NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Coronin is a conserved actin binding protein that promotes cellular processes that rely on rapid remodeling of the actin cytoskeleton, including endocytosis and cell motility. However, the exact mechanism by which coronin contributes to actin dynamics has remained elusive for many years. Here, we integrate observations from many groups and propose a unified model to explain how coronin controls actin dynamics through coordinated effects on Arp2/3 complex and cofilin. At the front end of actin networks, coronin protects new (ATP-rich) filaments from premature disassembly by cofilin and recruits Arp2/3 complex to filament sides, leading to nucleation, branching and network expansion. At the rear of networks, coronin has strikingly different activities, synergizing with cofilin to dismantle old (ADP-rich) filaments. Thus, coronin spatially targets Arp2/3 complex and cofilin to opposite ends of actin networks. The net effect of coronin's activities is acceleration of polarized actin subunit flux through filamentous arrays. This increases actin network plasticity and replenishes the actin monomer pool required for new filament growth.
Introduction
Dynamic remodeling of the actin cytoskeleton generates force and structural organization for diverse physiological processes, such as cell migration, endocytosis, cytokinesis and cell morphogenesis. 1 Optimal plasticity of cellular actin networks is achieved by maintaining actin filaments in a state of constant flux, where the subunits comprising filaments are turned over rapidly. There is a net addition of actin monomers at the available barbed ends of filaments and net loss of subunits from the pointed ends. New filament growth in cells is required for expansion of existing actin networks, construction of new actin arrays and force production, while the rapid disassembly of older filaments is necessary for sustaining network plasticity and replenishing the pool of assembly-competent actin monomers available for new growth. This dynamic flux (‘turnover’) of networks allows them to be reconfigured rapidly in response to spatial and temporal cues.
The growth and remodeling of actin networks in vivo is controlled with exquisite timing and precision through the concerted activities of numerous actin-associated proteins. Key points of control include filament nucleation, elongation, bundling, branching, capping and severing, in addition to monomer sequestration and recycling. Some actin-binding proteins are highly conserved across distant species and are ubiquitously expressed (e.g., cofilin, profilin, Arp2/3 complex and capping protein), thus defining a core set of actin-regulating proteins in eukaryotic cells. Other actin-binding proteins have more specific functions and tailor the properties of actin arrays to the unique requirements of each cell type or organism. While the functions of some core actin-regulating proteins have been studied extensively and their biochemical and cellular functions firmly established, others have only recently begun to be characterized. One of these ubiquitous yet elusive actin-binding proteins is coronin.
In this chapter, we discuss how coronin is thought to influence actin dynamics in cells. Included below are sections describing coronin domains and physical interactions, genetic evidence supporting coronin's importance in the actin cytoskeleton, coronin regulation of Arp2/3 complex-mediated actin assembly, and coronin regulation of coflin-mediated actin disassembly and turnover. We then integrate these functions into a unified model describing the overall effects of coronin on the dynamics of cellular actin networks.
Deconstructing Coronin: Isoforms, Domains and Interactions
Coronin was first identified in actin-myosin preparations isolated from Dictyostelium discoi- deum and was shown to bind directly to F-actin in vitro and colocalize with F-actin structures in vivo.2 Since then, a wide variety of coronins have been characterized. Some model organisms (e.g., Saccharomyces cerevisiae and Schizosaccharomyces pombe) have a single coronin gene, while others (e.g., Dictyostelium discoideum, Drosophila melanogaster and Caenorhabditis elegans) have 2-3 coronin genes and mammals have up to seven different coronin-related genes.3 Most coronins have a characteristic three-part domain layout, consisting of the -propeller domain, followed by a highly variable ‘unique’ segment and a C-terminal coiled-coil domain (Fig. 1).
The β-Propeller Domain
The signature domain of coronin family proteins is the WD-repeat region.4 Crystallization of murine Coronin 1A (lacking its coiled-coil domain) revealed that it forms a seven-bladed β-propeller structure assembled from five canonical WD repeats and two noncanonical repeats (Fig. 1).5 In addition, two tandem stretches of conserved residues located in the C-terminal extension of the WD-repeat region closely associate with the underside of the propeller, possibly providing additional structural integrity. Although in principle β-propeller structures can support multiple protein-protein interactions, in the eighteen years that coronin has been studied only one binding partner of its propeller domain has been identified, F-actin. Binding to F-actin was first demonstrated for Dictyostelium coronin,2 and later this activity was dissected for yeast coronin, where it was shown that an intact propeller domain is suffcient to bind actin filaments.6 More recently, a single conserved actin-binding residue was identified in the mouse Coronin 1B propeller domain.7 The next steps required to gain a deep understanding of coronin-F-actin interactions include: (1) identifying the entire actin-binding footprint on coronin, (2) defining the reciprocal coronin-binding surface(s) on F-actin and (3) determining how coronin binding might influence F-actin conformation and/or nucleotide binding state.
