NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Introduction
Telomeres, the ends of chromosomes, and their associated proteins, maintain genomic stability.1 Intact telomeres prevent the ends of chromosomes from being degraded by exonucleases, from illegitimately recombining to produce end-to-end fusions and from activating DNA damage checkpoints.2 In most eukaryotes, telomeres consist of tandem repeats of short G-rich sequences, such as d(TTAGGG)n in humans and d(TTGGGG) in the ciliated protozoan, Tetrahymena.3–5 In certain yeast, such asSaccharomyces cerevisiae and Kluyveromyces lactis, the telomeric sequences are irregular d(TG2-3(TG)1-3) or longer d(GGATTTGATTAGGTATGTGGTGTAC), respectively. Telomeres range insize from under fifty base pairs (bp) in some ciliated protozoa, a few hundred bp in yeast, 2-15 kilobase pairs (kbp) in human to 10-60 kbp in the common laboratory mouse, Mus musculus.3,6 If conventional DNA polymerases and primases were the only enzymes involved in replicating the terminal bases of a linear DNA molecule, then chromosomes would shorten with the removal of terminal RNA primers at each round of replication (known as the end replication problem).4,7,8 Telomerase is an enzyme that compensates for this loss, and thus defines one of the mechanisms for replicating and maintaining chromosomal termini.8,9
Telomerase activity was first identified in the ciliated protozoan, Tetrahymena thermophila, and is present in most eukaryotes studied with some exceptions such as Drosophila melanogaster.4,5,10 Telomerase activity has been biochemically and molecularly characterized most extensively in ciliated protozoa, and these studies have been reviewed in detail.11 Mammalian, ciliate and yeast telomerases have also been the subject of other recent reviews.5,12–14 The focus of this Chapter is to review and integrate the current knowledge of human, mouse and yeast telomerase biochemistry, with reference to seminal and recent studies of telomerase biochemistry in ciliated protozoa.
Telomerase Components
As a ribonucleoprotein (RNP), telomerase has a protein catalytic subunit (TERT), one or more associated proteins, and an integral RNA component that serves as a template for the synthesis of telomeric repeats (termed hTR for human telomerase RNA, mTR for mouse telomerase RNA, TLC1 for S. cerevisiae telomerase RNA).9 The in vitro reconstitution of human and Tetrahymena telomerase activities using rabbit reticulocyte lysate (RRL) coupled transcription/translation systems suggests that TERT (telomerase reverse transcriptase) and TR (telomerase RNA, also called TER) are the minimal components required for activity.15–17
Different reconstitution systems have been used to identify the regions of the telomerase RNA necessary for both TERT binding and telomerase activity, the regions of TERT essential for activity and binding to TR, and the regions of TERT implicated in multimerization with other TERT molecules. First reported is a system that reconstitutes telomerase activity by adding in vitro-transcribed telomerase RNAs to partially purified telomerase from Tetrahymena or human cells depleted of endogenous telomerase RNA by micrococcal nuclease treatment.18–22 The second system reconstitutes telomerase activity from recombinant TERT and RNA subunits using coupled in vitro transcription/translation systems (RRL).15,16,22,23 A third system consists of the addition of in vitro-transcribed hTR to extracts from the telomerase-negative VA13 cell line ectopically expressing human TERT (hTERT), or the expression of hTR or hTERT in 293 or VA13 cells.23–27 Other reconstitution systems that will be useful in the characterization of human telomerase are based on: 1) the expression of hTERT and hTR in yeast cells;28–30 2) the addition of hTR to recombinant hTERT expressed in insect cells;31,32 and 3) the coexpression of hTERT and hTR in insect cells (Huard, S., Bachand, F. and Autexier, C, unpublished). Recently, mTR was characterized by expressing mutant mTRs in mTR−/− cells.33
Telomerase activity can also be reconstituted in vitro from heterologous components by the expression of mTERT and hTR in RRL, or in vivo by the transfection of the mTR or hTR gene into mTR−/− cells.34–36 In contrast to the efficient reconstitution of telomerase activity by the expression in RRL of hTERT and hTR, or mTERT and hTR, mTR is unable to efficiently reconstitute telomerase activity when expressed in conjunction with hTERT or mTERT in RRL, or when added to micrococcal nuclease-treated human 293 cell extracts (Demers, J. and Autexier, C., unpublished).16,20 These results suggest that although the mouse and human telomerase complexes are not entirely conserved and interchangeable, hTR can functionally complement mTR in the reconstitution of telomerase activity and that murine mTERT is amenable to heterologous TR activation. These reconstitution systems will allow a detailed characterization of telomerases from different organisms and lead to a better understanding of the similarities and differences in telomerase and telomere biology between ciliates, yeasts, human and mouse.5,7,11,12,37,38
Telomerase Reverse Transcriptase
Catalytic reverse transcriptase components of telomerase have been identified in various organisms including ciliates, human (hTERT), mouse (mTERT) and yeast (EST2 for S. cerevisiae).17,35,36,39–48 The TERT proteins are large, ranging in size from 103 kDa in S. cerevisiae to 134 kDa in Tetrahymena17,39,40,45 Similar to the budding yeast TERT, the fission yeast protein is also smaller than most TERTs, at 116 kDa.41The human and mouse proteins are two of the largest members of the TERT family, at 127 kDa.35,36,41
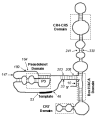
The TERT component of telomerase is a reverse transcriptase (RT), containing motifs (1 and 2, and A-E) that are common to RT enzymes, as well as N- and C-terminal regions which are not conserved among RTs (Fig.1).49 Conserved amino acids in the RT motifs of telomerase are required for the catalytic activity of telomerase in vitro and in vivo, indicating that many basic elements of TERT catalytic function are likely to be shared with the RTs.15–17,35,39,40,43,50,58 The TERT N-terminus constitutes a large part of the total protein, and varies in length in different organisms. The yeast TERT N-termini are considerably shorter than the N-terminal regions of mouse and human TERTs (Fig. 1). TERT N-terminus length varies in most organisms due to differing lengths of a very poorly conserved putative ‘linker’ region joining two subdomains of the N-terminus (Fig. 1).59 The N-terminus contains six identified regions, including the linker. These are: region N, and motifs GQ, CP, QFP and T (Fig. 1) (see below for a description of alternative nomenclature for these regions).59 A new ciliate-specific motif, CP2, has also been recently identified in the linker region of Tetrahymena TERT.56 The TERT N-terminus most likely mediates telomerase-specific catalytic functions, and it will be discussed in greater detail below. The TERT C-terminus is more poorly conserved within the TERT family, but may be functionally analogous to the processivity-associated ‘thumb’ region of the HIV-1 reverse transcriptase.57
Telomerase TERT as a Reverse Transcriptase
The HIV-1 reverse transcriptase has been successfully aligned with the RT portion of seven TERTs.13 HIV-1 RT is often used as a model for TERT RT function because its crystal structures in complex with template, primer, nucleotide and inhibitors have previously been solved.60–64 The basic catalytic activity of reverse transcriptase proteins such as HIV-1 RT consists of a number of events. The central catalytic event is the template-specified addition of a dNTP to the 3'OH group of a primer. This addition requires Mg2+-coordinating residues and a general architecture that are conserved in both RNA- and DNA-directed polymerases.65 Mutational analysis of certain conserved TERT RT motif residues suggests that RT enzyme structures implicated in dNTP recognition and addition are probably conserved in the TERTs.56,57 Following nucleotide addition, primers are elongated in a template-directed fashion by sequential dNTP additions. In HIV-1 RT, processivity is defined as the continuous elongation of the same primer along a lengthy template strand by sequential dNTP addition. Regions in the yeast telomerase RT (EST2) may play a role in processivity analogous to the ‘thumb’ region of the HIV-1 RT.57
In addition, the HIV-1 RT is an assymetric dimer that assembles in a head-to-tail fashion.60 Studies of S. cerevisiae and human telomerase complexes generated in vivo and in vitro, and from recombinant components, suggest that telomerase is also a multimeric enzyme, containing two copies of the telomerase RNA, and more than one, complementing copy of the TERT component.30,32,66,67 Experiments conducted with hTERT mutants expressed in vivo and in vitro implicate the RT domain as one of the regions participating in the association of TERT subunits, though the RT motifs alone are not sufficient.67 The stoichiometry of TERTs in telomerase complexes remains to be clearly established, as does the functional relevance of multimerization.
Telomerase-Specific Functions of TERT
Telomerases differ from the HIV-1 RT in several important ways. First, most RTs are associated with their template RNAs/DNAs in a fashion that promotes mobility along the entire length of the long template. However, the telomerase RNA is stably associated with the TERT reverse transcriptase, and only a small portion of this RNA is used as a template (see below). In fact, though the RT domains of TERT are essential for catalytic activity, they are not sufficient for high-affinity binding of the RNA components of Tetrahymena and human telomerases.25,54,68,69 Thus, TERT is likely to contain telomerase-specific elements required for stable association with the telomerase RNA component, and for appropriate copying of this RNA's small internal template.
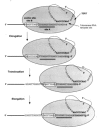
Second, in many organisms, telomerase exhibits a reiterative form of processivity in vitro, in which a single enzyme complex catalyzes the addition of multiple telomeric DNA repeats to the same telomeric DNA primer. Initially, the 3' terminus of the telomeric DNA primer aligns with the 3' end of the telomerase RNA template (template site) (Fig. 2).11 Elongation proceeds for a short distance to the 5' boundary of the template, followed by translocation/repositioning of the complex so that the 3' template region and the 3' terminus of the newly synthesized DNA are realigned (Fig. 2). This reiterative form of processivity is likely to require telomerase-specific elements, including a possible anchor site (Fig. 2). This site has been hypothesized to stabilize the association of 5' telomeric primer DNA with telomerase during repositioning of the DNA substrate and RNA template relative to each other (see section on the Mechanism of Telomere Synthesis by Telomerase).9,70 Additionally, as only a short region of the telomerase RNA serves as a template, 5' template boundaries must be strictly regulated (see section on Template boundary).
