NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The bacterial flagellum is a biological macromolecular nanomachine for locomotion. A membrane embedded molecular motor rotates a long helical filament that works as a propeller driving the bacterium through the liquid environment. The flagellum is composed of about 30 different proteins with copy numbers ranging from a few to a few thousands and is made by self-assembly of those proteins. The helical filament can be transformed into various distinct supercoiled forms by changes in chemical environment, single amino acid mutations, or mechanical forces. The axial portion of the flagellum involves several substructures: the rod, the hook, the hook-filament junction, the long helical filament and a cap at the filament tip. Although the axial component proteins are dissimilar at the primary sequence level, they share common structural characteristics. The central portions of their amino acid sequences are hypervariable whereas the amino- and carboxy-terminal parts contain highly conserved segments. In their monomeric form, they posses large, natively-disordered terminal regions that are essential in controlling and mediating intersubunit interactions. Recent structural studies—combining X-ray diffraction and single particle image analysis by cryo-EM—have given insights into the molecular mechanisms of self-assembly, supercoiling and polymorphic ability and open the way for construction of various flagella-based systems for applications in vaccination, bio- or nanotechnology.
Introduction
Flagella are the organelles for bacterial locomotion. These supramolecular structures extend from the cytoplasm to the cell exterior and are composed of three major structural elements, the basal body, the hook and the filament (Fig. 1).1,2 The swimming pattern of bacteria, such as Escherichia coli and Salmonella, consists of straight swimming for a few seconds and tumbling for a fraction of a second. During the straight swimming phase, the helical filaments form a bundle behind the cell body, where the filaments are all in a left-handed supercoiled form, each acting as a propeller driven by a rotary motor at the base of the flagellum. Bacteria tumble once every few seconds to change their swimming direction for the chemotactic behavior. The tumbling is triggered by quick reversal of motor rotation, which produces a twist in the filament structure and transforms it into right-handed supercoils momentarily. This allows the bundle to fall apart smoothly and then the uncoordinated propelling forces change the orientation of the cell quickly.3,4 The flagellar filaments can also be transformed into various but distinct polymorphic forms including two straight forms, in response to amino acid replacements in flagellin5 and to chemical changes in the environment6,7 as well as to mechanical forces.3,8
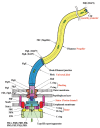
Figure 1
Schematic diagram of the bacterial flagellum. Different colors represent different protein components (Reprinted with permission from Yonekura et al. Res Microbiol 2002; 153:191-197. ©2002, Elsevier).
The long helical filament is connected to the basal structure via a short, highly curved segment called the hook.9 Both the hook and the helical filament are self-assembling macromolecular structures composed of the hook protein (FlgE) and flagellin (FliC), respectively. Each filament may comprise as many as ˜30000 flagellin subunits and can grow up to ˜15 μm. The hook is a helical assembly of ˜130 copies of FlgE subunits with a well regulated length of 55 nm ± 6 nm,10 capable of forming polymorphic supercoil structures.11 Bacterial flagellar hook acts as a molecular universal joint that can transmit the torque produced by the basal body, a rotary motor, to the flagellar filament.
Other components of the filamentous axial portion of the bacterial flagellum are the five rod proteins (FliE, FlgB, FlgC, FlgF, FlgG)12 and the hook associated proteins HAP1 (FlgK), HAP2 (FliD) and HAP3 (FlgL).13,14 HAP1 and HAP3 are junction proteins that connect the hook to the filament. HAP2 forms a capping structure at the distal end of the flagellar filament, which helps to incorporate flagellin monomers—transported through the central channel—into the filament at the tip.
The flagellar proteins forming the structures lying beyond the cytoplasmic membrane are synthesized in the cell and exported sequentially by the flagellum-specific protein export apparatus from the cytoplasm to the site of assembly at the distal end of the growing filament.15 They must be translocated through the narrow—20-25 Å wide—central channel of the flagellum in mostly unfolded conformation. The flagellar protein export system is thought to exist at the cytoplasmatic face of the basal body to distinguish flagellar proteins from other cytoplasmatic proteins and to facilitate their transportation. The flagellar protein export system is a specialized type III export machinery.1,15,16 The type III secretion systems include those for secretion of virulence factors by a wide variety of pathogenic bacteria.
In this review we will focus on the external filamentous portion of the bacterial flagellum, the structural characteristics of the axial component proteins, their role in filament assembly and polymorphism and applications in bio- and nanotechnology. Results obtained mainly with E. coli and Salmonella enterica serovar Typhimurium will be described. The flagellar motor, the flagellum-specific export machinery and several other interesting topics related to the flagellar system lie beyond the scope of this chapter. There are excellent recent reviews discussing these topics.15,17-19
Flagellar Filament Assembly
In Vivo Assembly
The assembly process of the bacterial flagellum starts from the formation of the FliF ring complex (also called the MS ring) of the basal body in the cytoplasmic membrane and proceeds in both inward and outward directions, as well as laterally.1,20,21 The inward assembly involves the formation of the C ring, which is also called the switch complex, in the cytoplasmic space and the flagellar export apparatus is formed within the C ring, getting ready to export flagellar axial proteins through its central channel.
For all axial structure formation, the flagellum-specific type III protein export system selectively binds and translocates flagellar axial proteins into the central channel of the flagellum.15 Since the identification and enzymatic characterization of FliI ATPase as a component of the flagellar protein export system,22 it had been thought that the flagellar protein export is driven by the energy of ATP hydrolysis. A recent study, however, clearly showed that the flagellar proteins are exported to form the flagellum even in the absence of FliI and that the proton motive force across the cytoplasmic membrane is responsible for driving most part of the export process that involves unfolding of export substrate proteins and translocation of the unfolded chains, with a help of the FliI hexamer ring complex for insertion of the NH2-terminal chain of the export substrate proteins (Minamino and Namba, Nature in press).
The axial proteins construct the rod, the hook, the hook-filament junction and the long filament in this order. The rod composed of five proteins is connected to the FliF ring at its proximal portion and with the hook at its distal end and traverses the periplasmic space through the LP ring, where the P ring is located within the peptidoglycan (PG) layer and the L ring within the outer membrane. The rod cap made of FlgJ not only facilitates rod protein assembly but also makes a local hole through the PG layer by its muramidase activity, thus permitting penetration of the rod through the PG layer.23 Then FlgE assembles in a helical manner to form the hook. The hook cap made of FlgD is attached at the distal end of the hook until the hook grows up to a length of about 55 nm.24 The FliK and FlhB proteins function together to control the hook length within 10% of the average length.25-27 The hook length is determined at the point that the export system switches its export specificity from recognition of early substrates called rod/hook type (rod proteins, FlgJ, hook protein, FlgD, FliK) to late substrates called filament type (FlgM, HAP proteins, flagellin).28 Then the FlgD cap falls off the tip of the hook and is replaced by HAP1 and then HAP3 and HAP2 are bound at the distal end in this order to form the basal body-hook-HAP1-HAP3-HAP2 complex momentarily before the initiation of flagellin assembly into the long helical filament.13,14 Flagellin monomers travel a long way through the narrow central channel,29 only 2 nm in diameter but up to 10-15 μm long,30 to the flagellar tip, presumably by a diffusion process.