Unique Region
The unique region of coronin is highly variable in length and sequence and its function(s) remains poorly understood. Interestingly, the unique regions of S. cerevisiae Crn1 and D. melano- gaster Dpod1 share noted sequence homology with the microtubule-binding region of mammalian MAP1B and the corresponding purified coronin proteins bind to microtubules and crosslink microtubules and actin filaments in vitro.6,8 Genetic analyses of these two coronins are also consistent with their regulation of microtubule-based cellular functions. In yeast, low penetrance phenotypes (aberrant cytoplasmic microtubules and short cell-cycle delays) suggest that Crn1 may help promote nuclear migration.6,9 This process involves coordinated interactions between polarized actin cables and cytoplasmic microtubules, which are required to translocate the nucleus to the mother-bud neck and orient the pre-anaphase mitotic spindle to ensure faithful segregation of chromosomes. In Drosophila, Dpod1 is required for proper axonal guidance. This process also depends on close physical interactions and regulatory feedback cues between microtubules and cortical actin networks, which are necessary for growth cone steering and navigation.8
Coiled- Coil Domain
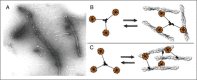
Remarkably, the smallest functional region of coronin, its C-terminal coiled-coil domain (~35-50 residues, 4-7 heptad repeats), mediates at least three different functional interactions (with itself, F-actin and Arp2/3 complex). The first interaction defined for the coiled-coil domain was homo-oligomerization (forming dimers or trimers), which is required for actin filament bundling by coronin (Fig. 2A).6,10-12 This observation has led to the widely accepted model that coronin bundles actin filaments through multimerization of its β-propeller actin binding site domain (Fig. 2B). However, oligomerization has only been demonstrated in solution in the absence of F-actin and thus alternative models for bundling remain possible. For instance, bundling may result from individual (non-oligomerized) coronin molecules utilizing two separate actin-binding sites to crosslink filaments (Fig. 2C). In support of this alternative model, deletion of the coiled-coil domain dramatically weakens the actin binding affinity of coronin.7,13 Although one study reported that the coiled-coil domain alone does not bind F-actin and suggested that the ability of the coiled-coil domain to increase actin-binding affinity is due to avidity (multimerization of the β-propeller domain),7 other studies have detected F-actin binding activity in the coiled-coil domain.13,14 The presence of a second actin binding site in the coiled-coil domain would allow coronin to bundle F-actin by a mechanism using two distinct actin-binding domains (one in the β-propeller, one in the coiled-coil domain). In this model, oligomers of coronin may represent an inactive molecular state, with distribution between oligomeric and non-oligomeric forms possibly being regulated by posttranslational modification of coronin (Fig. 2C; and see Section on Coronin-Arp2/3 Complex).
The coiled-coil domain of coronin also mediates direct interactions with the Arp2/3 complex both in vivo and in vitro.15-17 The specific effects of coronin on Arp2/3 complex activity and how this interaction contributes to the regulation of cellular actin dynamics are addressed in Section on Coronin-Arp2/3 Complex.
What remains unclear is how these three seemingly distinct functional roles of the coiled-coil domain are integrated. Can the coiled-coil domain mediate oligomerization, F-actin binding and/ or Arp2/3 complex binding simultaneously? Are there separable sites/residues in the coiled-coil domain that direct these different interactions? Answering these questions will require detailed structure-function analysis focused on the coiled-coil domain. However, regardless of the mechanism used, it is remarkable that this relatively small domain can serve as a multi-functional platform, whereas the much larger β-propeller domain has so far been implicated in binding only to F-actin.
Table 1 summarizes all of the known direct interactions and effects on actin of purified coronin proteins. From this compilation, it is apparent that F-actin binding and bundling are conserved functions of coronin. Arp2/3 complex and cofilin regulation by coronin may be equally well conserved, but this remains to be determined, as biochemical tests for these activities have been limited to a few studies so far.
Table 1
Biochemical activities of purified coronin proteins.
Life Without Coronin
All coronins examined to date (with the exception of mammalian coronin 7) bind to F-actin in vitro and localize to actin-rich cellular structures, underscoring the conservation and importance of the coronin-F-actin interaction. Genetic disruptions of coronin in S. cerevisiae, D. discoideum, C. elegans, D. melanogaster, X. laevis and mammals have further demonstrated the important roles coronins play in a wide variety of actin-based cellular processes (e.g., phagocytosis, endocytosis, cytokinesis, cell motility) and physiological functions (e.g., early embryonic development and lymphocyte function) (see Fig. 3).17-22,24,26
In S. cerevisiae, coronin colocalizes with F-actin at cortical sites of endocytosis.6,9 Although a deletion of the coronin gene (CRN1) causes no overt phenotypes in endocytosis or actin organization, crn1Δ mutants exhibit specific genetic interactions with other mutations (act1-159, cof1-22 and arp2-21), demonstrating the importance of coronin in regulating the actin cytoskeleton.6 Further, CRN1 overexpression is lethal in yeast cells and causes the formation of aberrant actin loops, which can be suppressed by specific alleles of Arp2/3 complex.6,15 In Dictyostelium, coronin localizes to crown-like cortical projections of cells and deletion of the coronin gene causes approximately 3-fold decreases in endocytosis and cell motility, defects in cytokinesis (which in Dictyostelium depends on efficient cell migration) and defects in phagocytosis.18,19 The C. elegans coronin Pod-1 colocalizes with actin-rich structures found at the cell cortex and a pod-1 gene deletion causes strong defects in anterior-posterior cell polarity and embryonic cell division.21 In D. melanogaster, coronin homozygous mutations (coro-/coro-) are lethal at early to late pupal stages, with about 10% escaping adults. These viable adults show severe defects in legs, wings and eyes, often accompanied by reduced F-actin staining in the affected cells.20 In addition, the Drosophila coronin Dpod1 is enriched in developing axons and is required for normal axonal guidance, possibly due to its above mentioned ability to crosslink actin and microtubules.8 In Xenopus, Xcoronin localizes to the actin-rich cell periphery and expression in cultured A6 Xenopus cells of a dominant negative construct (lacking the coiled-coil domain) causes defects in the formation of lamellipodia and cell spreading.22
In mammalian species, the seven different coronins have distinct expression profiles and/or subcellular localization patterns and possibly different functions (reviewed by ref 3). Genetic analysis is available for three mammalian coronins, including hematopoietic Coronin 1A and the more ubiquitously expressed Coronin 1B and Coronin 1C Coronin 1A localizes to the F-actin rich membrane protrusions of activated T-lymphocytes,23 and mouse coronin1–/– lymphocytes are impaired in cell motility and fail to develop uropods (retractile structures at the rear of the cell).17 In addition, expression of a dominant negative Coronin 1A construct in human neutrophils leads to impaired phagocytosis and reduced cell spreading and adhesion.24,25 Coronin 1B localizes to the leading edge of fibroblasts and RNAi-mediated depletion reduces retrograde actin flow, causes thinner and more densely branched actin filament networks at the leading edge and decreases membrane ruffling and cell motility26 Coronin 1C localizes to lamellipodia and membrane ruffles in fibroblasts and HEK293 cells10 and RNAi-mediated depletion causes defects in wound healing and cell migration.54
Collectively, these genetic observations from diverse organisms point to conserved and important roles for coronin in regulating in vivo actin dynamics. In the sections below, we explore the mechanisms underlying these cellular functions.