Candidate TERT regions for telomerase-specific functions are the N- and C-termini of the proteins. These regions bear no substantial homology to the RT-flanking domains of the reverse transcriptases. Most of the C-terminus of Tetrahymena and human TERTs is required for in vitro catalytic activity, though this region is not required for high-affinity binding of the telomerase RNA.25,68,69 Yeast expressing C-terminally-truncated EST2 exhibit a significant processivity defect which correlates with stable, short telomere lengths that permit cell survival.57,71 The C-terminus of hTERT may also be implicated in functional multimerization with other hTERT molecules,67 in recruitment of the enzyme to telomeres,72 and in nuclear localization.73 C. elegans TERT lacks several motifs found in all other telomerases, including the C-terminus, suggesting an atypical telomerase or nonfunctional enzyme.47
The TERT N-Terminus
The TERT N-terminus is much larger than the C-terminus and is more highly conserved among the TERTs (Fig. 1).57,59 Essential regions of the EST2 N-terminus have been mapped by unigenic evolution.71 Multiple sequence alignment using Hidden Markov Modelling (HMM)-based alignment has revealed substantial overlap between alignment-identified motifs and essential, hypomutable regions identified by unigenic evolution.59,71 Alignment of multiple sequences identified six major regions in the N-terminus of the TERTs (Fig. 1): the non-conserved extreme N-terminus (N), motif GQ (also identified as motif N, T2 and Region 1:,47,56,59,71 respectively), motif CP,45 a poorly conserved putative ‘linker’ region between motif GQ and motif CP,59 motif QFP59 and motif T.39,41 The CP motif, first identified in ciliates, corresponds to Region II in EST2,71 and contains residues that are conserved in non-ciliate TERTs.59 An additional, ciliate-specific motif, CP2, has been identified in the Tetrahymena TERT ‘linker’ region,56 whereas the hTERT linker contains a catalytically-essential vertebrate-specific motif, the VSR motif.30 Motifs QFP and T overlap with hypomutable regions III and IV in EST2.71 A DAT (dissociates activities of telomerase) domain required for telomere length maintenance but not catalytic activity has also been identified in the hTERT N-terminus.74 Mutation or deletion of specific residues in the TERT N-terminus has identified functions for this region in the reconstitution of telomerase activity in vitro and in vivo, and in telomerase RNA interactions, template definition, telomere maintenance, non-specific nucleic acid binding, and functional multimerization with other TERT molecules.15,25,30,54,56,59,67–69,71,74
The extreme N-terminal region of TERT (Fig. 1: N) is not conserved among the telomerase reverse transcriptases, but is required for appropriate human and yeast telomerase activity in vitro and telomere maintenance in vivo;30,59,74 this region of EST2 is also implicated in non-specific binding of single-stranded nucleic acids in vitro.59 A small 10 amino acid deletion at the extreme N-terminal portion of hTERT substantially reduces telomerase activity in vitro and causes a slight loss in binding of the human telomerase RNA component.30 Therefore, this poorly conserved region is nevertheless required for in vivo and in vitro telomerase activity. Interestingly, though vertebrate TERTs diverge in sequence at several places in the N-terminus, they exhibit 75% sequence identity in the ‘N’ region.30 Thus, the extreme N-terminal region may be implicated in organism- or vertebrate-specific telomerase functions.
The first conserved motif of the TERT N-terminus, GQ (Fig. 1), contains residues and subregions that are critical for in vitro telomerase activity in Tetrahymena, S. cerevisiae and human30,56,59,71,74 and which are implicated in telomerase RNA-binding by hTERT and EST2.30,71 Small deletions within the GQ motif of hTERT result in a small, 10–15% reduction in hTR binding.30 However, N-terminal truncations encompassing the GQ motif of Tetrahymena and human TERTs do not substantially affect high affinity binding of the telomerase RNA component, though they abolish or nearly eliminate telomerase activity in vitro.25,68,69 Thus, as for the extreme N-terminus, mutations in the GQ motif of yeast and human TERTs are associated with minor defects in telomerase RNA binding, but neither of these regions appears to be the primary site for high affinity binding of the telomerase RNA.30,69 hTERT truncation and deletion analysis also suggests that the GQ motif is not required for functional multimerization of hTERT molecules, in vitro or in vivo.25,30–67 However, some mutations in the GQ motif of EST2 and hTERT cause the uncoupling of in vitro telomerase activity and in vivo telomere length maintenance,59,74 suggesting that this region of the TERT N-terminus may be involved in interactions with proteins that regulate the proper subcellular and telomeric localization of telomerase. In hTERT, the N-terminal region required for telomere length maintenance but not in vitro activity is the DAT domain.74
The TERT “linker” region (Fig. 1) exhibits low sequence complexity and a large degree of variability among the TERTs in terms of length and sequence composition.59 The S. cerevisiae and Schizosaccharomyces pombe linkers are 78 and 98 amino acids long, respectively, whereas the human and mouse TERT linkers contain 200 and 204 residues, respectively.59 Though this region is poorly conserved among the TERTs, mutagenesis and deletion analysis have demonstrated that parts of it are required for functional rescue of senescing S. cerevisiae strains lacking est2,71 and for in vitro reconstitution of wild-type levels of telomerase activity using Tetrahymena and human TERTs.30,56 The Collins group has recently identified a second, ciliate-specific motif (CP2) in the linker region, and mutation of specific residues in this motif indicates that it is required for proper definition of the template boundary in Tetrahymena TERT.56 More recently, one of two RNA-binding regions in the hTERT N-terminus has been identified at the C-terminal border of the hTERT linker region.30 Interestingly, this region is poorly conserved within the TERTs in general; however it is very well conserved among the vertebrate TERTs, suggesting an organism- or vertebrate-specific function in RNA binding.30 Deleting the portions of the hTERT linker implicated in RNA binding also abrogates the ability of hTERT to functionally complement another full-length, catalytically-inactive hTERT.30 Based on these observations, it is tempting to speculate that the highly variable linker region may in fact interact in a sequence-, size- and/or structure-specific fashion with the highly variable telomerase RNAs from different organisms.
The CP motif (Fig. 1) was first identified in the ciliates,45 but multiple sequence alignment has demonstrated that certain residues in this motif are in fact conserved among all of the TERTs.59 In yeast, Tetrahymena and human TERTs the CP motif is implicated in telomerase RNA binding.30,54,71 Deletions in the hTERT CP motif that confer an RNA-binding defect are also associated with defects in functional association with full-length hTERT molecules.30 As for the CP motif, the QFP motif (Figure 1) has been implicated in telomerase RNA binding and functional multimerization with other TERT molecules.30,71
The hTERT T motif (telomerase-specific) (Fig. 1) was identified when the telomerase reverse transcriptases were first cloned from a number of organisms.39,41 Conserved amino acids in the T-motif are required for the catalytic activity of telomerase.15 In addition, the T-motif has long been considered a candidate region for telomerase-specific functions, such as telomerase RNA binding.15 Indeed, mutagenesis of specific residues in Tetrahymena and human TERTs reveals that elements of the T-motif are required for RNA binding.30,54 Therefore, the TERT N-terminal T, QFP and CP motifs, in addition to portions of the ‘linker’ region, appear to be required for binding of TERT to the telomerase RNA component.
In the last two years, analysis of the N-terminus of yeast, Tetrahymena and human TERTs has identified a role for this region in telomerase activity in vitro and in vivo, and in telomerase RNA binding, template definition, telomere maintenance, non-specific nucleic acid binding, and functional multimerization with other TERT molecules. Evidently, the N-terminus of the telomerase reverse transcriptases is essential for telomerase-specific functions.
Telomerase RNA
The RNA component of telomerase has been identified in ciliates, yeast, and 32 vertebrate species including human and mouse.9,75,76 The telomerase RNAs range in size from 146 nucleotides (nt) in the ciliated protozoan, Tetrahymena paravorax, to 1544 nt in the budding yeast Candida albicans,77,78 and contain template sequences that are complementary to the species-specific telomeric repeats. hTR and mTR (451 and 408 nt, respectively) contain the sequences 5'-CUAACCCUAAC-3' and 5'-CUAACCCU-3', respectively, which direct the synthesis of the TTAGGG telomeric repeats in both of these organisms. The mTR template region is shorter than the hTR template region, which may explain the lack of processivity of the mouse telomerase enzyme under standard telomerase assay conditions in vitro (see section on processivity).79,80 The 5' ends of the human and mouse telomerase RNA transcripts are 45 and 2 nt upstream of the template, respectively.81 hTR is transcribed by RNA polymerase II, unlike ciliate telomerase RNAs which are RNA polymerase III transcripts, and the 3' end of hTR has been mapped 3 nt downstream of the ACA motif (see below).82,83 S. cerevisiae telomerase may be assembled as a small nuclear RNP (snRNP), supporting a model in which telomerase RNA is transcribed by RNA polymerase II, 7-methylguanosine-capped, polyadenylated and processed.84,85
The ciliate telomerase RNAs share little primary sequence homology, with the exception of the template region and a conserved region upstream of the template (CU)GUCA.11 However, experimental evidence suggests that the ciliate telomerase RNAs fold into a similar secondary structure.11 The murine and human telomerase RNA subunits share 65% identity.80 The secondary structure of vertebrate telomerase RNAs displays a similar topology to the ciliate structure and contains eight highly conserved regions (CRs), including a pseudoknot domain, the CR4-CR5 domain, a Box H/ACA domain characteristic of small nucleolar RNPs (snoRNPs), and the CR7 domain (Fig. 3).76 To date, limited phylogenetic analyses and predicted secondary structures for yeast telomerase RNAs have been reported.78,86,87 No significant sequence homology exists between the telomerase RNA components of the yeasts S. cerevisiae and K. lactis.88,89
Characterization of the Tetrahymena, human, mouse and yeast telomerase RNAs, using in vitro and in vivo reconstitution systems, has identified: 1) specific template residues that play an active role in enzymatic function, in addition to specifying the incorporation of nucleotides into DNA products; and 2) specific non-template regions of the RNA that are important for activity, RNP assembly and binding to the telomerase reverse transcriptase.15,16,18–24,26,33,41,43,68,69,85,86,90–97 Most studies addressing the catalytic function of the template nucleotides of telomerase RNAs have been previously reviewed in detail.5,11 One recent study of particular interest analyzed the effects of complete in vivo replacement of a Tetrahymena telomerase RNA template with sequences encoding nontelomeric repeats.98 Suprisingly, these mutant telomerases were catalytically active in vitro, albeit with some alteration in enzymatic properties such as processivity. These results suggest that base-specific interactions between the RNA template and the telomerase reverse transcriptase are not required for nucleotide addition and base discrimination.98
Human and Mouse Telomerase RNAs
One of the highly conserved regions of vertebrate telomerase RNAs is the H/ACA box.76 The H/ACA box is a characteristic feature of snoRNPs, which are implicated in the pseudouridinylation of ribosomal RNAs.97 In vivo, the Box H/ACA domain of hTR (Fig. 3) is essential for hTR accumulation, hTR 3' end processing and telomerase activity.97 The Box H/ ACA and CR7 domains of mTR also affect its accumulation and stability in the cell.33 However, the Box H/ACA and CR7 domains of hTR (Fig. 3) are not required for reconstitution of telomerase activity in vitro, or for hTERT binding,16,20,23,25,68 suggesting that these motifs play specific essential roles in vivo.