Filament assembly in vivo requires the presence of the HAP2 pentamer complex capping the distal end of the filaments where flagellin monomers are assembled.14,29 The cap permits assembly of flagellin and prevents its excretion. The simplest role of the cap is to prevent subunits from diffusing away from the tip but it may also induce a conformational change in flagellin required for polymerization. Interactions between the cap and distal end of the filament seem to have unique characteristics. The binding is supposed to be very stable so that dissociation of the cap rarely occurs and yet flagellin monomers can be easily inserted between the HAP2 cap and the end of the filament.
In Vitro Filament Reconstruction
The flagellar filaments can be easily detached from cells and depolymerized into monomeric flagellin by either heat treatment (60°C for 10 minutes) or acid treatment (around pH 2.5). The flagellin monomers thus obtained are capable of self-assembly. They can be reconstituted into the filaments under physiological conditions by adding short filaments as seeds or a precipitant such as ammonium sulfate.32 The hook can also be reconstituted from monomeric hook proteins under appropriate conditions in vitro.33 Moreover, it has been demonstrated that the hook-filament complex can also be formed by sequential addition of HAP1, HAP3, flagellin and HAP2 to the hook.34
Overall Structural Characteristics of Axial Proteins
Sequential Similarities
Flagellar axial proteins can be divided into three major families.35,36 Hook protein shows sequential similarities, especially in its terminal regions, to the rod proteins (FliE, FlgB, FlgC, FlgF, FlgG), which compose the drive shaft of the flagellar motor and also to the proximal hook associated protein, HAP1. Flagellin is not similar to the hook family, but appears to be related to HAP3. HAP2 is the most dissimilar among the axial proteins; it shows no significant sequential homology to flagellin or hook protein.
Conserved Terminal Regions and Variable Central Portions
In spite of the sequential dissimilarities, comparison of amino acid sequences from different bacterial sources reveals common motifs for flagellar axial proteins. Their terminal regions are highly conserved whereas the central portion contains highly variable segments. For example, flagellins have widely differing molecular masses in the range from 27 kDa to 67 kDa.37,38 The difference is in the central part of the molecule demonstrating that large deletions and insertions can be introduced into the central region of flagellin without destroying its ability to form the flagellar filaments. The variable central part is known to be the major determinant for flagellar antigenicity.
Heptad Repeats of Hydrophobic Amino Acids in the Terminal Regions
The NH2- and COOH-terminal regions of the axial components of bacterial flagella possess heptad repeats of hydrophobic amino acid residues,36 a characteristic of sequences that fold into α-helical coiled coils. It was suggested that the terminal regions of axially neighboring subunits interact to form coiled-coil bundles upon polymerization. This interlocking organization has been postulated to be the common motif by which the axial proteins form a continuous, mechanically stable structure36 and it has been visualized in the atomic model of one of the two straight-type flagellar filament.30
Disordered Terminal Regions and Compact Central Portions
Proteolytic, calorimetric and NMR studies revealed that terminal disorder is a common structural feature of the axial proteins of the bacterial flagellum.12,39-42 Representative members from all the major groups of the axial protein family have been demonstrated to possess disordered terminal regions. For example, the disordered terminal regions of Salmonella flagellin from the SJW1103 strain span the first 66 and the last 44 amino acid residues.39 The disordered regions belong to the most conserved part of flagellin, indicating their important structural and functional roles. The disordered terminal regions possess a high α-helix forming potential and assemble into helical bundles upon polymerization. The other parts of flagellin form compact, well-folded domains, which involve conserved and variable segments as well.41,43,44
The disordered terminal regions possess multiple roles. Polymerization properties of all axial proteins seem to be similarly controlled by their disordered terminal regions.44-46 Subunits are synthesized in the cytoplasm in large amounts and therefore it is essential to prevent their polymerization within the cell. Monomeric flagellin stays monomeric even at high protein concentrations since subunits with large disordered regions can not effectively bind to each other. The self-assembly of these proteins requires the presence of template structures, therefore polymerization occurs only at the distal end of the growing filament.29
The disordered terminal regions are also used to mediate interactions with multiple partners. Terminal regions of flagellin are not only involved in flagellin-flagellin subunit interactions but also in interactions with the hook-associated proteins, HAP3 at the base and HAP2 at the tip, in flagellar assembly in vivo.
Terminal disorder also plays a role in fine-tuning of intersubunit interactions. Conformational entropy of the disordered leg domains of the HAP2 pentamer cap moderates the otherwise too tight HAP2-filament interactions to allow insertion of flagellin subunits below the cap.41 Disordered terminal regions of flagellin are also involved in sliding and switching interactions essential for polymorphic supercoiling of flagellar filaments.48 Finally, terminal disorder as a common structural feature of axial proteins seems to serve as the recognition signal for the flagellum-specific export machinery.49,50
The Helical Filament
Structural Organization of Flagellin
The flagellar filament is composed of a single protein, flagellin. Flagellin from a wild-type strain of Salmonella, SJW1103, is composed of 494 amino acids.5 The amino acid sequences of the terminal regions of flagellin, including about 180 NH2-terminal and 100 COOH-terminal residues, are known to be well conserved from species to species of bacteria, while the central region is highly variable.37,38
Proteolytic experiments revealed that both terminal regions of flagellin are highly susceptible to even a mild proteolytic treatment. The readily hydrolysed terminal portions span approximately the first 65 and the last 45 residues of the molecule.39,51 These experiments suggested that there are no significant structural constraints in the terminal regions to limit the accessibility of the peptide bonds, i.e., these regions have no ordered tertiary structure and are in extensive contact with the surrounding media. NMR experiments provided direct evidence for disorder and mobility in the terminal regions of the monomer.52 In contrast to the poorly ordered terminal regions, the central part of the flagellin molecule is quite stable against proteolysis and has a cooperatively ordered structure.39 Calorimetric experiments demonstrated that thermal denaturation of the compact part of flagellin is highly reversible and it is composed of three autonomous folding domains.43
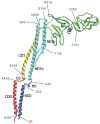
Far-UV CD and NMR studies demonstrated that the disordered regions become ordered upon filament formation into a largely α-helical domain.52,53 Electron cryomicroscopy and X-ray diffraction studies have revealed the subunit structure within the filament.30,54 The overall shape of flagellin looks like an upper case Greek gamma (Γ) with a vertical dimension of about 140 Å and a horizontal dimension of about 110 Å. Flagellin consists of four linearly connected domains labelled D0, D1, D2 and D3, which are arranged from the inside to outside of the filament. The ribbon diagram of the polypeptide backbone in Figure 2 shows the chain folding. The NH2-terminal chain starts from D0, going through D1, D2 and reaches D3 and then comes back through D2 and D1 and the COOH-terminal chain ends in D0. The terminal chains form an α-helical coiled coil in domain D0. Domain D1 comprises an NH2-terminal segment from Ala 44 to Gln 176 and a COOH-terminal segment from Asn 406 to Glu 454. Domain D2 also comprises two segments: Lys 177 to Gly 189 and Ala 284 to Glu 405. A central segment from Tyr 190 to Val 283 makes up domain D3. All three domain connections are formed by pairs of short antiparallel chains. The one that connects domains D0 and D1 is longer than the other two and therefore it is called the spoke region.