Coronin-Arp2/3 Complex: Conditional Inhibition and Recruitment Drive Front-End Assembly
We still have much to learn about the intricacies of how coronin controls actin assembly in cells, but solid footholds have been gained in recent years. One major advance was identifying the Arp2/3 complex as a binding partner of coronin, then determining the activities of purified coronin on Arp2/3 complex and how genetic disruption of this interaction influences in vivo actin dynamics and organization.
The Arp2/3 complex is a seven subunit complex conserved in all eukaryotes. It is composed of two actin-related protein (Arp) components, Arp2 and Arp3 and five non-Arp components, p41/ ARPC1, p34/ARPC2, p21/ARPC3, p20/ARPC4 and p16/ARPC5.27 In all eukaryotic organisms and cell types examined, the Arp2/3 complex localizes to sites of dynamic actin assembly, such as the leading edge of motile fibroblasts, the dynamic cortical projections of Dictyostelium cells and yeast cortical endocytic actin patches.28-30 At these sites, Arp2/3 complex performs two apparently coupled functions, actin filament nucleation and branching.31 Nucleation involves Arp2 and Arp3 forming a pseudo-actin dimer that seeds the polymerization of actin. Branching involves association of the Arp2/3 complex with the side of an existing (mother) actin filament and nucleation of a new (daughter) filament at a 70° angle.32 Alone, the Arp2/3 complex has inherently weak nucleation activity, but can be activated and transformed into a strong nucleator. Activation requires direct binding of Arp2/3 complex to a nucleation-promoting factor (NPF), such as a WASp/SCAR/ WAVE family protein and possibly interaction with the side of a pre-existing mother filament.31 The potent nucleation and branching activities of Arp2/3 complex must be tightly regulated in cells. This is achieved by multiple cell signaling pathways, which converge on WASp/SCAR/WAVE proteins and other NPFs to direct spatial and temporal activation of Arp2/3 complex.
In addition to localized activation by NPFs, Arp2/3 complex is regulated through direct association with coronin. The first clue to coronin-Arp2/3 functional interactions was the observation that Coronin 1A cofractionates with Arp2/3 complex isolated from neutrophil cell lysates through multiple chromatography steps.33 Subsequently, a direct interaction between yeast Crn1 and Arp2/3 complex was reported, which was shown to depend on the Crn1 coiled-coil domain.15 Purified Crn1 directly inhibited the nucleation activity of WASp-stimulated Arp2/3 complex in vitro (Fig. 4A). However, inhibition occurred specifically in the absence of pre-existing actin filaments and was relieved fully by the addition of preformed filaments. In the presence of filaments, full-length Crn1, which has a high-affinity interaction with F-actin,6 recruited Arp2/3 complex to the sides of mother filaments. These observations led to a model suggesting that coronin has two distinct effects on Arp2/3 complex. It inhibits Arp2/3 complex nucleation activity in regions of the cell where pre-existing filaments are sparse, suppressing spontaneous and/or unbranched nucleation events. On the other hand, in regions where filaments are abundant (e.g., leading edge networks), coronin recruits Arp2/3 complex to the sides of existing filaments, thereby promoting rather than inhibiting nucleation and branching (i.e., coronin reinforces the expansion of existing filament networks)15. By acting as a spatial regulator of Arp2/3 complex activity, coronin has a net positive effect on the assembly of Arp2/3-dependent filament networks.
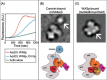
The molecular mechanism for coronin inhibition of Arp2/3 complex was later addressed using electron microscopy and single particle analysis to solve the structures of free and Crn1-bound yeast Arp2/3 complexes.34 Free Arp2/3 complex was evenly distributed among three separate conformations—‘open’, ‘intermediate’ and ‘closed’. Mutational analysis and docking of the crystal structure of inactive Arp2/3 complex showed that the open conformation of the complex is inactive, whereas the closed conformation is active (primed for nucleation). Consistent with these assignments, 100% of Crn1-bound Arp2/3 complexes were in the open (inactive) conformation, whereas 100% of WASp-bound Arp2/3 complexes were in the closed (active) conformation (Fig. 4B and 4C). From these results, it was suggested that Crn1 inhibits Arp2/3 complex by stabilizing the open (inactive) conformation. These effects appear to be mediated by a direct interaction between the coronin coiled-coil domain and the p35/ARPC2 subunit of Arp2/3 complex. Tree separate lines of evidence suggest this: (1) a two-hybrid interaction between the C-terminus of Crn1 and p35/ARPC2 was reported;15 (2) electron micrographs of Crn1-bound Arp2/3 complex revealed a 40 Ã… diameter mass (likely to represent the - -propeller of Crn1) positioned close to p35/ ARPC2;34 (3) point mutations at a conserved solvent-exposed surface on p35/ARPC2 have been shown to abolish Crn1 interactions with Arp2/3 complex (Daugherty and Goode, unpublished data). By interacting with p35/ARPC2, coronin is in a prime position to influence Arp2/3 complex activity. The p35/ARPC2 subunit is part of the structural/functional hub of the complex, making direct contacts with three other subunits (Arp2, Arp3 and p19/ARPC4) and the sides of mother filaments. p35/ARPC2 also plays an essential role in relaying activation signals from WASP (and possibly filament side binding) to other subunits in the complex that lead to actin nucleation.34 What remains unclear and needs to be addressed in future studies is how binding of the coronin coiled-coil domain to p35/ARPC2 affects interactions of this subunit with the sides of mother filaments and affects filament branch formation and turnover.