The hTR CR4-CR5 domain (Fig. 3) is essential for telomerase activity, reconstituted both in vitro and in vivo.23,68,97 The pseudoknot and CR4/CR5 domains of mTR are required for telomerase activity, but are not essential for mTR stability or accumulation in the cell.33 However, the CR4-CR5 domain of mTR is important for mTR processing, and the CR4-CR5 domain of hTR is important for both hTR accumulation and processing in vivo.33,97 The essential role of the hTR CR4-CR5 domain in enzyme activity, specifically residues 208–330 or 241–330, is apparently mediated through interactions with hTERT.24,68,99 Previous reports that the minimal hTR region required for telomerase activity maps to nucleotides 10–159 or 44–203 do not support an essential role for the CR4-CR5 domain.16,20,25 Discrepancies among these findings might be attributable to: 1) differences in cycle number and other conditions during PCR amplification of telomerase elongation products; 2) the addition of the telomerase RNA either during or after the translation of hTERT in RRL; 3) the presence or absence of different cellular cofactors; 4) selection for stable hTERT-hTR complexes during immunoprecipitation reactions; and 5) the ability, under certain reconstitution conditions, of a second hTERT binding site (see below) to compensate for the absence of a CR4-CR5 domain.
Mutational analysis of hTR by two groups has identified two distinct regions that can independently bind hTERT, one region spanning nucleotides 33–147 or 1–209 and the other spanning nucleotides 208–330 or 241–330.24,68 Independently, neither region is competent to reconstitute telomerase activity. However, residues 33–147 or 1–209 of hTR can reconstitute telomerase activity in the presence of hTR164–330 or 241–330, respectively.23,24,68 The complementation between two independent hTR domains to reconstitute human telomerase activity may be due to the separate contributions of a template domain by hTR33–147, and an hTERT-binding site (between nucleotides 208–330) by hTR 164–330.68
The telomerase RNA pseudoknot (Figure 3) is required for reconstitution of telomerase activity. Interestingly, both mTR pseudoknot topology and sequence are critical for telomerase activity reconstituted by expressing mTR in mTR−/− cells, whereas only the structure, but not the sequence, of the Tetrahymena telomerase pseudoknot is essential for telomerase activity reconstituted by overexpressing TER in Tetrahymena.33,95 Mutations in the pseudoknot of hTR (specifically in the P3 helix) (Fig. 3) also significantly alter the reconstitution of a catalytically active telomerase in vitro, though the mutated RNAs can bind hTERT.20,68,76,82,99 In Tetrahymena, however, deletion of the stem III/pseudoknot or mutations predicted to destabilize the pseudoknot structure do not significantly perturb telomerase activity in vitro.21,22 Thus, certain conditions in vitro may compensate for the lack of a pseudoknot, though it is also possible that the mutated RNAs fold into structures that mimic the structure and/or function of the pseudoknot.21 Chemical modification studies have previously indicated a role for the Tetrahymena telomerase RNA pseudoknot in protein binding.100 However, the Tetrahymena telomerase RNA pseudoknot appears to bind TERT in vivo, but not in vitro. A study performed using Tetrahymena telomerase reconstituted in vitro identified a four-nucleotide region 5' to the template (CAUU15-18) that is critical for TERT binding in Tetrahymena; moreover the 5' end of the telomerase RNA, excluding the pseudoknot, is sufficient for TERT binding.22,69 The recent demonstration that RNase VI cleavage of the Tetrahymena telomerase RNA stem III/pseudoknot region is enhanced upon TERT binding in RRL further supports the observation that the pseudoknot is not implicated in TERT binding.101 However, Tetrahymena telomerase complexes reconstituted in vivo with a wild-type telomerase RNA contain TERT, as determined by Western analysis, while a complex reconstituted with an RNA mutated in the pseudoknot region does not, suggesting that the pseudoknot is essential for assembly of the complex in vivo.95 Therefore, though the Tetrahymena telomerase RNA pseudoknot appears dispensable for TERT binding and telomerase activity in vitro, it may modulate some yet uncharacterized regulatory function in vivo.21,69,95,101
The effects of template-mutated hTR have also been studied in vitro and in vivo.20,26,27,82,102 In vitro, template-mutated human telomerase RNAs can reconstitute mutant telomerase activity; however, only short products are synthesized by a template-mutated hTR specifying the synthesis of TTGGGG sequences.20,82 The expression of mutant human telomerases upon transfection of template-mutated hTRs (that specify TTTGGG, TTGGGG, TTAAGG or TGAGGG sequences) into either immortal, telomerase-positive (HT-1080) or hTR-negative cells (VA13 expressing hTERT) results in cell cycle deregulation, rapid loss of viability, and nuclear and chromosomal abnormalities.26,27 Similarly, low expression of different template-mutated telomerase RNAs (UAUAUAUAUAA or CUAAAACCCUAAC instead of the wild-type template, CUAACCCUAAC) in prostate and breast cancer cell lines also decreases cellular viability and increases apoptosis.102 These studies clearly demonstrate the importance of correct telomere sequences at chromosome ends.
Yeast Telomerase RNAs
Initially, studies of yeast telomerase RNAs focused on the template region. K. lactis telomerase RNA template mutations lead to the addition of mutated telomeric DNA sequences at the telomeres, followed by decreased cell viability, and an increase in telomere length to approximately 100 times normal length.89 Expression of template-mutated telomerase RNAs in K. lactis is also associated with telomere shortening.103 In S. cerevisiae, four of five trinucleotide substitutions in 11 of the potential 17 telomerase RNA template residues (positions 467-483) affect template function and lead to shorter telomeres, but do not affect haploid cell growth.92 However, in vitro, these mutated RNAs are active, albeit at slightly reduced levels compared to wild-type enzyme. One specific trinucleotide substitution at positions 474-476 (476GUG) does not reconstitute telomerase activity in vitro or in vivo, and leads to progressive telomere shortening, slow growth and eventual cellular senescence typical of yeast lacking either the telomerase RNA or the EST2 catalytic component.5,92 Interestingly, the 476GUG mutant RNA is stable, since its levels in yeast cells are comparable to wild-type telomerase RNA levels, and it assembles into a normal telomerase RNP, as characterized by fractionation on DEAE-agarose and migration through native gels.92 Six single- and double-base changes within positions 474-476 result in: 1) telomere elongation; 2) progressive telomere shortening that leads to cellular senescence; or 3) telomere shortening that establishes a new, shorter telomere length setpoint that is associated with normal cell growth.104 All six mutant telomerases are enzymatically active in vitro. Telomere elongation phenotypes correlate with a reduction in Rap1p binding to the mutated telomeric sequences specified by mutant telomerase RNAs in both S. cerevisiae and K. lactis.89,104,105 However, the telomere shortening phenotypes of some template mutants also correlate with decreased Rap1p binding, suggesting that the binding of factors that mediate telomere addition, either directly (telomerase) or indirectly, (Est1p, Cdc13p), may also be affected.103,104 Specifically, K. lactistelomerases synthesizing Rap1p binding sites mutated on the right side or on both sides lead to telomere shortening, and mutations on the left side lead to telomere elongation.103–105 Thus the authors propose that, normally, Rap1 binding to the left side of the Rap1 binding site negatively regulates telomere length and the right side is bound by positive regulators of telomere length such as telomerase, Est1p or Cdc13p.103
As in ciliates, nontemplate residues in the yeast telomerase RNA are important for telomerase function.21,86,93 Detailed mutagenesis of the telomerase RNA of K. lactis reveals that more than half of the RNA is dispensable for telomerase activity in vivo. However, deletion of four regions outside the template (435–464), that comprise nucleotides 20–60, 493–580, 630–730 and 980–987, abolishes telomerase function in vivo and leads to telomere shortening, without affecting RNA synthesis or stability.86 Telomerase activity is undetectable in extracts made from cells expressing the mutated RNAs, indicating defects in catalysis. Specifically, two of the mutant enzymes reconstituted with RNA deleted at residues 493–580 and 630–730 are defective in telomerase RNP assembly, and the RNA deleted at residues 980–987 appears to have an altered conformation that affects RNP assembly.86 Interestingly, nucleotides 980–987 fall within a predicted stem-loop structure in the K. lactis telomerase RNA. Secondary structure prediction and analysis of mutations and compensatory mutations in nucleotides 288–335 of the S. cerevisiae telomerase RNA indicate that a stem loop structure forms in this region of the RNA.87 This S. cerevisiae telomerase RNA stem-loop structure appears to interact with telomere-bound Ku, suggesting a mechanism by which telomerase can be retained at the telomere.87 Nontemplate residues, specifically residues 662–917 of the S. cerevisiae telomerase RNA, also contribute to the high affinity and sequence-specific recognition of DNA substrates characteristic of yeast telomerase (see section on Primer substrates and specificity).106 Protein-independent binding of the telomerase RNA to telomeric DNA has previously been reported for Euplotes telomerase.70,107
Template Boundary
Specific sequences and structures in ciliate and yeast telomerase RNAs define the 5' boundary of the template, preventing the synthesis of nontelomeric repeats.19,78,92 In Tetrahymena, yeast and human, altered telomere sequences can result in deregulation of telomere length, nuclear and chromosomal abnormalites, altered growth, loss of viability and eventual cell death, indicating that it is essential to prevent the copying of telomerase RNA sequences not complementary to the telomeric sequences.27,89,92,94,102–104,108,109 In ciliates, a conserved (CU)GUCA sequence in the telomerase RNA, two nucleotides upstream of the template, has been proposed to regulate the template boundary.110 Telomerase reconstituted in vitro with RNAs mutated in this conserved domain have an altered template boundary.19 In S. cerevisiae, mutation of residues 465–467 alters the 5' boundary of the template (435–464).92 In K. lactis, disruption of a conserved secondary structure adjacent to the template allows the use of nontemplate residues in DNA synthesis, resulting in altered telomere sequences, telomere shortening and cellular growth defects.78 Interestingly, the E. aediculatus telomerase can use two different regions of the RNA as templates to mediate the correct addition of telomere sequences.111 It has been proposed that telomerase proteins such as TERT, rather than an RNA sequence, define the 5' template boundary in E. aediculatus.111 In support for a role of telomerase proteins in the regulation of the 5' template RNA boundary, TERT sequences in the CP2 motif have been identified that regulate template boundary in Tetrahymena telomerase.56 Sequences or structures that regulate the template boundary have not yet been identified in vertebrate telomerase RNAs. However, at least for mTR, where the template is 2 nt downstream of the 5' end of the RNA, template boundary regulation is unlikely to be mediated by RNA sequences or structures upstream of the template.81
Characterization of the telomerase RNA component since it was first identified over 10 years ago in Tetrahymena has revealed unexpected roles for this component. The RNA not only serves as a template for telomere synthesis, but it plays critical roles in mediating or influencing telomerase processivity and fidelity, endonucleolytic cleavage, regulation of the template boundary, RNP assembly, hTERT binding, binding to associated proteins (see below), telomere length and sequence regulation, chromosomal stability and cell viability. The influence of the telomerase RNA on telomerase processivity, fidelity and endonucleolytic cleavage is discussed further in the section “Mechanism of Telomere Synthesis by Telomerase.”