Domain D1 comprises three α-helices and a strand of a unique β-turn/β-hairpin/α-helix motif. This rod-shaped domain forms an extensive hydrophobic core along its central axis in a similar way to four-helix bundles in other protein structures. Because of an irregularity in one of the four strands involving the β-turn/β-hairpin/α-helix in series, however, the packing of hydrophobic side chains is relatively loose in the middle portion. In domain D2, the Cα backbone trace shows a previously undescribed fold. The fold comprises a series of β-hairpins all pointing away from one another like three thin leaves spread out or like a curve of cubic polynomial called the ‘folium of Descartes’. A common feature found in these folds called β-folium is that most of the tips of β-hairpins are either bent or twisted and so some of them are better described as a β-finger with a claw. Flagellin can have domain D3 deleted without losing its ability to form filaments.55,56 The relatively independent arrangement of this domain from the rest explains why its deletion has a minimal effect on the structural integrity of flagellin.
Filament Structure
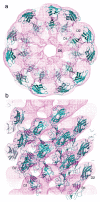
The flagellar filament is composed of 11 strands of protofilaments, which are nearly longitudinal arrays of subunits (Fig.3). The densely packed central core of filament consists of a concentric double tubular structure made of domains D0 and D1.30 Domains D2 and D3, which project out from the filament core, are relatively well separated from one another. The diameter of the filament is approximately 230 Å and that of the central channel is about 20 Å. α-helical coiled-coils in domains D0 and D1 stabilize the filament structure by making intimate intersubunit interactions both axially and laterally.30,57
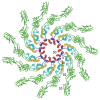
Figure 3
Structure of the flagellar filament viewed from the distal end. Flagellin molecules are shown in the same way as in Figure 2 (Reprinted with permission from Yonekura et al. Nature 2003; 424:643-650. ©2003, Macmillan Publishers Ltd: Nature).
The disordered terminal regions are involved in the inner tube of the concentric tubular structure forming long helical bundles and their direct interaction is responsible for the proper folding the filament core. There is no absolute requirement for either the NH2- or the COOH-terminal disordered region to be present for filament formation; fragments lacking either the whole disordered COOH-terminal region or most of the disordered NH2-terminal region are still capable of polymerization, although high precipitant concentrations are required for the filament formation and stability.45 However, both disordered terminal regions cannot be completely removed simultaneously without losing the ability to form the filaments. The basic subunit packing in the filaments made from truncated flagellin fragments was found to be compatible with that of the normal filaments.46,47 The polymorphic ability of the filaments of the fragments is also largely reduced, but it is not lost completely. Given the result that removal of about 80 of the total 110 disordered terminal residues does not destroy the filament structure, it is reasonable to assume that the main driving force behind filament formation is the interaction of the nondisordered conserved segments. These experiments suggest that the major part of the disordered terminal regions of flagellin is not essential for filament formation, but it does play an important role in stabilizing the filament structures and influence their polymorphic conformation.
Polymorphism of the Helical Filaments
The flagellar filaments may adopt a number of distinct helical forms and can be interconverted between these forms by mechanical force8 or changes in the environmental conditions.6,7 Molecular events behind the swimming-tumbling pattern of bacterial motility involve such polymorphic transitions of normally left-handed supercoiled filaments into right-handed forms upon quick reversal of motor rotation that drives the filament.3,4
It is not a simple task for chemically identical subunits to construct supercoiled filaments by bonding to each other in a regular manner. Polymorphic supercoiling of the filament is reasonably well explained by a bistable protofilament model in which subunit conformation is assumed to be the same within each protofilament and the protofilament conformation, including interactions with neighboring protofilaments, is classified into two distinct forms.32,58,59 The model assumes that conformational changes occur cooperatively along the protofilament. When all the protofilaments have an identical conformation the filament morphology is straight. There are actually two types of straight filaments with distinct helical symmetries; one is called the L-type, in which the protofilaments are tilted to the left and the other called the R-type with the protofilaments tilted to the right.60 The L-type protofilament has a slightly longer intersubunit distance along the protofilament than that of the R-type.57 If some of the 11 protofilaments are in the L-state and the others in the R-state, this gives rise to curvature, which, together with an inclination of the protofilaments from the filament axis, results in a supercoiled form. When the majority of protofilaments are in the L-type, the resulting supercoils are left-handed. When the number of protofilaments in the R-state increases the resulting supercoils can be right-handed onesdue to right-handed inclination of the protofilaments.
The straight filaments isolated from the mutant strains SJW1160 and SJW1655, both of which are point mutants of the wild type strain SJW1103, have the helical symmetry of the L- and R-type respectively57 and these two types of flagellins can be copolymerized to form different types of supercoiled filaments depending on the mixing ratio.61 Therefore, the two conformations of these flagellin mutants are thought to represent the two that coexist in the supercoiled filaments.
Although the bistable protofilament model gives a reasonable static and mechanical view of polymorphic supercoiling, it cannot give insights into mechanistic details at the molecular level. Recently, a massive molecular dynamics simulation was performed to further elucidate the detailed underlying molecular mechanisms of the polymorphic supercoiling.48 The following three types of subunit interactions were found to be essential: “permanent” interactions, which are always maintained between subunits in the various supercoil structures, “sliding” interactions which are formed between variable hydrophilic or hydrophobic residue pairs, allowing intersubunit shear without large changes in energy, while the formation and breakage of “switch” interactions stabilize inter- and intrasubunit interactions, respectively.