In vivo regulation of Arp2/3 complex by yeast coronin was suggested by two separate genetic observations. Combining crn1Δ and arp2-21 mutations caused synthetic defects in cell growth and overexpression of Crn1 was lethal and led to the formation of aberrant actin loop structures decorated with Arp2/3 complex. It was suggested that these defects arise from coronin mis-regulation of Arp2/3 complex, because they are suppressed by specific alleles of p35/Arc35 or by deleting the Arp2/3-interacting coiled-coil domain of Crn1.15
More recently, it has been found that coronin-Arp2/3 complex interactions and inhibitory effects are conserved in mammals.16,17,25,26 These studies have provided important new insights into how Coronin-Arp2/3 complex interactions affect the assembly of cellular actin networks and actin-based processes such as cell motility and phagocytosis. One study showed that mouse Coronin 1B coimmunoprecipitates with Arp2/3 complex and that this interaction is important for motility.16 Further, RNAi-mediated depletion of Coronin 1B in Rat2 fibroblast cells decreased lamellipodial advancement and protrusion persistence, leading to a decrease in whole-cell motility.26 Interestingly, the barbed end zone at the leading edge of the motile cells, which normally grows very rapidly, was significantly narrowed after Coronin 1B depletion. This suggests that loss of cororin may have caused a reduction in barbed end generation at the leading edge, consistent with the model described above for spatial control of Arp2/3 complex regulation by coronin. In another study, it was suggested that mouse Coronin 1A controls steady-state F-actin levels via an Arp2/3 complex-dependent mechanism in T-lymphocytes.17 Phalloidin staining revealed that F-actin levels are elevated in cororin1–/– T-cells. Further, expression of wild type Coronin 1A in the mutant cells restored F-actin levels back to the wild type state, but expression of Coronin 1A mutants impaired in Arp2/3 complex interactions (see below) failed to restore F-actin levels. The implication of these observations is that interactions between Coronin 1A and Arp2/3 complex are important for maintaining the F-actin equilibrium in T-cells. However, the truncation made in coronin for this study not only disrupts binding to Arp2/3 complex, but also deletes an important actin binding domain that influences cofilin activity (see below). Thus, it is equally plausible that disruption of coronin and cofilin synergy by this mutant leads to elevated steady state F-actin.
Coronin-Arp2/3 complex interactions also can be regulated by posttranslational modification. Protein kinase C (PKC) phosphorylation of Serine 2 on Coronin 1B weakens coronin-Arp2/3 complex interactions and reduces the speed of fibroblast migration in single cell tracking assays, but has no effect on Coronin 1B localization.16 Further, a phospho-mimetic allele of Coronin 1B (S2D) shows weakened interactions with Arp2/3 complex, whereas a nonphosphorylatable Coronin 1B mutant (S2A) shows strengthened interactions with Arp2/3 complex. Coronin 1A interactions with Arp2/3 complex may also be regulated by coronin phosphorylation. This is suggested by the observation that wild type Coronin 1A reverses the elevated F-actin levels in cororin1–/– T-cells, while Coronin 1A (S2D, or lacking coiled-coil domain) mutants that fail to interact with Arp2/3 complex are unable to do so. It is not yet clear how a phosphorylated residue located in the coronin β-propeller domain affects Arp2/3 complex interactions that are mediated by the more distal coiled-coil domain. However, one possibility is that the β-propeller domain, when phosphorylated, binds to the coiled-coil domain to disrupt interactions with Arp2/3 com- plex. It also waits to be seen whether other coronins are phosphorylated at or near Serine 2 and this regulatory mechanism is conserved. There is strong evidence for coronin phosphorylation in other cell types and organisms. Two-dimensional gel analysis suggests that Coronin 1C expressed in HEK293 fibroblasts is phosphorylated.10 In murine macrophages, the biochemical distribution of Coronin 1A between ~200 kDa and ~400-600 kDa complexes is regulated by PI-3 kinase.35 The Golgi membrane associated fraction of human Coronin 7 expressed in NIH 3T3 fibroblasts is phosphorylated at tyrosine residues.56 Further, yeast Crn1 is phosphorylated at multiple sites by Pho85/Cdk5 kinase (C. Humphries and B. Andrews, personal communication) and coronin and Cdk5 have been copurified with ubiquitin ligase Mib1 from neuronal postsynaptic densities.36
In summary, the coronin-Arp2/3 complex interaction is highly conserved across distantly related species and depends on a direct physical interaction between the coronin coiled-coil domain and a conserved surface on the p35/ARPC2 subunit of Arp2/3 complex. This interac- tion is regulated by coronin phosphorylation at Serine 2 in mammalian cells. Binding of coronin stabilizes the open (inactive) conformation of Arp2/3 complex, suppressing actin nucleation until these inhibitory effects are overridden by association of coronin: Arp2/3 with pre-existing filaments. Coronin actually assists in recruiting Arp2/3 complex to the sides of pre-existing fila- ments, thereby promoting actin nucleation and branching. Thus, coronin has the unique ability to spatially control Arp2/3 complex activity, selectively promoting the growth and expansion of existing networks. How coronin may affect actin filament branch turnover and Arp2/3 complex recycling remains to be determined.