Telomerase-Associated Proteins
Telomerase-associated proteins have been identified in Tetrahymena (p95, p80), Euplotes (p43), yeast (Est1p, Est3p, Cdc13p, the Sm proteins), humans (TEP1, hStau, L22 RNA binding protein, dyskerin), mouse (TP1) and rat (TLP1) (Table 1).9,85,112–119 The proteins associated with telomerase are not essential for catalytic activity in vitro; however, some likely regulate telomerase activity, assembly and function in vivo. Human, mouse and rat homologues of the Tetrahymena telomerase RNA-binding p80 protein are referred to as TP1, TEP1 or TLP1 (see above). TP1, like p80, associates with the telomerase RNA.16,17,116,120 but it is not required for telomerase function in vitro or in vivo.25,121 The genes encoding p80 and p95 in Tetrahymena are not essential for cell viability, nor are the proteins core telomerase components; however, loss of p80 and p95 results in telomere lengthening.122,123 Other proteins that have been identified on the basis of their interaction with the telomerase RNA are the hStau double-stranded RNA binding protein, the L22 RNA binding protein, dyskerin, and the Sm proteins.85,117,118 These proteins are implicated in the processing and stability of ribonucleoproteins. Human telomerase is also associated with the molecular chaperones hsp90/p2322,124,125 which are likely required for assembly and stability of the telomerase complex. The yeast proteins Est1p, Est3p and Cdc13p regulate access of telomerase to the telomere, through direct and indirect associations with the telomerase RNA and Est2p components.126–130 No homologues of these proteins have been discovered in other organisms.
Table 1
Yeast, human and mouse proteins associated with telomerase.
In S. cerevisiae, the telomerase ribonucleoprotein complex is greater than 669 kDa, which is larger than the combined mass of the two essential telomerase components, Est2p (103 kDa) and the TLC1 RNA (430 kDa).39 Thus the telomerase ribonucleoprotein probably contains more than one copy of each component, and/or a number of associated proteins.39 There is evidence that the yeast telomerase ribonucleoprotein complex contains two copies of the telomerase RNA.66 The approximate molecular mass of the purified human telomerase complex is 550–1000 kDa, suggesting that other components are associated with hTERT (127 kDa) and hTR (147 kDa),32,75,131 and/or that multiple copies of each component are present. In fact, as in yeast, human telomerase contains two cooperating telomerase RNA molecules.32 The molecular mass (600 kDa) of endogenous and recombinant human telomerases purified under highly stringent conditions suggests that the minimal telomerase complex may be composed of two copies of the TERT catalytic component, in addition to two copies of the telomerase RNA.32 This hypothesis is supported by the functional evidence that two separately inactive hTERT proteins can complement in vitro and in vivo to reconstitute telomerase activity.25,30,67 As for human telomerase complexes purified under less stringent conditions, the mouse and rat telomerase ribonucleoprotein complexes have a large molecular mass of 1000 kDa or more.75,119 This increase in molecular mass compared to purified human complexes suggests that additional telomerase-associated components may be present in rodent telomerases that are not present in the human holoenzyme. In Tetrahymena, the telomerase RNA is found in two distinct RNP complexes resolved by nondenaturing gel electrophoresis, one containing p95 and p80, and one without either of these associated proteins.93,95 This observation, and data demonstrating that many telomerase-associated proteins are involved in ribonucleoprotein assembly and processing suggest that in vivo differences in telomerase complex composition, even within one organism, are also likely to reflect the natural ‘life cycle’ of the enzyme. This may specifically be true of ciliate telomerases that are developmentally regulated (see section on Recombinant versus endogenous telomerases). For example, the Euplotes p43 protein is a component of the telomerase complex with homology to the La autoantigen, and therefore may function in the assembly and nuclear retention of telomerase.113,132 Other proteins that can interact or associate with telomerase are hnRNPA1, hnRNPC1 and C2, snoRNP proteins such as Cbf5p, Nhp2p, Nop10p, hNop10, and hNHP2, and 14–3–3.73,133,137 Possible functions of these proteins include recruitment of telomerase to telomeres (hnRNPs), accumulation of mature telomerase RNA (snoRNPs) and nuclear localization (14–3–3).
Mechanism of Telomere Synthesis by Telomerase
In the following section, a general model for the synthesis of telomeric DNA by telomerase is described. The influence of substrate sequence and length, nucleotide concentration and the telomerase RNA on processivity and synthesis of telomeric repeats is briefly presented, as is a discussion of the possible role of telomerase-mediated endonucleolytic cleavage. In vitro, telomerase preferentially binds and elongates oligodeoxynucleotide primers that are complementary to the RNA template and that mimic the sequence and single-stranded end of a chromosome (for example (TTAGGG)3 for human).138,139 In a conventional (also called standard or direct) telomerase assay, incubation of human telomerase in the presence of [α-32P]dGTP, dATP, dTTP, Mg2+ and a telomeric primer results in the synthesis of radiolabelled products that show a distinct six-base periodicity. This periodicity is produced by enzyme pausing and dissociation, which occurs predominantly at the first G within the sequence GGGTTA of each repeat (Fig. 2).138,140 A routinely used modification to the telomerase assay is the amplification of human or mouse telomerase products by PCR, known as the TRAP (telomeric repeat amplification protocol) assay.141,142
Primer Substrates and Specificity
The principal rules that determine whether a primer can serve as a substrate for telomerase in vitro are similar among telomerases from ciliates and human.11,75 Telomeric sequences of approximately 18 nt are preferred (at concentrations ranging from 2.5 nM to 1 mM), yet nontelomeric sequences can be elongated if oligonucleotides are provided at concentrations greater than 3 mM.75,143 Interestingly, although G-rich primers are preferred substrates, extension of AT-rich primers at low concentrations (25 nM) by Tetrahymena telomerase occurs efficiently if Mg2+ concentrations are increased from 0.5 mM to 1.25 mM.144 High concentrations of primer (1–5 mM) are required if primers are short (6 nt).143,145 Primer binding and elongation are more efficient if the 3' end of the primer is complementary to the telomerase RNA template site.138,143,145,146
Telomerase contains a second distinct primer-binding domain, called the anchor site.139,143,145,146 Binding at the anchor site is more efficient with telomeric or G-rich sequences. Similarly to RNA polymerase, this second site is critical in maintaining contact with the growing nucleic acid chain while the active site is repositioned for another round of elongation.145,147 In ciliates and human, the anchor site is critical in the recognition and elongation of nontelomeric substrates.139,146,148,149 Evidence supporting the existence of a protein anchor site came from the demonstration that the 5'-end of a primer, 20–22 nucleotides from the 3'-end, can be crosslinked to a telomerase protein and the telomerase RNA.70 Primer binding and elongation are also more efficient if telomeric or G-rich sequences towards the 5' end of the primer are available for interaction with the anchor site.138,143,145,146 Binding studies suggest that Tetrahymena telomerase has low substrate sequence specificity.150 Interestingly, Tetrahymena telomerase can also elongate RNA-containing oligonucleotides and incorporate rGTP into elongation products.151
S. castellii and S. cerevisiae telomerases share the telomeric primer specificity characteristic of other telomerases.152 In contrast to Tetrahymena telomerase, S. cerevisiae telomerase recognizes and binds the dG-rich strand of yeast telomeres with high affinity and specificity.153 In vitro, high affinity primer binding does not correlate with primer elongation, as nontelomeric primers with low affinity binding are efficiently elongated by telomerase.153 Primers with high affinity binding are less efficiently elongated by yeast telomerase, suggesting that high affinity binding may be a mechanism that negatively regulates telomerase.153 High affinity primer binding does, however, correlate with the ability of the sequence to seed new telomere formation, or to be elongated in vivo, suggesting that binding may be a limiting step for extension in vivo.153 Thus, at least in vivo, high affinity substrate binding does correlate with substrate elongation by S. cerevisiae telomerase.