Permanent interactions determine the overall architecture of the flagellar filament. Many residues of domain D1 are involved in permanent interactions that are mainly responsible for geometrical constraints of the tubular structure, but many others of domain D1 are involved in sliding and switch interactions. Sliding interactions provide a mechanism that allows large flexibility in the intersubunit interface without large changes in the interaction energy. Switch interactions play essential roles in locking each protofilament interface to be in either the R- or L-type state. The D0 domain was found to be preferentially involved in sliding and switching interactions. Some experimental results suggest that the flagellin monomer tends to be in the R-type conformation if the constraints to form the native tubular structures are weakened. MD simulation showed that the formation of switch salt bridges and hydrogen bonds actually drives the subunit to take the L-type conformation in the filament. This result indicates that switch interactions are formed at the expense of intrasubunit interaction energy, leading to coexistence of the R- and L-type protofilaments in the polymorphic supercoil structures.
MD simulation also shed light on the mechanism of polymorphic transitions from a right- to left-handed supercoil and vice versa, which were driven by the torque applied on domains D0 and D1 to simulate the torque produced by the rotary motor.48 Here, the transitions clearly show hysteresis. This hysteresis indicates that the transitions should occur in two steps. The initial step mainly involves twist changes while roughly maintaining curvature and accompanying these twist changes, the filament is transformed between right- and left-handed helices. It was observed that the number of switch interactions remained almost constant during the first step. The first step is therefore a geometrical response of the filament helix to external torque. After the first step, the filament structure relaxes slowly into a new metastable state involving changes in switch and other interactions. In this second step, curvature changes while the handedness of the filament is maintained. This two-step mechanism was designated as the “transform and relax” mechanism. More coarse-grained model simulations are under way to understand the dynamic behavior of the polymorphic supercoil transitions in more details.
The Hook
The FlgE Subunit
The polypeptide chain of Salmonella hook protein contains 402 amino acid residues. Proteolytic and NMR studies demonstrated that both terminal regions of hook protein, involving about 70 NH2- terminal and 30 COOH-terminal residues are disordered in solution.40 Upon polymerization, the disordered terminal regions fold into a predominantly α-helical domain. It has been demonstrated that polymerization ability is lost completely upon removal of both disordered terminal regions.44
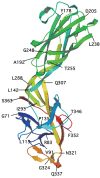
Attempts to crystallize intact hook protein have been unsuccessful since it readily assembles into filamentous structures under crystallization conditions, instead of forming 3D crystals. However, the FlgE31 fragment corresponding to residues 71-369 out of 402 and having a molecular mass of 31 kDa was successfully expressed in E. coli, purified and crystallized. This fragment lacks both terminal regions that are unfolded in the monomeric form in solution.44 The Cα backbone trace of FlgE31 (Fig. 4) obtained by X-ray crystallography shows two domains, D1 and D2, connected by a short stretch of two-stranded anti-parallel β-sheet.62 The longest dimensions of domain D1 and D2 are about 50 Å and 45Å, respectively and these two domains are connected along their long axes with an angle of about 70°. The amino-terminal segment from Gly 71 to Ala 144 and the carboxy-terminal segment from Pro 285 to Ser 363 comprise D1 and a central segment from Ala 145 to Lys 284 makes up domain D2. Both domains have an oval shape and are made mostly of β-structures. Domain D2 is a flattened, eight-strand β-barrel but with significant irregularity and extra loops. Domain D1 has a rather complex, unique fold composed of many different folding motifs: a stack of four β-hairpins one above another, alternating their orientations with crossing angles of about 120°; a triangular loop; a four-stranded and a two-stranded β-sheet; two consecutive β-turns; and a vertically extended chain, which appears to be the backbone around which the other motifs assemble.
Structure and Polymorphic Supercoiling of Hook
The bacterial flagellar hook acts as a molecular universal joint, transmitting torque produced by the rotary motor to the filament. The hook is much more flexible than the filament.63 The hook, like the helical filament, is also an assembly of 11 circularly arranged protofilaments and shows polymorphic supercoiling, which is only visible in polyhook structures.11 A specific aspect of the hook structure is its very small radius of curvature and short helical pitch. The most plausible mechanical explanation of how the hook works as a universal joint is that a supercoiled form is converted repeatedly into an identical form by permutation of the protofilament conformations from one to the next along the circumference in the direction opposite to its rotation, keeping the helical axis of the supercoil fixed in the same position and orientation.
It is curious how hook protein and flagellin with very different structural characteristics form tubular structures with basically the same architecture and helical symmetry. Low resolution EM analysis revealed that the hook subunit is composed of three morphological domains: the rod shaped inner domain (D1), the middle spherical domain (D2) and the outer ellipsoidal domain.64 The outer domain, D2, hangs out off the middle domain D1. There is an axial gap between the D2 domains, which provides freedom for the hook to curve. On the inner side of D1 are the rod-like features that form the inner tube with a narrow central channel. This design allows the structure to bend without breaking. In the case of the hook, the design represents a way to maintain structural integrity while still permitting a more flexible and more highly curved structure than the filament, which has a more rigid, double-tube design. The central channel of the hook has a diameter similar to that of the filament.
Higher resolution structural information for the hook was obtained by fitting the atomic structure of the FlgE31 fragment determined by X-ray crystallography to a density map of a straight hook reconstructed from images obtained by electron cryomicroscopy (Fig. 5).65 The two domains of FlgE31 were docked into the middle and outer domains of the map. The missing terminal sequences, which are unfolded in the monomer and are stabilized into α-helical bundles upon polymerization, were supposed to form the innermost rod-shaped domain in the hook.
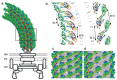
Based on the obtained structure, the molecular mechanism of the universal joint function of the hook has been investigated by a ˜ two-million-atom molecular dynamics simulation.66 On the inner side of the supercoil, protein subunits are highly packed along the protofilament and no gaps remain for further compression, whereas subunits are largely apart and slightly separated with hydrogen bonding through one layer of water molecules on the outer side (Fig. 6). As for the intersubunit interactions between protofilaments, subunits are packed along the 6-start helix in a left-handed supercoil whereas they are highly packed along the 5-start helix in a right-handed supercoil. It was concluded that the supercoiled structures of the hook in the left- and right-handed forms make maximal use of the gaps between subunits, which was called as the gap compression/extension mechanism. Mutual sliding of subunits at the subunit interface accompanying rearrangements of intersubunit hydrogen bonds is interpreted as a mechanism to allow continuous structural change of the hook during flagellar rotation at low energy cost.