Coronin Influence on Cofilin Activity: Protect the Front, Dismantle the Rear?
While actin nucleation represents one key control point in determining the dynamic be- havior of cellular actin networks, an equally important point of control is filament disassembly. Only by maintaining actin polymers in a state of rapid turnover can cells maintain a pool of assembly-competent actin subunits for new growth and reorganize their networks rapidly in response to signals. Replenishment of subunits is accelerated by cellular factors that selectively destabilize and depolymerize the older (ADP-bound) filaments in networks. Cofilin (also called ADF) plays a central role in this process and recently it has emerged that coronin assists cofilin in driving these events.
Cofilins are a widely conserved family of proteins that accelerate actin network disassembly and are required for dynamic actin-based processes, including cell motility, endocytosis and cytokinesis.37 Cofilin binds to the sides of actin filaments in a cooperative manner and increases the twist of filaments, leading to filament severing and disassembly. Cofilin promotes filament disassembly in concert with several other conserved actin-binding proteins, each of which makes a mechanistically distinct contribution to turnover. These include actin-interacting protein-1 (Aip1),38-40 cyclase-associated protein (CAP),41-44 twinfilin45,46 and now coronin.
The first clue to the coronin-cofilin functional connection came from genetic interaction studies in yeast, where combining a crn1Δ mutation with a hypomorphic cofilin allele (cof1-22) caused synthetic defects in cell growth and actin organization.6 Similar genetic interactions were observed between crn1Δ and act1-159, an allele of actin with decreased rates of actin turnover. More recently, it was found that crn1Δ causes a 4-fold reduction in rate of actin filament turnover in cells and that purified Crn1 synergizes biochemically with cofilin in severing and disassembling actin filaments.13 This functional interaction between coronin and cofilin appears to be conserved in mammals, as Brieher et al47 biochemically isolated mammalian Coronin 1A and Aip1 as cellular factors required (together with cofilin) to promote the rapid disassembly of Listeria actin tails and sustain Listeria motility. Together, Coronin 1A and Aip1 accelerated cofilin-mediated disassembly of tails by ~10-fold, with Coronin 1A contributing about ~3-fold to this effect. These observations also agree with an earlier report showing that coronin and cofilin are abundant components of isolated Listeria tails.48
Further in vivo evidence for coronin function in promoting F-actin disassembly comes from a study on T-lymphocytes, where Coronin 1A genetic disruption led to an increase in F-actin levels and a concomitant decrease in G-actin levels.17 In addition, mammalian Coronin 1B has been shown to regulate cofilin activity in migrating fibroblasts by recruiting Slingshot phosphatase to the lamellipodium. Phosphorylated (inactive) cofilin is then dephosphorylated (activated) by Slingshot, leading to filament severing.26 Accordingly, cells depleted of Coronin 1B by RNAi have higher levels of phospho-cofilin and decreased rates of retrograde actin flow and cell motility. Although Slingshot is not conserved in S. cerevisiae and C. elegans, it is possible that other phosphatases perform this function and analogously control coronin and/or cofilin activities in these organisms.
While a conserved cellular function for coronin in regulating cofilin-dependent actin disassembly is clear, the mechanism underlying these effects is only just emerging. To describe the current understanding of coronin mechanism in actin disassembly, it is beneficial to first consider the separate effects of its β-propeller and coiled-coil domains on cofilin activity and then combine this knowledge to arrive at a more complete picture of this mechanism.
Recent analyses using purified proteins have demonstrated that a fragment of yeast Crn1 that includes the β-propeller domain but lacks the coiled-coil domain biochemically synergizes with cofilin in severing and disassembling actin.13 The same fragment of Crn1 genetically complements loss of CRN1 in a cof1-22 background, suggesting that the coronin-cofilin functional synergy occurs in vivo. Further, this synergy depends on direct binding of the Crn1 β—propeller to F-actin, an interaction that alters the twist of actin filaments, possibly predisposing them for cofilin binding (B. Goode and A. McGough, unpublished data). A second possibility, not mutually exclusive from the first, is that coronin more directly recruits cofilin to actin filaments. While there is no evidence available to suggest a direct physical interaction between coronin and cofilin in solution, it remains possible that they associate when bound to actin filaments.
In contrast to the stimulatory effects of the β-propeller domain of Crn1 on cofilin activity, the coiled-coil domain of Crn1 inhibits filament binding and severing by cofilin.13 Moreover, full-length yeast and mammalian coronin proteins (which include the coiled-coil domain) inhibit filament binding and severing by coronin.7,13 Thus, the coiled-coil domain is both required and sufficient for cofilin inhibition and the inhibitory effects of the coiled-coil domain appear to dominate over the positive synergistic effects of the β-propeller domain with cofilin. Mechanistically, the inhibitory effect of the coiled-coil domain stems from its ability to bind F-actin and competitively displace cofilin from filaments.13 These effects may be conserved, because F-actin-binding activity has been reported for a coiled-coil domain containing fragment of mammalian Coronin 1A.14 In addition, full-length mammalian Coronin 1B competitively displaces cofilin from F-actin and reciprocally, cofilin binding to F-actin displaces Coronin 1B from filaments.7 Thus, not only does the coiled-coil domain function to directly regulate Arp2/3 complex, it also protects actin filaments from the effects of cofilin.