The 3' ends of telomeric oligonucleotides that are elongated by yeast telomerase apparently play a role in alignment with the RNA template, as point mutations in the 3'-most primer positions (−1 to −3) reduce the level of nucleotide incorporation.154 These results indicate that the interaction of the 3' end of DNA substrates with the telomerase RNA template site is a common feature of telomerases.11 The 5' region of the primer (−16 to −21; Site B, Fig. 2) appears important for enzyme binding, but less so for polymerization, and most probably plays a role in the previously defined anchor function of telomerase.154 Mutations in the middle of yeast substrate primers (between the −4 and −14 position) (Fig. 2, Site A) affect primer binding and elongation, distinct from mutations that affect the 3' and 5' primer regions. Such mutations enhance primer extension, although binding affinity is reduced.154 Thus the identification of an additional interaction site (site A) between yeast telomerase and its DNA substrates supports a tripartite model of interaction between yeast telomerase and its DNA substrates.154 Observations from previous studies in Tetrahymena and Oxytricha are also consistent with a tripartite model of enzyme interaction with telomeric DNA. For Tetrahymena telomerase, Vmax values are greater for a 10 nt primer, that may potentially interact with site A, than for longer and shorter primers, and the rate of synthesis by Oxytricha telomerase is higher with an 8 nt primer than a 16 nt primer.143,154,155 Subsequent studies have revealed that the high sequence specificity of yeast telomerase is mediated largely through primer interactions with the telomerase RNA component.106 In contrast, Tetrahymena TERT alone appears to be sufficient for specific telomeric primer binding, suggesting that in vitro, the Tetrahymena telomerase RNA does not significantly contribute to primer binding.101 Differences in mechanisms of substrate recognition in ciliates and yeast may be due to different in vivo requirements for de novo telomere synthesis.154
Processivity and Nucleotide Requirements
In vitro, human and ciliate telomerases are generally processive, while mouse, hamster and Xenopus telomerases produce only one or two repeats under conventional assay conditions.79,138,156–158 Nonprocessive enzymes can be stimulated to synthesize up to 10 repeats by increasing dGTP concentration 10- to 100-fold.75,158 Multiple-round primer elongation products generated by mouse telomerase can also be detected by PCR amplification.80,157 S. castelli telomerase is moderately processive, synthesizing five to seven 8-nt repeats, whereas S. cerevisiae telomerase is much less processive.152 K. lactis telomerase catalyzes only one round of repeat synthesis under all in vitro conditions tested.159
The processivity of human telomerase can be regulated by temperature, by primer and dNTP concentrations, and by G-quadruplex-interacting agents such as potassium ions (K).160 Specifically, processivity decreases with increasing temperature, primer and K concentration. Using a 5' biotinylated (TTAGGG)3 primer, the optimal temperature for processivity is 20°C, versus 37°C for enzyme activity.160 Maximal telomerase activity occurs with the addition of 1 mM biotinylated substrate, however, the proportion of shorter products increases, and processivity decreases as primer concentration increases. In the presence of excess dATP and dTTP (1 μM each), both telomerase activity and processivity reach a maximum at approximately 15 μM dGTP (highest concentration tested is 18 μM),160 though the pattern of elongation products does not change when dGTP, dATP or dTTP concentrations are varied. By contrast, E. aediculatus telomerase becomes less processive as the concentration of dGTP is increased above 100 μM.111 High dGTP concentrations affect not only processivity, but also the pattern of elongation products and the selection of nucleotides that serve as template residues.111 Previous studies also indicate that primers that form G-quadruplex structures (stabilized by K) cannot be elongated by human telomerase.155 Interestingly, the inhibition of telomerase activity and processivity by increasing K concentrations occurs after primers have been extended to six repeats, indicating that inhibition is dependent on the length of the substrate and probably on the formation of quadruplex structures.160,161
In vitro conditions that could modulate yeast telomerase processivity have not been characterized in detail. However, using fractionation and reaction conditions optimized for detecting the synthesis of long elongation products by S. castelli telomerase, only short products, characteristic of a nonprocessive telomerase, are synthesized by S. cerevisiae telomerase.152 S. cerevisiae telomeres are irregular, while those of S. castelli consist of TCTGGGTG repeats interspersed with TCTGGG(TG)2–4 repeats. The inherently high stalling frequency of S. cerevisiae telomerase may account for its nonprocessive properties both in vitro and in vivo, and could account for the irregularity of S. cerevisiae telomere sequences.88,152 Nonprocessive synthesis is commonly explained as the dissociation of telomerase from its substrate or elongation product. However, both S. cerevisiae and K. lactis telomerases can exist in a stalled elongation-incompetent complex stably bound to telomeric primer or elongation product.66,159 S. cerevisiae telomerase containing telomerase RNAs mutated in the template region are active in the presence of wild-type telomerase RNA and synthesize a mixture of wild-type and mutant telomeric repeats onto the extreme ends of chromosomes, confirming that S. cerevisiae acts nonprocessively in vivo.92 Although Tetrahymena telomerase is highly processive in vitro, in vivo its mode of DNA synthesis appears distributive, or nonprocessive, during de novo telomere formation.162
Nonprocessive S. cerevisiae and K. lactis telomerase enzymes, which complete at most one cycle of template copying (type I translocation/processivity), are functional in maintaining physiologic telomere lengths.152,159 Type II translocation has been defined as translocation of the enzyme after each cycle of template copying, and is more commonly described for the telomerase enzyme as processivity.57 However, disrupting the translocation of the enzyme during the first cycle of template copying (type I translocation) results in shortened telomeres, suggesting that the limiting factor for telomere maintenance in S. cerevisiae is enzyme type I ranslocation/processivity.57
Endonucleolytic Cleavage
A telomerase-mediated substrate DNA cleavage activity, similar to the cleavage activity of DNA-dependent RNA polymerases, has been characterized in T. thermophila, Euplotes crassus, S. cerevisiae, S. pombe, and Saccharomyces castelli, but not yet in vertebrates.92,145,149,152,154,163,164 In both E. crassus and S. cerevisiae, the nuclease acts through an endonucleolytic mechanism.149,165 The function of nucleotide cleavage by telomerase is unknown, but by analogy with RNA polymerases, may serve a proofreading role, to enhance processivity, or to ensure that nontemplate residues are not copied into DNA.11 DNA substrates that: 1) align at, or beyond, the 5' end of the template; 2) form a mismatch with the 5' end of the template; or 3) contain nontelomeric 3' sequences that extend past the second-to-last 5' template residue, are cleaved.145,149 However, the nuclease active site in E. crassus telomerase is not restricted to the 5' end of the template, but stretches over a 4-nt region, beginning at the last 5' template residue.164 Cleavage activity can also be reconstituted with wild-type and mutant Tetrahymena telomerases altered in the template region, except when template mutations prevent the alignment of the DNA substrate with the extreme end of the template.18,19 The replacement of the template with sequences encoding nontelomeric sequences does not abolish, but strongly affects, the cleavage activity of Tetrahymena telomerase.98 One mutant, in which the template sequence is altered to 5'-UAUAUAUAA-3' from 5'-CAACCCCAA-3', mediates cleavage at a position facing the middle of the template, supporting a role for specific template RNA nucleotides in cleavage.98 The Glaucoma chattoni and T. thermophila RNAs have 23 identical residues in the template and immediately adjacent regions. The cleavage activity of Glaucoma RNA-substituted telomerase extracted from Tetrahymena cells cuts a bound telomeric primer at a position complementary to the center of the template.93 These results suggest that the identity of RNA bases outside the template, which differ between Glaucoma and Tetrahymena, can influence cleavage activity.5 The endonucleolytic activity likely resides within the catalytic component of telomerase, and not an accessory factor, as Tetrahymena TERT and telomerase RNA components expressed in RRL are sufficient to reconstitute cleavage activity.17
Nucleolytic activity is more evident for S. cerevisiae telomerase than for S. castellii telomerase.152 A detailed characterization of telomerase cleavage activity in S. cerevisiae reveals that cleavage site selection is not stringent, since loss of one nuclease site in the DNA substrate by the introduction of a nonhydrolyzable linkage leads to the utilization of other sites.165 Interestingly, both fragments generated by cleavage can be elongated by S. cerevisiae telomerase, suggesting that the enzyme contains two distinct active sites for the reverse transcriptase and nuclease activities, or that the enzyme is a multimer, with each monomer containing a single active site that mediates both elongation and cleavage.165 Previous experiments with E. crassus telomerase have demonstrated elongation of only the 5' fragment generated by cleavage; however, the 3' fragments contained nontelomeric DNA and were likely to provide inefficient substrates for elongation.149,165 Primer/template mismatches can also enhance primer cleavage by yeast telomerase, specifically mismatches between the −4 to −6/−4 to −14 region of the primer and the template RNA.92,154
Recombinant Versus Endogenous Telomerase
In certain organisms, like E. crassus, telomerase processivity is developmentally regulated.166 During macronuclear development, larger telomerase complexes of 500 kD, 1600 kDa and 5-MDa form that processively elongate non-telomeric substrates more efficiently than the smaller 280 kDa complex present during vegetative growth.166,167 S. cerevisiae telomerase may also contain protein subunits that affect processivity and nuclease activity.168 These data suggest that in some organisms, associated factors may regulate the biochemical properties of telomerase. It is expected that recombinant telomerase assembled simply from TERT and TR would not recapitulate all the properties of endogenous telomerase from these organisms. To date, only recombinant Tetrahymena telomerase has been characterized in any detail.169 Recombinant Tetrahymena telomerase expressed in RRL demonstrates a more limited repeat addition processivity than endogenous telomerase.17 The recombinant enzyme requires micromolar concentrations of dGTP for processive elongation, in contrast to the submicromolar concentrations of dGTP required by endogenous telomerase.169 The dGTP-dependence of processive elongation is also a feature of endogenous E. aediculatus, human and hamster telomerases.111,158,160 It has been hypothesized that recombinant Tetrahymena telomerase lacks certain of the anchor sites integral to the endogenous enzyme.169 Using a modified version of a standard telomerase assay, partially purified RRL-synthesized human telomerase RNA and protein components appear to be as processive as partially purified endogenous human telomerase.15 Comparable elongation products analyzed by a standard telomerase assay are also generated by affinity-purified recombinant human telomerase expressed in insect cells, and by affinity-purified telomerase from HeLa cells.32
Conclusions
In the 16 years since the first identification of telomerase activity from the ciliate Tetrahymena, we have gained much information about this essential ribonucleoprotein. Future studies using recombinant components of telomerase will add to our knowledge of its mechanistic properties, and the characterization of its associated components will also be critical to understand the regulation of telomerase. Moreover, certain differences in telomerase and telomere biology between ciliates, yeasts, human and mouse will likely be explained by species-specific differences in telomerase enzyme activity and regulation.
