madame Curie Bioscience Database
- Shh Expression in Pulmonary Injury and Disease - Madame Curie Bioscience Databas...Shh Expression in Pulmonary Injury and Disease - Madame Curie Bioscience Database
- Preimplantation Genetic Diagnosis (PGD): Screening for Aneuploidy in Human Oocyt...Preimplantation Genetic Diagnosis (PGD): Screening for Aneuploidy in Human Oocytes and Polar Bodies - Madame Curie Bioscience Database
- Experimental Evidence for Immunomodulatory Effects of Opioids - Madame Curie Bio...Experimental Evidence for Immunomodulatory Effects of Opioids - Madame Curie Bioscience Database
- Cytokines and Chemokines in Autoimmune Disease: An Overview - Madame Curie Biosc...Cytokines and Chemokines in Autoimmune Disease: An Overview - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...