How can the observation that full-length yeast Crn1 and mammalian Coronin 1B inhibit coflin in vitro be reconciled with genetic observations showing that both of these coronins increase (rather than decrease) rates of cofilin-mediated actin turnover in vivo? Further, how can biochemical inhibition by these two coronins be reconciled with data from Brieher et al47 showing that Coronin 1A enhances (rather than inhibits) cofilin-mediated disassembly of Listeria tails? We postulate that coronin has highly distinct effects on older (ADP-bound) versus newer (ATP-bound) actin filaments. Specifically, presense of the coiled-coil domain enables cororin to bind to and selectively protect ATP-rich F-actin from cofilin, thus protecting newly assembled actin filaments at the front end of networks. In contrast, ADP-rich filaments at the rear of networks are not protected by coronin and thus are vulnerable to cofilin attack. By this mechanism, nucleotide-dependent interactions of cororin with F-actin would switch coronin's influence on cofilin from prohibitive to stimulatory. This model is supported by a recent study showing that full-length Coronin 1B binds to ATP/ADP+Pi-F-actin with 47 times greater afnity compared to ADP-F-actin.7 It is also consistent with the ADP-F-actin binding affinity of full-length Coronin 1B (Kd = 8 μM) matching the ATP-F-actin binding affinity of a truncated Crn1 lacking the coiled-coil domain. Suggesting that the coiled-coil domain does not contribute substantially to ADP-F-actin binding.13 Thus, filaments comprised of ADP-actin would be rapidly dismantled by the synergistic effects of the coronin β-propeller domain and cofilin. These unique abilities of coronin to act differentially on new versus old filaments would intensify the inherent binding preference of cofilin for ADP-F-actin compared to ATP-actin55 and sharpen the contrast in polarized behavior between actin dynamics at the front and the rear of networks.
Such a model would also explain some of the observations from Brieher et al47 that had been seemingly at odds with observations from other studies. They showed that Coronin 1A enhances (rather than inhibits) cofilin-mediated actin disassembly. We suggest that this difference is due to the substrate used for actin disassembly, Listeria tails, which are likely to have large regions of ADP-actin at the rear of the tails, where the coronin-cofilin synergy was observed. Further, Brieher et al47 observed that Coronin 1A substantially (4-5 fold) increased cofilin recruitment to Listeria tails, but only modestly (1.5-fold) increased cofilin recruitment to purified F-actin. Again, this difference may be explained by Listeria tails being richer in ADP-actin compared to purified actin filaments. Consistent with this possibility, a close examination of the data in this study suggests that cofilin is recruited preferentially to one end of the Listeria tail (Fig. 6B in Brieher et al47), presumably the ADP-actin-rich trailing end.
An Integrated Working Model for Coronin Mechanism and Function
In the previous two sections, we have described how coronin regulates Arp2/3 complex-mediated actin assembly and cofilin-mediated actin disassembly. How are these apparently separate functions of coronin coordinated spatially and temporally and what is the integrated effect of these two activities on actin networks ? A working model is presented in Figure 5. At the front end of the actin network, coronin binds with high affinity to the ATP/ADP+Pi-rich actin filaments using two separate actin-binding sites (in the β-propeller domain and coiled-coil domain). These interactions recruit Arp2/3 complex to the sides of filaments to promote nucleation and branching. Although coronin inhibits Arp2/3 complex activity in the absence of filaments (perhaps suppressing nucleation in cytoskeleton-free regions of the cytoplasm), it actually recruits Arp2/3 complex to preexisting filaments, where coronin inhibition is overridden and nucleation and branching proceed. A second important effect of coronin binding to ATP-F-actin at the front end of the network is protection from cofilin attack. Binding of the coiled-coil domain of coronin to F-actin blocks the ability of cofilin to bind and sever ATP-actin filaments. Thus, the net effect of coronin activities at the leading edge is protection of newly formed actin filaments from cofilin and enhancement of Arp2/3 complex-mediated assembly and expansion of branched networks.
In this model, the effects of coronin at the rear of the actin network are strikingly different. Filaments are rich in ADP-actin, which dramatically weakens coronin binding to F-actin. The coiled-coil domain no longer inhibits cofilin attack, allowing the β-propeller to recruit cofilin to F-actin and synergize with cofilin in severing and depolymerizing filaments. In this manner, coronin and cofilin selectively target older ADP-rich actin filaments for demolition and thereby promote a high rate of polarized actin network turnover. As filaments are being disassembled, Arp2/3 complex and coronin dissociate and then diffuse to the front end of the network where they are employed for new rounds of actin assembly and branching. This model is consistent with the treadmilling model for branched actin arrays, where Arp2/3 complex-dependent nucleation occurs at the front of the lamellipodia and cofilin-dependent depolymerization of filaments occurs at the rear.49
For simplicity, we have not included in the model how the changing phosphorylation states of coronin and cofilin infuence the mechanism. However, the studies of Bear and coworkers show convincingly that coronin interactions with Arp2/3 complex are governed by phosphorylation and that coronin recruitment of Slingshot phosphatase leads to cofilin-mediated F-actin disassembly.16,26 Thus, it will be intriguing to define the precise timing and order of coronin and cofilin phosphory- lation and dephosphorylation events and how they relate to the inherent differential affnity and biochemical effects of coronin and cofilin on new versus old actin filaments.