Acknowledgments
The authors wish to thank Lea Harrington and Tara Beattie for communicating results prior to publication and F. Bachand, J. Demers, S. Dupuis and S. Huard for comments on the manuscript. Work in the lab of C. Autexier is supported by the Canadian Institutes of Health Research and the Cancer Research Society, Inc.
References
- 1.
- Price CM. Telomeres and telomerase: broad effects on cell growth. Curr Opin Genet Dev. 1999;9:218–224. [PubMed: 10322143]
- 2.
- Vaziri H, Benchimol S. From telomere loss to p53 induction and activation of a DNA-damage pathway at senescence: The telomere loss/DNA damage model of cell aging. Exp Gerontol. 1996;31:295–301. [PubMed: 8706799]
- 3.
- Wellinger RJ, Sen D. The DNA structures at the ends of eukaryotic chromosomes. Eur J Cancer. 1997;33:735–749. [PubMed: 9282112]
- 4.
- Pardue M -L, DeBaryshe PG. Telomeres and telomerase: more than the end of the line. Chromosoma. 1999;108:73–82. [PubMed: 10382069]
- 5.
- Blackburn E. Telomerases In: Gestland, RF, Cech, TR Atkins, JF, ed.The RNA World 2nd Edition. Cold Spring Harbor: Cold Spring Harbor Laboratory Press,1999609–635.
- 6.
- Zijlmans JM, Martens UM, Poon S S S. et al. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad USA. 1997;94:7423–7428. [PMC free article: PMC23837] [PubMed: 9207107]
- 7.
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. [PubMed: 8811183]
- 8.
- Colgin LM, Reddel RR. Telomere maintenance mechanisms and cellular immortalisation. Curr Opin Genet Dev. 1999;9:97–103. [PubMed: 10072358]
- 9.
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes & Dev. 1998;12:1073–1085. [PubMed: 9553037]
- 10.
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. [PubMed: 3907856]
- 11.
- Collins K. Ciliate telomerase biochemistry. Annu Rev Biochem. 1999;68:187–218. [PubMed: 10872448]
- 12.
- Collins K. Mammalian telomeres and telomerase. Curr Opin Cell Biol. 2000;12:378–383. [PubMed: 10801465]
- 13.
- O'Reilly M, Teichmann SA, Rhodes D. Telomerases. Curr Opin Struct Biol. 1999;9:56–65. [PubMed: 10047586]
- 14.
- Weilbaecher RG, Lundblad V. Assembly and regulation of telomerase. Curr Opin Chem Biol. 1999;3:573–577. [PubMed: 10508671]
- 15.
- Weinrich SL, Pruzan R, Ma L. et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. [PubMed: 9398860]
- 16.
- Beattie TL, Zhou W, Robinson MO. et al. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. [PubMed: 9443919]
- 17.
- Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymenatelomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. [PMC free article: PMC21102] [PubMed: 9671704]
- 18.
- Autexier C, Greider CW. Functional reconstitution of wild type and mutant Tetrahymena telomerase. Genes & Dev. 1994;8:563–575. [PubMed: 7523243]
- 19.
- Autexier C, Greider CW. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes & Dev. 1995;15:2227–2239. [PubMed: 7557377]
- 20.
- Autexier C, Pruzan R, Funk WD. et al. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article: PMC452365] [PubMed: 8918470]
- 21.
- Autexier C, Greider CW. Mutational analysis of the Tetraymena telomerase RNA: identification of residues affecting telomerase activity in vitro. Nucleic Acids Res. 1998;26:787–795. [PMC free article: PMC147331] [PubMed: 9443971]
- 22.
- Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes & Dev. 1999;13:1116–1125. [PMC free article: PMC316941] [PubMed: 10323863]
- 23.
- Tesmer VM, Ford LP, Holt SE. et al. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol Cell Biol. 1999;19:6207–6216. [PMC free article: PMC84565] [PubMed: 10454567]
- 24.
- Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell. 2000;6:361–371. [PubMed: 10983983]
- 25.
- Beattie TL, Zhou W, Robinson MO. et al. Polymerization defects within human telomerase are distinct from telomerase RNA and Tep1 binding. Mol Biol Cell. 2000;11:3329–3340. [PMC free article: PMC14995] [PubMed: 11029039]
- 26.
- Marusic L, Anton M, Tidy A. et al. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol Cell Biol. 1997;17:6394–6401. [PMC free article: PMC232491] [PubMed: 9343401]
- 27.
- Guiducci C, Cerone MA, Bacchetti S. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene. 2001;20:714–725. [PubMed: 11314005]
- 28.
- Bachand F, Autexier C. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J Biol Chem. 1999;274:38027–38031. [PubMed: 10608871]
- 29.
- Bachand F, Kukolj G, Autexier C. Expression of hTERT and hTR in cis reconstitutes an active human telomerase ribonucleoprotein. RNA. 2000;6:778–784. [PMC free article: PMC1369957] [PubMed: 10836798]
- 30.
- Moriarty T, Huard S, Dupuis S. et al. Functional multimerization of human telomerase requires an RNA interaction domain in the N-terminus of the catalytic subunit. Mol Cell Biol. 2001;22:1253–1265. [PMC free article: PMC134651] [PubMed: 11809815]
- 31.
- Masutomi K, Kaneko S, Hayashi N. et al. Telomerase activity reconstituted in vitro with purified human telomerase reverse transcriptase and human telomerase RNA component. J Biol Chem. 2000;275:22568–22573. [PubMed: 10811633]
- 32.
- Wenz C, Enenkel B, Amacker M. et al. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–3534. [PMC free article: PMC125520] [PubMed: 11432839]
- 33.
- Martin-Rivera L, Blasco MA. Identification of functional domains and dominant negative mutations in vertebrate telomerase RNA using an in vivo reconstitution system. J Biol Chem. 2001;276:5856–5865. [PubMed: 11056167]
- 34.
- Blasco MA, Lee H -W, Hande MP. et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. [PubMed: 9335332]
- 35.
- Greenberg RA, Allsopp RC, Chin L. et al. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. [PubMed: 9582020]
- 36.
- Martin-Rivera L, Herrera E, Albar JP. et al. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc Natl Acad Sci USA. 1998;95:10471–10476. [PMC free article: PMC27918] [PubMed: 9724727]
- 37.
- Artandi SE, DePinho RA. Mice without telomerase: what can they teach us about human cancer? Nat Med. 2000;6:852–855. [PubMed: 10932211]
- 38.
- Wright WE, Shay JW. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse biology. Nat Med. 2000;6:849–851. [PubMed: 10932210]
- 39.
- Lingner J, Hughes TR, Shevchenko A. et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. [PubMed: 9110970]
- 40.
- Counter CM, Meyerson M, Eaton EN. et al. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94:9202–9207. [PMC free article: PMC23115] [PubMed: 9256460]
- 41.
- Nakamura TM, Morin GB, Chapman KB. et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. [PubMed: 9252327]
- 42.
- Meyerson M, Counter CM, Eaton EN. et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. [PubMed: 9288757]
- 43.
- Harrington L, Zhou W, McPhail T. et al. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes & Dev. 1997;11:3109–3115. [PMC free article: PMC316744] [PubMed: 9389643]
- 44.
- Kilian A, Bowtell D D L, Abud HE. et al. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Gen. 1997;6:2011–2019. [PubMed: 9328464]
- 45.
- Bryan TM, Sperger JM, Chapman KB. et al. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. [PMC free article: PMC21101] [PubMed: 9671703]
- 46.
- Fitzgerald M, Riha K, Gao F. et al. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96:14813–14818. [PMC free article: PMC24730] [PubMed: 10611295]
- 47.
- Malik HS, Burke WD, Eickbush TH. Putative telomerase catalytic subunits from Giardia lamblia and Caenorhabditis elegans. Gene. 2000;251:101–108. [PubMed: 10876087]
- 48.
- Guo W, Okamoto M, Park NH. et al. Cloning and expression of hamster telomerase catalytic subunit cDNA. Int J Mol Med. 2001;8:73–78. [PubMed: 11408953]
- 49.
- Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998;92:587–590. [PubMed: 9506510]
- 50.
- Bodnar AG, Ouellette M, Frolkis M. et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. [PubMed: 9454332]
- 51.
- Nakayama J, Tahara H, Tahara H. et al. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat Genet. 1998;18:65–68. [PubMed: 9425903]
- 52.
- Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. [PubMed: 9501072]
- 53.
- Counter CM, Meyerson M, Eaton EN. et al. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. [PubMed: 9528864]
- 54.
- Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell. 2000;6:493–499. [PubMed: 10983995]
- 55.
- Bryan TM, Goodrich KJ, Cech TR. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J Biol Chem. 2000;275:24199–24207. [PubMed: 10807925]
- 56.
- Miller MC, Liu JK, Collins K. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 2000;19:4412–4422. [PMC free article: PMC302041] [PubMed: 10944124]
- 57.
- Peng Y, Mian SI, Lue NF. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol Cell. 2001;7:1201–1211. [PubMed: 11430823]
- 58.
- Haering CH, Nakamura TM, Baumann P. et al. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 2000;97:6367–6372. [PMC free article: PMC18609] [PubMed: 10829083]
- 59.
- Xia J, Peng Y, Mian S. et al. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol Cell Biol. 2000;20:5196–5207. [PMC free article: PMC85968] [PubMed: 10866675]
- 60.
- Kohlstaedt LA, Wang J, Freidman JM. et al. Crystal structure at 3.5A resolution of HIV reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. [PubMed: 1377403]
- 61.
- Jacobo-Molina A, Ding J, Nanni RG. et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. [PMC free article: PMC46920] [PubMed: 7687065]
- 62.
- Huang H, Chopra R, Verdine GL. et al. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. [PubMed: 9831551]
- 63.
- Jaeger J, Restle T, Steitz TA. The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 1998;17:4535–4542. [PMC free article: PMC1170784] [PubMed: 9687519]
- 64.
- Ren J, Esnouf R, Hopkins A. et al. The structure of the HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design. Structure. 1995;3:915–926. [PubMed: 8535785]
- 65.
- Sousa R. Structural and mechanistic relationships between nucleic acid polymerases. Trends Biochem Sci. 1996;21:186–190. [PubMed: 8871404]
- 66.
- Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes & Dev. 1997;11:2790–2800. [PMC free article: PMC316652] [PubMed: 9353249]
- 67.
- Beattie TL, Zhou W, Robinson MO. et al. Functional multimerization of the human telomerase reverse transcriptase. Mol Cell Biol. 2001;21:6151–6160. [PMC free article: PMC87332] [PubMed: 11509658]
- 68.
- Bachand F, Autexier C. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol Cell Biol. 2001;21:1888–1897. [PMC free article: PMC86762] [PubMed: 11238925]
- 69.
- Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. [PMC free article: PMC99554] [PubMed: 11158287]
- 70.
- Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. [PMC free article: PMC231754] [PubMed: 8972210]
- 71.
- Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes & Dev. 1999;13:2863–2874. [PMC free article: PMC317136] [PubMed: 10557213]
- 72.
- Counter CM, Hahn WC, Wei W. et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. [PMC free article: PMC24516] [PubMed: 9843956]
- 73.
- Seimiya H, Sawada H, Muramatsu Y. et al. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–2661. [PMC free article: PMC212742] [PubMed: 10835362]
- 74.
- Armbruster BN, Banik S S R, Guo C. et al. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol. 2001;21:7775–7786. [PMC free article: PMC99947] [PubMed: 11604512]
- 75.
- Greider CW, Collins K, Autexier C. Telomerases In: DePamphlis, M, ed.DNA replication in eukaryotic cells Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press,1996619–638.
- 76.
- Chen J -L, Blasco M, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. [PubMed: 10721988]
- 77.
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. [PubMed: 1840508]
- 78.
- Tzfati Y, Fulton TB, Roy J. et al. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. [PubMed: 10797010]
- 79.
- Prowse KR, Avilion AA, Greider CW. Identification of a nonprocessive telomerase activity from mouse cells. Proc Natl Acad Sci USA. 1993;90:1493–1497. [PMC free article: PMC45900] [PubMed: 8434010]
- 80.
- Blasco M, Funk W, Villeponteau B. et al. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. [PubMed: 7544492]
- 81.
- Hinkley CS, Blasco MA, Funk WD. et al. The mouse telomerase RNA 5'-end lies just upstream of the telomerase template sequence. Nucleic Acids Res. 1998;26:532–536. [PMC free article: PMC147299] [PubMed: 9421511]
- 82.
- Feng J, Funk WD, Wang S -S. et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. [PubMed: 7544491]
- 83.
- Zaug AJ, Lingner J, Cech TR. Method for determining RNA 3' ends and application to human telomerase RNA. Nucleic Acids Res. 1996;24:532–533. [PMC free article: PMC145649] [PubMed: 8602368]
- 84.
- Chapon C, Cech TR, Zaug AJ. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article: PMC1369572] [PubMed: 9409624]
- 85.
- Seto AG, Zaug AJ, Sobel SG. et al. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. [PubMed: 10490028]
- 86.
- Roy J, Fulton TB, Blackburn EH. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes & Dev. 1998;12:3286–3300. [PMC free article: PMC317211] [PubMed: 9784502]
- 87.
- Peterson SE, Stellwagen AE, Diede SJ. et al. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. [PubMed: 11138000]
- 88.
- Singer MS, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. [PubMed: 7545955]
- 89.
- McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. [PubMed: 7630414]
- 90.
- Gilley D, Lee M, Blackburn EH. Altering specific telomerase RNA template residues affects active site function. Genes & Dev. 1995;9:2214–2226. [PubMed: 7557376]
- 91.
- Gilley D, Blackburn EH. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol. 1996;16:66–75. [PMC free article: PMC230979] [PubMed: 8524330]
- 92.
- Prescott J, Blackburn EH. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 1997;11:528–540. [PubMed: 9042865]
- 93.
- Bhattacharyya A, Blackburn EH. A functional telomerase RNA swap in vivo reveals the importance of nontemplate RNA domains. Proc Natl Acad Sci USA. 1997;94:2823–2827. [PMC free article: PMC20280] [PubMed: 9096304]
- 94.
- Kirk KE, Harmon BP, Reichardt IK. et al. Block in anaphase chromosome separation caused by a telomerase template mutation. Science. 1997;275:1478–1481. [PubMed: 9045613]
- 95.
- Gilley D, Blackburn EH. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc Natl Acad Sci USA. 1999;96:6621–6625. [PMC free article: PMC21964] [PubMed: 10359761]
- 96.
- Autexier C, Triki I. Tetrahymena telomerase ribonucleoprotein RNA-protein interactions. Nucleic Acids Res. 1999;27:2227–2234. [PMC free article: PMC148444] [PubMed: 10219097]
- 97.
- Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3' end. Mol Cell Biol. 1999;19:567–576. [PMC free article: PMC83914] [PubMed: 9858580]
- 98.
- Ware TL, Wang H, Blackburn EH. Three telomerases with completely non-telomeric template replacements are catalytically active. EMBO J. 2000;19:3119–3131. [PMC free article: PMC203363] [PubMed: 10856255]
- 99.
- Bachand F, Triki I, Autexier C. Human telomerase RNA-protein interactions. Nucleic Acids Res. 2001;29:3385–3393. [PMC free article: PMC55859] [PubMed: 11504876]
- 100.
- Zaug AJ, Cech TR. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, self-splicing rRNA and U2 snRNA. RNA. 1995;1:363–374. [PMC free article: PMC1482408] [PubMed: 7493315]
- 101.
- Sperger JM, Cech TR. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–7016. [PubMed: 11401544]
- 102.
- Kim MM, Rivera MA, Botchkina IL. et al. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc Natl Acad Sci USA. 2001;98:7982–7987. [PMC free article: PMC35454] [PubMed: 11438744]
- 103.
- McEachern MJ, Iyer S, Boswell Fulton T. et al. Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc Natl Acad Sci USA. 2000;97:11409–11414. [PMC free article: PMC17213] [PubMed: 11016977]
- 104.
- Prescott JC, Blackburn EH. Telomerase RNA template mutations reveal sequence-specific requirements for the activation and repression of telomerase action at telomeres. Mol Cell Biol. 2000;20:2941–2948. [PMC free article: PMC85540] [PubMed: 10733598]
- 105.
- Krauskopf A, Blackburn EH. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature. 1996;383:354–357. [PubMed: 8848051]
- 106.
- Lue NF. Sequence-specific and conformation-dependent binding of yeast telomerase RNA to single-stranded telomeric DNA. Nucleic Acids Res. 1999;27:2560–2567. [PMC free article: PMC148461] [PubMed: 10352186]
- 107.
- Hammond PW, Cech TR. Euplotes telomerase: evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry. 1998;37:5162–5172. [PubMed: 9548747]
- 108.
- Yu G -L, Bradley JD, Attardi LD. et al. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. [PubMed: 1689810]
- 109.
- Smith CD, Blackburn EH. Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J Cell Biol. 1999;145:203–214. [PMC free article: PMC2133106] [PubMed: 10209018]
- 110.
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes & Dev. 1994;8:1984–1998. [PubMed: 7958872]
- 111.
- Hammond PW, Cech TR. dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res. 1997;25:3698–3704. [PMC free article: PMC146957] [PubMed: 9278493]
- 112.
- Collins K, Kobayashi R, Greider CW. Purification of Tetrahymena telomerase and cloning of the genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. [PubMed: 7774009]
- 113.
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a 3' overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. [PMC free article: PMC38220] [PubMed: 8855245]
- 114.
- Steiner BR, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. [PMC free article: PMC39716] [PubMed: 8610124]
- 115.
- Lingner J, Cech TR, Hughes TR. et al. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. [PMC free article: PMC23412] [PubMed: 9326584]
- 116.