The HAP2 Cap
HAP2 Monomer and Self-Association
HAP2 appears to be a distinct member among the axial proteins, which does not show any significant sequential similarity to other components of the flagellar system.36 Inspite of its dissimilarity, HAP2 also possesses disordered terminal regions like flagellin and hook protein. HAP2 from Salmonella SJW1103 is composed of 466 amino acid residues and its highly mobile parts span about 42 NH2-terminal and 51 COOH-terminal residues.41 Secondary structure prediction for Salmonella HAP2 suggested that the terminal regions of HAP2 are dominated by α-helical structure, which is a general characteristic for the flagellar axial proteins. Removal of the disordered terminal portions by trypsin results in a fragment of 40 kDa (HP40), which is quite stable against proteolysis, indicating that it is composed of compact structural units. Unlike flagellin and hook protein, which have a central part composed of β-structural domains, the central portion of the HAP2 molecule appears to contain comparable amounts of α and β-structure.41
The purified HAP2 protein has been found to readily self-assemble into annular structures in vitro, which are composed of ten subunits and have a pentagonal shape.67,68 The HP40 fragments do not form the decamer, but they can assemble into the pentamer.41 The disordered terminal regions of monomeric HAP2 are stabilized and protected in the decameric form. The three-dimensional structure reconstructed from electron micrographs of negatively stained HAP2 decamers showed a bipolar pair of pentamers (Fig. 7a), where each pentamer was composed of a pentagonal plate and five leg domains.69
HAP2 performs its physiological capping function as a pentamer.69,70 The disordered terminal regions are not essential for the formation of HAP2 pentamers, which is governed by interactions between the compact central portions of the subunits.41 The terminal regions of HAP2 in the pentamers are exposed and disordered, like in the case of monomeric HAP2 and are readily degraded by proteases. It seems that the major role of the highly conserved and mobile terminal regions is to participate in capping interactions and to moderate the affinity of the HAP2 cap for the filament by imposing an entropic penalty for binding.
Structure of the Cap-Flagellar Filament Complex
The three-dimensional density map of the cap-filament complex was reconstructed by single particle image analysis (Fig. 7b).69 There is a lid-like plate or cap at the top of filaments which is 120 Å wide and 25 Å thick; the diameter roughly corresponds to that of the outer tube of the filament. Its pentagonal shape in the end-on view indicates that the cap is a pentamer of HAP2. Inside the filament just below the cap plate, there is a cavity roughly 40 Å wide and 70 Å deep. Presumably, flagellin subunits are exported through the narrow central channel in a partially unfolded state. The cavity appears to have the right size to accommodate only one flagellin subunit at a time, allowing its refolding without aggregation with other subunits.
The cap is attached to the distal end of the filament at five positions through its anchor domains, where these anchor domains show significant deviation from the fivefold symmetry. The five anchor domains are well separated from one another, forming five gaps with different sizes and shapes between the plate and the filament end. One of the gaps is distinctly larger than the other four, having an inverted L shape and its size corresponds to the size of domain D1 of flagellin. This is most likely to be the site of flagellin assembly.
Mechanism of Flagellin Assembly
Because of the axial stagger by half a subunit in the lateral subunit packing, the end of the filament is not flat but has five indentations around its circumference, where one of them is a double indentation. It appears that four of the five indentations are filled with the HAP2 leg domains, while the one with double indentation is not filled by the last leg domain.69,70 This leaves an inverted L-shaped opening fully open for flagellin assembly. This opening has the right size and shape and probably an appropriate atomic surface structure, for domain D1 of the newly arriving flagellin subunit to insert and bind.
There is a mismatch in the rotational symmetry between the cap and the filament structures. While the 11 protofilaments are arranged into the tubular structure of the filament with the axial stagger to have a nearly 5.5-fold circular symmetry, the cap has a 5-fold circular symmetry. Because of this symmetry mismatch between the filament and the cap, the five leg domains of the cap must have significantly different conformations from one another for interaction with flagellin subunits.41,71 The highly flexible nature of the leg domain, as partly indicated by the disordered terminal regions in the isolated cap, allows such polymorphic interactions, reducing conformational strains and permitting stable interactions between the two structures in spite of the symmetry mismatch. Upon every subunit incorporation, four legs of the cap rearrange their conformations and the last one changes its binding partner; namely these legs walk along the helical steps of the filament end.69,71 Every insertion of a flagellin subunit is likely to force the cap to move into the next stable position energetically equivalent to the current position. The cap moves up 4.7 Å and rotates 6.5 degree along the left-handed 5-start helix of the filament. This would result in a roughly complete rotation of the cap by the assembly of 55 flagellin subunits. The energy for the cap rotation with the conformational rearrangements is presumably supplied by the binding energy of the newly incorporated flagellin subunit at the distal end of the filament.41 The binding of flagellin subunits are due to their high local concentration. The flagellar type III protein export apparatus presumably located on the cytoplasmic face of the basal body translocates flagellin subunits into the export channel to keep the effective concentration of flagellin at the distal end of the export channel above the critical concentration. The energy source for the flagellar protein export is maily proton motive force with a help of FliI ATPase to make the export process more efficient. (Minamino and Namba, Nature in press) The energy transduction network formed here is unique and interesting, because the energy produced in the cytoplasmic space is transmitted quite a long distance (10-15 μm) through flagellin export to be used for the helical movement of the cap at the distal growing end.
Applications
Flagella Display Technology
Kuwajima et al56 reported that various deletions can be introduced into the hypervariable central region of E. coli flagellin (497 residues) without destroying its ability to form the flagellar filaments. The smallest deletion mutant lacked 187 residues and retained only the NH2-terminal 193 residues and the COOH-terminal 117 residues. Since then, the surface-exposed dispensable central region of the fliC gene has been the target position for insertions of foreign peptide/protein gene sequences to be displayed on flagella. The use of recombinant microorganisms with surface exposed protein structures has gained a lot of interest during the last decade since this type of research holds great promise in different areas such as biotechnology, microbiology and biomedicine.