Conclusions and Perspective
In a cellular milieu of actin-binding proteins, where each makes a highly specific contribution to actin dynamics, coronin is somewhat unique in its ability to influence two separate and crucial control points, actin assembly and disassembly. These seemingly distinct aspects of actin regulation are actually tightly interwoven. While actin polymerization provides the force and directionality for many cellular processes (e.g., endocytosis, cytokinesis and cell motility), actin disassembly is required to an equal extent for replenishing the pool of monomers used for new rounds of filament assembly and allowing responsive and dynamic reorganization of networks. Coronin has properties that allow it to coordinate events in both actin assembly and disassembly.
We have integrated the observations from many groups and proposed a unified model to explain how coronin might regulate actin dynamics in a wide variety of organisms and cell types. This model suggests that coronin controls activities of the Arp2/3 complex and cofilin, spatially directing them to opposite ends of filament networks. This would have an overall effect of driving polar flux of actin subunits through filamentous networks, thereby increasing actin dynamics, consistent with genetic and biochemical analyses of many different coronins. Importantly, this is only one of many possible models, and still requires rigorous experimental testing. While it is appealing to suggest that coronins from diverse organisms share a common mechanism for controlling actin dynamics, it is also possible that their mechanisms and functions have diverged. Resolving this issue will require new biochemical studies on coronins from different species and characterization of their in vivo binding partners and functional cofactors.
There are also many other open questions and unresolved mysteries surrounding coronin function that require future attention. It is uncertain if or how coronins that lack a coiled-coil domain (e.g., Pod-1) affect cytoskeletal dynamics and/or organization, as there are only limited reports on their biochemical and cellular activities. Microtubule functions suggested for some coronins also remain poorly defined and it is unclear how interactions with microtubules and actin are coordinated. In addition, we have only a limited view of how coronin is regulated by phosphorylation. Recently, important breakthroughs were made on this front illustrating how phosphorylation of coronin can affect its interactions with Arp2/3 complex. This likely represents only the tip of the iceberg on coronin phospho-regulation and thus more intense future investigations are needed to define coronin post-translational modifications and effects. Last, there is the question of how diverse mammalian coronin isoforms are targeted to different subcellular locations (e.g., lamellipodia, stress fibers and Golgi) and whether they have distinct roles in controlling different actin structures at these locations.
Acknowledgements
We are indebted to Melissa Chesarone, Karen Daugherty, Eugeno de Hostos, Amy Grace DuPage, Avital Rodal and Elsie Yu for comments on the chapter and valuable discussions. This work was supported in part by a grant from the NIH to B.G. (GM063691). Owing to space restrictions, we were unable to include all published works on coronin and we apologize to the authors of papers omitted.
References
- 1.
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. [PubMed: 12600310]
- 2.
- de Hostos EL, Bradtke B, Lottspeich F. et al. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta subunits. EMBO J. 1991;10(13):4097–4104. [PMC free article: PMC453159] [PubMed: 1661669]
- 3.
- Uetrecht AC, Bear JE. Coronins: the return of the crown. Trends Cell Biol. 2006;16(8):421–426. [PubMed: 16806932]
- 4.
- de Hostos EL. The coronin family of actin-associated proteins. Trends Cell Biol. 1999;9(9):345–350. [PubMed: 10461187]
- 5.
- Appleton BA, Wu P, Wiesmann C. The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes. Structure. 2006;14(1):87–96. [PubMed: 16407068]
- 6.
- Goode BL, Wong JJ, Butty AC. et al. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J Cell Biol. 1999;144(1):83–98. [PMC free article: PMC2148128] [PubMed: 9885246]
- 7.
- Cai L, Makhov AM, Bear JE. F-actin binding is essential for coronin 1B function in vivo. J Cell Sci. 2007;120(Pt 10):1779–1790. [PubMed: 17456547]
- 8.
- Rothenberg ME, Rogers SL, Vale RD. et al. Drosophila pod-1 crosslinks both actin and microtubules and controls the targeting of axons. Neuron. 2003;39(5):779–791. [PubMed: 12948445]
- 9.
- Heil-Chapdelaine RA, Tran NK, Cooper JA. The role of Saccharomyces cerevisiae coronin in the actin and microtubule cytoskeletons. Curr Biol. 1998;8(23):1281–1284. [PubMed: 9822583]
- 10.
- Spoerl Z, Stumpf M, Noegel AA. et al. Oligomerization, F-actin interaction and membrane association of the ubiquitous mammalian coronin 3 are mediated by its carboxyl terminus. J Biol Chem. 2002;277(50):48858–48867. [PubMed: 12377779]
- 11.
- Oku T, Itoh S, Ishii R. et al. Homotypic dimerization of the actin-binding protein p57/coronin-1 mediated by a leucine zipper motif in the C-terminal region. Biochem J. 2005;387(Pt 2):325–331. [PMC free article: PMC1134960] [PubMed: 15601263]
- 12.
- Gatfield J, Albrecht I, Zanolari B. et al. Association of the leukocyte plasma membrane with the actin cytoskeleton through coiled coil-mediated trimeric coronin 1 molecules. Mol Biol Cell. 2005;16(6):2786–2798. [PMC free article: PMC1142424] [PubMed: 15800061]
- 13.
- Gandhi M, Goode BL. Coronin promotes in vivo turnover of actin networks by nucleotide-sensitive regulation of cofilin effects on F-actin. submitted.
- 14.
- Liu CZ, Chen Y, Sui SF. The identification of a new actin-binding region in p57. Cell Res. 2006;16(1):106–112. [PubMed: 16467882]
- 15.
- Humphries CL, Balcer HI, D’Agostino JL. et al. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol 23. 2002;159(6):993–1004. [PMC free article: PMC2173993] [PubMed: 12499356]
- 16.