- Harrington L, McPhail T, Mar V. et al. A mammalian telomerase-associated protein. Science. 1997;275:973–977. [PubMed: 9020079]
- 117.
- Le S, Sternglanz R, Greider CW. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol Biol Cell. 2000;11:999–1010. [PMC free article: PMC14826] [PubMed: 10712515]
- 118.
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. [PubMed: 10591218]
- 119.
- Nakayama J, Saito M, Nakamura H. et al. TLP1: A gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. [PubMed: 9118230]
- 120.
- Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes & Dev. 1998;12:721–733. [PMC free article: PMC316588] [PubMed: 9499406]
- 121.
- Liu Y, Snow BE, Hande PM. et al. Telomerase-associated protein TEP1 is not essential for telomerase activity or telomere length maintenance in vivo. Mol Cell Biol. 2000;20:8178–8184. [PMC free article: PMC86427] [PubMed: 11027287]
- 122.
- Miller MC, Collins K. The Tetrahymena p80/p95 complex is required for proper telomere length maintenance and micronuclear genome stability. Molecular Cell. 2000;6:827–837. [PubMed: 11090621]
- 123.
- Mason D, Autexier C, Greider CW. Tetrahymena proteins p90 and p95 are not core telomerase components. Proc Natl Acad Sci USA. 2001;98:12368–12373. [PMC free article: PMC60060] [PubMed: 11592988]
- 124.
- Holt SE, Aisner DL, Baur J. et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes & Dev. 1999;13:817–826. [PMC free article: PMC316592] [PubMed: 10197982]
- 125.
- Forsythe HL, Jarvis JL, Turner JW. et al. Stable association of hsp90 and p23, but not hsp70 with active human telomerase. J Biol Chem. 2001;276:15571–15574. [PubMed: 11274138]
- 126.
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. [PubMed: 10506558]
- 127.
- Hughes TR, Evans SK, Weilbaecher RG. et al. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000;10:809–812. [PubMed: 10898986]
- 128.
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. [PubMed: 11239396]
- 129.
- Chandra A, Hughes TR, Nugent CI. et al. Cdc13 both positively and negatively regulates telomere replication. Genes & Dev. 2001;15:404–414. [PMC free article: PMC312634] [PubMed: 11230149]
- 130.
- Grandin N, Damon C, Charbonneau M. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol. 2000;20:8397–8408. [PMC free article: PMC102147] [PubMed: 11046137]
- 131.
- Schnapp G, Rodi H -P, Rettig WJ. et al. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 1998;26:3311–3313. [PMC free article: PMC147663] [PubMed: 9628936]
- 132.
- Aigner S, Lingner J, Goodrich KJ. et al. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 2000;19:6230–6239. [PMC free article: PMC305813] [PubMed: 11080168]
- 133.
- LaBranche H, Dupuis S, Ben-David Y. et al. Telomere elongation by hnRNPA1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. [PubMed: 9620782]
- 134.
- Fiset S, Chabot B. hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res. 2001;29:2268–2275. [PMC free article: PMC55710] [PubMed: 11376145]
- 135.
- Ford LP, Suh JM, Wright WE. et al. Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol Cell Biol. 2000;20:9084–9091. [PMC free article: PMC86561] [PubMed: 11074006]
- 136.
- Dez C, Henras A, Faucon B. et al. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 2001;29:598–603. [PMC free article: PMC30409] [PubMed: 11160879]
- 137.
- Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol. 2000;20:9028–9040. [PMC free article: PMC86556] [PubMed: 11074001]
- 138.
- Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. [PubMed: 2805070]
- 139.
- Morin GB. Recognition of a chromosome truncation site associated with a-thalassaemia by human telomerase. Nature. 1991;353:454–456. [PubMed: 1896089]
- 140.
- Greider CW. Telomerase is processive. Mol Cell Biol. 1991;11:4572–4580. [PMC free article: PMC361337] [PubMed: 1875940]
- 141.
- Kim NW, Piatyszek MA, Prowse KR. et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. [PubMed: 7605428]
- 142.
- Kim NW, Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res. 1997;25:2595–2597. [PMC free article: PMC146790] [PubMed: 9185569]
- 143.
- Lee MS, Blackburn EH. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–6599. [PMC free article: PMC364717] [PubMed: 8413255]
- 144.
- Wang H, Blackburn EH. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–879. [PMC free article: PMC1169687] [PubMed: 9049315]
- 145.
- Collins K, Greider CW. Tetrahymena telomerase catalyzes nucleolytic cleavage and non-processive elongation. Genes & Dev. 1993;7:1364–1376. [PubMed: 8330740]
- 146.
- Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. [PubMed: 1896088]
- 147.
- Chamberlin MJ. New models for the mechanism of transcription elongation and its regulation. The Harvey Lectures. 1993;88:1–21. [PubMed: 1285418]
- 148.
- Melek M, Davis BT, Shippen DE. Oligonucleotides complementary to the Oxytricha nova telomerase RNA delineate the template domain and uncover a novel mode of primer utilization. Mol Cell Biol. 1994;14:7827–7838. [PMC free article: PMC359322] [PubMed: 7969123]
- 149.
- Melek M, Greene EC, Shippen DE. Processing of nontelomeric 3' ends by telomerase: default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. [PMC free article: PMC231338] [PubMed: 8668159]
- 150.
- Harrington LH, Hull C, Crittenden J. et al. Gel shift and UV crosslinking analysis of Tetrahymena telomerase. J Biol Chem. 1995;270:8893–8901. [PubMed: 7721797]
- 151.
- Collins K, Greider CW. Elongation of RNA primers and incorporation of ribonucleotides by Tetrahymena telomerase. EMBO J. 1995;14:5422–5432. [PMC free article: PMC394651] [PubMed: 7489731]
- 152.
- Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. [PubMed: 7618104]
- 153.
- Lue NF, Xia J. Species-specific and sequence-specific recognition of the dG-rich strand of telomeres by yeast telomerase. Nucleic Acids Res. 1998;26:1495–1502. [PMC free article: PMC147437] [PubMed: 9490797]
- 154.
- Lue NF, Peng Y. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 1998;26:1487–1494. [PMC free article: PMC147436] [PubMed: 9490796]
- 155.
- Zahler AM, Williamson JR, Cech TR. et al. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. [PubMed: 2023635]
- 156.
- Mantell LL, Greider CW. Telomerase activity in germline and embryonic cells of Xenopus. EMBO J. 1994;13:3211–3217. [PMC free article: PMC395213] [PubMed: 8039513]
- 157.
- Prowse KR, Greider CW. Developmental and tissue specific regulation of mouse telomerase and telomere length. Proc Natl Acad Sci USA. 1995;92:4818–4822. [PMC free article: PMC41798] [PubMed: 7761406]
- 158.
- Maine IP, Chen S -F, Windle B. Effect of dGTP concentration on human and CHO telomerase. Biochemistry. 1999;38:15325–15332. [PubMed: 10563818]
- 159.
- Boswell-Fulton T, Blackburn EH. Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol Cell Biol. 1998;18:4961–4970. [PMC free article: PMC109080] [PubMed: 9710579]
- 160.
- Sun D, Lopez-Guajardo CC, Quada J. et al. Regulation of catalytic activity and processivity of human telomerase. Biochemistry. 1999;38:4037–4044. [PubMed: 10194316]
- 161.
- Fletcher TM, Sun D, Salazar M. et al. Effect of DNA secondary structure on human telomerase activity. Biochemistry. 1998;37:5536–5541. [PubMed: 9548937]
- 162.
- Yu G -L, Blackburn EH. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. [PubMed: 1934071]
- 163.
- Lue NF, Peng Y. Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25:4331–4337. [PMC free article: PMC147048] [PubMed: 9336465]
- 164.
- Greene EC, Bednenko J, Shippen DE. Flexible positioning of the telomerase-associated nuclease leads to preferential elimination of nontelomeric DNA. Mol Cell Biol. 1998;18:1544–1552. [PMC free article: PMC108869] [PubMed: 9488471]
- 165.
- Niu H, Xia J, Lue NF. Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol Cell Biol. 2000;20:6806–6815. [PMC free article: PMC86210] [PubMed: 10958677]
- 166.
- Greene EC, Shippen DE. Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes & Dev. 1998;12:2921–2931. [PMC free article: PMC317169] [PubMed: 9744868]
- 167.
- Bednenko J, Melek M, Greene EC. et al. Developmentally regulated initiation of DNA synthesis by telomerase: Evidence for factor-associated de novo telomere formation. EMBO J. 1997;16:2507–2518. [PMC free article: PMC1169850] [PubMed: 9171363]
- 168.
- Petrov AV, Dokudovskaya SS, Sokolov KA. et al. Telomerase from Saccharomyces cerevisiaecontains several protein subunits and may have different activities depending on the protein content. FEBS Lett. 1998;436:35–40. [PubMed: 9771889]
- 169.
- Hardy CD, Schultz CS, Collins K. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J Biol Chem. 2001;276:4863–4871. [PubMed: 11096070]
- Human, Mouse and Yeast Telomerase - Madame Curie Bioscience DatabaseHuman, Mouse and Yeast Telomerase - Madame Curie Bioscience Database
- In Situ Activation of Caspases Revealed by Affinity Labeling Their Enzymatic Sit...In Situ Activation of Caspases Revealed by Affinity Labeling Their Enzymatic Sites - Madame Curie Bioscience Database
- The Origins of Species - Madame Curie Bioscience DatabaseThe Origins of Species - Madame Curie Bioscience Database
- Telomerase Activity as a Marker of Tumor Cell Survival to Evaluate Sensitivity o...Telomerase Activity as a Marker of Tumor Cell Survival to Evaluate Sensitivity of Neoplastic Cells to Cancer Treatment - Madame Curie Bioscience Database
- Nuclear Envelope Dynamics in Drosophila Pronuclear Formation and in Embryos - Ma...Nuclear Envelope Dynamics in Drosophila Pronuclear Formation and in Embryos - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...