The most frequent application of flagella display is the construction of novel recombinant vaccines.72-75 Live bacterial vaccine delivery vehicles have been developed through the surface display of foreign antigens on the flagellar filament surface. Flagella display has also been used for the display of a random peptide library as an alternative to the phage display technique.76,77 Lu et al78 constructed a fusion protein (FliTrx) in which the entire coding sequence of E. coli thioredoxin was inserted into the dispensable region of the flagellin gene fliC. Flagella display has proven to be applicable also in bacterial adhesion technology since large fragments of bacterial adhesins up to 302 amino acid residues can be functionally expressed as fusions to flagellin.79
Flagella Nanotubes
Kumara et al80-81 demonstrated the versatility of employing bioengineered flagella for the generation of a variety of nanoparticle arrays and nanotubes. The FliTrx fusion protein was genetically engineered to display rationally designed histidine, arginine-lysine and aspartic acid-glutamic acid peptide loops on the solvent-accessible outer domain region. The resulting flagellin monomers were polymerized to obtain corresponding oligomeric flagellar bionanotubes in which the peptide loops were 5 nm apart along each protofilament. These flagella nanotubes were able to bind various metal ions. Controlled reduction of these metal ions yielded ordered arrays of nanoparticles and nanotubes. Covalent attachment of separately synthesized gold nanoparticles to the flagella scaffold was also demonstrated. Flagellar nanotubes were also used to initiate the formation of silica and titania bionanotubes.
Also, E. coli flagellin was functionalized to form tailored nanotubes that bind single types or pairs of ligands, including divalent cations, fluorescent antibodies, or biotin-avidin-linked moieties such as ferritins.82 Tobacco Etch Virus (TEV) protease site-containing flagellins were cleaved by the cognate protease without filament disintegration, potentiating their use as removable nanolithography masks to deposit attached ligands by protease cleavage.
Conclusion
Formation of complex biological structures such as cells and cellular organelles is based on self-assembly of component macromolecules such as proteins and nucleic acids either in solution or within lipid membranes. The self-assembly processes are all driven by precise recognition of template structures by assembling molecules, both of which have well-defined three-dimensional atomic arrangements for specific binding and interactions and yet extensive flexibility of these molecules including disordered conformation plays essential roles in formation and stabilization of complex structures that are otherwise impossible to form. The bacterial flagellum is one of the typical examples as described in this article. The well-established abilities of biological macromolecules to self-organize into complex three-dimensional architectures therefore give us a wonderful opportunity to use them in nanotechnology applications such as design of self-assembling nanostructures for nano-devices and nano-systems.
Acknowledgements
We thank F. Oosawa, S. Asakura, H. Hotani, P. Závodszky, F. Friedler and D.L.D. Caspar for continuous support and encouragement. This work was supported in part by Grants-in-Aid for Scientific Research to K.N. (16087207) from the Ministry of Education, Science and Culture of Japan.
References
- 1.
- Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. [PubMed: 12730325]
- 2.
- Namba K, Vonderviszt F. Molecular architecture of bacterial flagellum. Q Rev Biophys. 1997;30:1–65. [PubMed: 9134575]
- 3.
- Macnab RM, Ornston MK. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J Mol Biol. 1977;112:1–30. [PubMed: 328893]
- 4.
- Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. [PMC free article: PMC101988] [PubMed: 10781548]
- 5.
- Kanto S, Okino H, Aizawa SI. et al. Amino acids responsible for flagellar shape are distributed in terminal regions of flagellin. J Mol Biol. 1991;219:471–480. [PubMed: 2051483]
- 6.
- Kamiya R, Asakura S. Helical transformations of Salmonella flagella in vitro. J Mol Biol. 1976;106:167–186. [PubMed: 9518]
- 7.
- Kamiya R, Asakura S. Flagellar transformations at alkaline pH. J Mol Biol. 1977;108:513–518. [PubMed: 13224]
- 8.
- Hotani H. Micro-video study of moving bacterial flagellar filaments. III. Cyclic transformation induced by mechanical force. J Mol Biol. 1982;156:791–806. [PubMed: 7120394]
- 9.
- DeRosier DJ. Spinning tails. Curr Opin Struct Biol. 1995;187-193 [PubMed: 7648320]
- 10.
- Hirano T, Yamaguchi S, Oosawa K. et al. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. [PMC free article: PMC196732] [PubMed: 8071222]
- 11.
- Kagawa H, Aizawa SI, Asakura S. Transformations in isolated polyhooks. J Mol Biol. 1979;129:333–336. [PubMed: 39175]
- 12.
- Saijo-Hamano Y, Uchida N, Namba K. et al. In vitro characterization of FlgB, FlgC, FlgF, FlgG and FliE, flagellar basal body proteins of Salmonella. J Mol Biol. 2004;339:423–435. [PubMed: 15136044]
- 13.
- Ikeda T, Homma M, Iino T. et al. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J Bacteriol. 1987;169:1168–1173. [PMC free article: PMC211915] [PubMed: 3546266]
- 14.
- Ikeda T, Asakura S, Kamiya R. Cap on the tip of Salmonella flagella. J Mol Biol. 1985;184:735–737. [PubMed: 4046031]
- 15.
- Macnab RM. Type III flagellar protein export and flagellar assembly. BBA. 2004;1694:207–217. [PubMed: 15546667]
- 16.
- Blocker A, Komoriya K, Aizawa S-I. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci USA. 2003;100:3021–3030. [PMC free article: PMC152238] [PubMed: 12631703]
- 17.
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. [PubMed: 12500982]
- 18.
- Kojima S, Blair DF. The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol. 2004;233:93–134. [PubMed: 15037363]
- 19.
- Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002;277:9625–9628. [PubMed: 11779877]
- 20.
- Yonekura K, Maki-Yonekura S, Namba K. Growth mechanism of the bacterial flagellar filament. Res Microbiol. 2002;153:191–197. [PubMed: 12066889]
- 21.
- Kubori T, Shimamoto N, Yamaguchi S. et al. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. [PubMed: 1640458]
- 22.
- Fan F, Macnab RM. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. [PubMed: 8943245]
- 23.
- Nambu T, Minamino T, Macnab RM. et al. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181:1555–1561. [PMC free article: PMC93546] [PubMed: 10049388]
- 24.
- Onishi K, Ohta Y, Aizawa S-I. et al. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176:2272–2281. [PMC free article: PMC205349] [PubMed: 8157595]
- 25.
- Williams AW, Yamaguchi S, Togashi F. et al. Mutations in FliK and FlhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. [PMC free article: PMC178035] [PubMed: 8631688]
- 26.
- Shibata S, Takahashi N, Chevance FF. et al. FliK regulates flagellar hook length as an internal ruler. Mol Microbiol. 2007;64:1404–1415. [PubMed: 17542929]
- 27.
- Moriya N, Minamino T, Hughes KT. et al. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J Mol Biol. 2006;359:466–477. [PubMed: 16630628]
- 28.
- Ferris HU, Minamino T. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 2006;14:519–526. [PubMed: 17067800]
- 29.
- Iino T. Assembly of Salmonella flagellin in vitro and in vivo. J Supramol Str. 1974;2:372–384. [PubMed: 4612254]
- 30.
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. [PubMed: 12904785]
- 31.
- Ikeda T, Yamaguchi S, Hotani H. Flagellar growth in a filament-less Salmonella fliD mutant supplemented with purified hook-associated protein 2. J Biochem. 1993;114:39–44. [PubMed: 8407873]
- 32.
- Asakura S. Polymerization of flagellin and polymorphism of flagella. Adv Biophys. 1970;1:99–155. [PubMed: 4950148]
- 33.
- Kato S, Aizawa SI, Asakura S. Reconstruction in vitro of the flagellar polyhook from Salmonella. J Mol Biol. 1982;161:551–560. [PubMed: 7154090]
- 34.
- Ikeda T, Asakura S, Kamiya R. Total reconstitution of Salmonella flagellar filaments from hook and purified flagellin and hook-associated proteins. J Mol Biol. 1989;209:109–114. [PubMed: 2810363]
- 35.
- Homma M, Kutsukake K, Hasebe M. et al. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990;211:465–477. [PubMed: 2129540]
- 36.
- Homma M, DeRosier DJ, Macnab RM. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other components of the flagellum. J Mol Biol. 1990;213:819–832. [PubMed: 2193164]
- 37.
- Wilson DR, Beveridge TJ. Bacterial flagellar filaments and their component flagellins. Can J Microbiol. 1994;39:67–71. [PubMed: 8330258]
- 38.
- Beatson SA, Minamino T, Pallen MJ. Variation in bacterial flagellins: from sequence to structure. Trends Microbiol. 2006;14:151–155. [PubMed: 16540320]
- 39.
- Vonderviszt F, Kanto S, Aizawa SI. et al. Terminal regions of flagellin are disordered in solution. J Mol Biol. 1989;209:127–133. [PubMed: 2810365]
- 40.
- Vonderviszt F, Ishima R, Akasaka K. et al. Terminal disorder: a common structural feature of the axial proteins of bacterial flagellum? J Mol Biol. 1992;226:575–579. [PubMed: 1507216]
- 41.
- Vonderviszt F, Imada K, Furukawa Y. et al. Mechanism of self-association and filament capping by flagellar HAP2. J Mol Biol. 1998;284:1399–1416. [PubMed: 9878359]
- 42.
- Furukawa Y, Imada K, Vonderviszt F. et al. Interactions between bacterial flagellar axial proteins in their monomeric state in solution. J Mol Biol. 2002;318:889–900. [PubMed: 12054831]
- 43.
- Vonderviszt F, Uedaira H, Kidokoro SI. Structural organization of flagellin. J Mol Biol. 1990;214:97–104. [PubMed: 2196378]
- 44.
- Vonderviszt F, Zavodszky P, Ishimura M. et al. Structural organization and assembly of flagellar hook protein from Salmonella typhimurium. J Mol Biol. 1995;251:520–532. [PubMed: 7658470]
- 45.
- Vonderviszt F, Aizawa SI, Namba K. Role of the disordered terminal regions of flagellin in filament formation and stability. J Mol Biol. 1991;221:1461–1474. [PubMed: 1942062]
- 46.
- Mimori-Kiyosue Y, Vonderviszt F, Namba K. Locations of terminal segments of flagellin in the filament structure and their roles in polymerization and polymorphism. J Mol Biol. 1997;270:222–237. [PubMed: 9236124]
- 47.
- Mimori-Kiyosue Y, Vonderviszt F, Yamashita I. et al. Direct interaction of flagellin termini essential for polymorphic ability of flagellar filament. Proc Natl Acad Sci USA. 1996;93:15108–15113. [PMC free article: PMC26364] [PubMed: 8986772]
- 48.
- Kitao A, Yonekura K, Maki-Yonekura S. et al. Switch interactions control energy frustration and multiple flagellar filament structures. Proc Natl Acad Sci USA. 2006;103:4894–4899. [PMC free article: PMC1458766] [PubMed: 16549789]
- 49.
- Vegh BM, Gal P, Dobo J. et al. Localization of the flagellum-specific secretion signal in Salmonella flagellin. Biochem Biophys Res Commun. 2006;345:93–98. [PubMed: 16674914]
- 50.
- Kornacker MG, Newton A. Information essential for cell-cycle-dependent secretion of the 591-residue Caulobacter hook protein is confined to a 21-amino-acid sequence near the N-terminus. Mol Microbiol. 1994;14:73–85. [PubMed: 7830563]
- 51.
- Kostyukova AS, Pyatibratov MG, Filimonov VV. et al. Flagellin parts acquiring a regular structure during polymerization are disposed on the molecule ends. FEBS Letters. 1988;241:141–144. [PubMed: 3058510]
- 52.
- Aizawa SI, Vonderviszt F, Ishima R. et al. Termini of Salmonella flagellin are disordered and become organized upon polymerization into flagellar filament. J Mol Biol. 1990;211:673–677. [PubMed: 2313691]
- 53.
- Uratani Y, Asakura S, Imahori K. A circular dichroism study of Salmonella flagellin: evidence for conformational change on polymerization. J Mol Biol. 1972:85–98. [PubMed: 5042466]
- 54.
- Samatey FA, Imada K, Nagashima S. et al. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410:331–337. [PubMed: 11268201]
- 55.
- Mimori-Kiyosue Y, Yamashita I, Fujiyoshi Y. et al. Role of the outermost subdomain of Salmonella flagellin in the filament structure revealed by electron cryomicroscopy. J Mol Biol. 1997;270:222–237. [PubMed: 9813134]
- 56.
- Kuwajima G. Construction of a minimum-size functional flagellin of Eschericia coli. J Bacteriol. 1988;170:3305–3309. [PMC free article: PMC211289] [PubMed: 3290204]
- 57.
- Yamashita I, Hasegawa K, Suzuki H. et al. Structure and switching of bacterial flagellar filaments studied by X-ray fiber diffraction. Nat Struct Biol. 1998;5:125–132. [PubMed: 9461078]
- 58.
- Calladine CR. Construction of bacterial flagella. Nature. 1975;255:121–124. [PubMed: 1128677]
- 59.
- Calladine CR. Change of waveform in bacterial flagella—role of mechanics at molecular level. J Mol Biol. 1978;118:457–479.
- 60.
- Kamiya R, Asakura S, Wakabayashi K. et al. Transition of bacterial flagella from helical to straight forms with different subunit arrangements. J Mol Biol. 1979;131:725–742. [PubMed: 41952]
- 61.
- Kamiya R, Asakura S, Yamaguchi S. Formation of helical filaments by copolymerization of two types of ‘straight’ flagellins. Nature. 1980;286:628–630. [PubMed: 7402342]
- 62.
- Samatey FA, Matsunami H, Imada K. et al. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature. 2004;431:1062–1068. [PubMed: 15510139]
- 63.
- Kato S, Okamoto M, Asakura S. Polymorphic transition of the flagellar polyhook from Eschericia coli and Salmonella typhimurium. J Mol Biol. 1984;173:463–476. [PubMed: 6368839]
- 64.
- Morgan DG, Macnab RM, Francis NR. et al. Domain organization of the subunit of the Salmonella typhimurium flagellar hook. J Mol Biol. 1993;229:79–84. [PubMed: 8421316]
- 65.
- Shaikh TR, Thomas DR, Chen JZ. et al. A partial atomic structure for the flagellar hook of Salmonella typhimurium. Proc Natl Acad Sci USA. 2005;102:1023–1028. [PMC free article: PMC545859] [PubMed: 15657146]
- 66.
- Furuta T, Samatey FA, Matsunami H. et al. Gap compression/extension mechanism of bacterial flagellar hook as the molecular universial joint. J Struct Biol. 2007;157:481–490. [PubMed: 17142059]
- 67.
- Ikeda T, Oosawa K, Hotani H. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J Mol Biol. 1996;259:679–686. [PubMed: 8683574]
- 68.
- Imada K, Vonderviszt F, Furukawa Y. et al. Assembly characteristics of flagellar cap protein HAP2 of Salmonella: decamer and pantamer in the pH-sensitive equilibrium. J Mol Biol. 1998;277:883–891. [PubMed: 9545379]
- 69.
- Yonekura K, Maki S, Morgan DG. et al. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290:2148–2152. [PubMed: 11118149]
- 70.
- Maki S, Imada K, Furukawa Y. et al. Plugging interactions of HAP2 pentamer into the distal end of flagellar filament revealed by electron microscopy. J Mol Biol. 1998;277:771–777. [PubMed: 9545371]
- 71.
- Maki-Yonekura S, Yonekura K, Namba K. Domain movements of HAP2 in the cap-filament complex formation and growth process of the bacterial flagellum. Proc Natl Acad Sci USA. 2003;100:15528–15533. [PMC free article: PMC307601] [PubMed: 14673116]
- 72.
- Wu JY, Newton S, Judd A. et al. Expression of immunogenic epitopes of hepatitis B surface antigen with hybrid flagellin proteins by a vaccine strain of Salmonella. Proc Natl Acad Sci USA. 1989;86:4726–4730. [PMC free article: PMC287346] [PubMed: 2471978]
- 73.
- Newton SMC, Joys TM, Anderson SA. Expression and immunogenicity of an 18-residue epitope of HIV1 gp41 inserted in the flagellar protein of a Salmonella live vaccine. Res Microbiol. 1995;146:203–216. [PubMed: 7569315]
- 74.
- Stocker BA, Newton SM. Immune responses to epitopes inserted in Salmonella flagellin. Int Rev Immunol. 1994;11:167–178. [PubMed: 7519231]
- 75.
- Cattozo EM, Stocker BAD, Radaelli A. et al. Expression and immunogenicity of V3 loop epitopes of HIV-1, isolates SC and WMJ2, inserted in Salmonella flagellin. J Biotech. 1997;56:191–203. [PubMed: 9304878]
- 76.
- Westerlund-Wikstrom B. Peptide display on bacterial flagella:principles and applications. Int J Med Microbiol. 2000;290:223–230. [PubMed: 10959724]
- 77.
- Westerlund-Wikstrom B, Tanskanen J, Virkola R. et al. Functional expression of adhesive peptides as fusions to Eschericia coli flagellin. Prot Eng. 1997;10:1319–1326. [PubMed: 9514121]
- 78.
- Lu Z, Murray KS, Van Cleave V. et al. Expression of thioredoxin random peptide libraries on the Eschericia coli cell surface as functional fusions to flagellin: a system designed for exploring protein-protein interactions. Biotechnology. 1995;13:366–372. [PubMed: 9634778]
- 79.
- Tanskanen J, Korhonen TK, Westerlund-Wikstrom B. Construction of a multihybrid display system: flagellar filaments carrying two foreign adhesive peptides. Appl Env Microbiol. 2000;66:4152–4156. [PMC free article: PMC92277] [PubMed: 10966447]
- 80.
- Kumara MT, Muralidharan S, Tripp BC. Generation and characterization of inorganic and organic nanotubes on bioengineered flagella of mesophilic bacteria. J Nanosci Nanotechnol. 2007;7:2260–2272. [PubMed: 17663239]
- 81.
- Kumara MT, Srividya N, Muralidharan S. et al. Bioengineered flagella protein nanotubes with syteine loops: self-assembly and manipulation in an optical trap. Nano Lett. 2006;6:2121–2129. [PubMed: 16968037]
- 82.
- Woods RD, Takahashi N, Aslam A. et al. Bifunctional nanotube scaffolds for diverse ligands are purified simply from Eschericia coli strains coexpressing two functionalized flagellar genes. Nano Lett. 2007;7:1809–1816. [PubMed: 17489638]
- Structure, Function and Assembly of Flagellar Axial Proteins - Madame Curie Bios...Structure, Function and Assembly of Flagellar Axial Proteins - Madame Curie Bioscience Database
- The ø29 DNA Packaging Motor: Seeking the Mechanism - Madame Curie Bioscience Dat...The ø29 DNA Packaging Motor: Seeking the Mechanism - Madame Curie Bioscience Database
- Molecular Phylogeny and Evolution of the Coronin Gene Family - Madame Curie Bios...Molecular Phylogeny and Evolution of the Coronin Gene Family - Madame Curie Bioscience Database
- Cell-Cell Fusion: Transient Channels Leading to Plasma Membrane Merger - Madame ...Cell-Cell Fusion: Transient Channels Leading to Plasma Membrane Merger - Madame Curie Bioscience Database
- Replacement of Specific Neuronal Populations in the Spinal Cord - Madame Curie B...Replacement of Specific Neuronal Populations in the Spinal Cord - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...