- Cai L, Holoweckyj N, Schaller MD. et al. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J Biol Chem. 2005;280(36):31913–31923. [PubMed: 16027158]
- 17.
- Foger N, Rangell L, Danilenko DM. et al. Requirement for coronin 1 in T-lymphocyte trafficking and cellular homeostasis. Science. 2006;313(5788):839–842. [PubMed: 16902139]
- 18.
- de Hostos EL, Rehfuess C, Bradtke B. et al. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J Cell Biol. 1993;120(1):163–173. [PMC free article: PMC2119478] [PubMed: 8380174]
- 19.
- Maniak M, Rauchenberger R, Albrecht R. et al. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell. 1995;83(6):915–924. [PubMed: 8521515]
- 20.
- Bharathi V, Pallavi SK, Bajpai R. et al. Genetic characterization of the Drosophila homologue of coronin. J Cell Sci. 2004;117(Pt 10):1911–1922. [PubMed: 15090595]
- 21.
- Rappleye CA, Paredez AR, Smith CW. et al. The coronin-like protein POD-1 is required for anterior-posterior axis formation and cellular architecture in the nematode caenorhabditis elegans. Genes Dev. 1999;13(21):2838–2851. [PMC free article: PMC317117] [PubMed: 10557211]
- 22.
- Mishima M, Nishida E. Coronin localizes to leading edges and is involved in cell spreading and lamellipodium extension in vertebrate cells. J Cell Sci. 1999;112(Pt 17):2833–2842. [PubMed: 10444378]
- 23.
- Nal B, Carroll P, Mohr E. et al. Coronin-1 expression in T-lymphocytes: insights into protein function during T-cell development and activation. Int Immunol. 2004;16(2):231–240. [PubMed: 14734608]
- 24.
- Yan M, Collins RF, Grinstein S. et al. Coronin-1 function is required for phagosome formation. Mol Biol Cell. 2005;16(7):3077–3087. [PMC free article: PMC1165393] [PubMed: 15829569]
- 25.
- Yan M, Di Ciano-Oliveira C, Grinstein S. et al. Coronin function is required for chemotaxis and phagocytosis in human neutrophils. J Immunol. 2007;178(9):5769–5778. [PubMed: 17442961]
- 26.
- Cai L, Marshall TW, Uetrecht AC. et al. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128(5):915–929. [PMC free article: PMC2630706] [PubMed: 17350576]
- 27.
- Machesky LM, Gould KL. The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol. 1999;11(1):117–121. [PubMed: 10047519]
- 28.
- Welch MD, DePace AH, Verma S. et al. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138(2):375–384. [PMC free article: PMC2138188] [PubMed: 9230079]
- 29.
- Winter D, Podtelejnikov AV, Mann M. et al. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7(7):519–529. [PubMed: 9210376]
- 30.
- Insall R, Muller-Taubenberger A, Machesky L. et al. Dynamics of the Dictyostelium Arp2/3 complex in endocytosis, cytokinesis and chemotaxis. Cell Motil Cytoskeleton. 2001;50(3):115–128. [PubMed: 11807934]
- 31.
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7(10):713–726. [PubMed: 16990851]
- 32.
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95(11):6181–6186. [PMC free article: PMC27619] [PubMed: 9600938]
- 33.
- Machesky LM, Reeves E, Wientjes F. et al. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 1997;328(Pt 1):105–112. [PMC free article: PMC1218893] [PubMed: 9359840]
- 34.
- Rodal AA, Sokolova O, Robins DB. et al. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12(1):26–31. [PubMed: 15592479]
- 35.
- Didichenko SA, Segal AW, Thelen M. Evidence for a pool of coronin in mammalian cells that is sensitive to PI 3-kinase. FEBS Lett. 2000;485(2-3):147–152. [PubMed: 11094157]
- 36.
- Choe EA, Liao L, Zhou JY. et al. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci. 2007;27(35):9503–9512. [PMC free article: PMC6673137] [PubMed: 17728463]
- 37.
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. [PubMed: 10611961]
- 38.
- Aizawa H, Katadae M, Maruya M. et al. Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells. 1999;4(6):311–324. [PubMed: 10421841]
- 39.
- Okada K, Obinata T, Abe H. XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J Cell Sci. 1999;112 (Pt 10):1553–1565. [PubMed: 10212149]
- 40.
- Rodal AA, Tetreault JW, Lappalainen P. et al. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145(6):1251–1264. [PMC free article: PMC2133144] [PubMed: 10366597]
- 41.
- Moriyama K, Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J Cell Sci. 2002;115(Pt 8):1591–1601. [PubMed: 11950878]
- 42.
- Balcer HI, Goodman AL, Rodal AA. et al. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin and Aip1. Curr Biol. 2003;13(24):2159–2169. [PubMed: 14680631]
- 43.
- Bertling E, Hotulainen P, Mattila PK. et al. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol Biol Cell. 2004;15(5):2324–2334. [PMC free article: PMC404026] [PubMed: 15004221]
- 44.
- Mattila PK, Q uintero-Monzon O, Kugler J. et al. A high-affinity interaction with ADP-actin monomers underlies the mechanism and in vivo function of Srv2/cyclase-associated protein. Mol Biol Cell. 2004;15(11):5158–5171. [PMC free article: PMC524793] [PubMed: 15356265]
- 45.
- Goode BL, Drubin DG, Lappalainen P. Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J Cell Biol. 1998;142(3):723–733. [PMC free article: PMC2148182] [PubMed: 9700161]
- 46.
- Moseley JB, Okada K, Balcer HI. et al. Twinfilin is an actin-filament-severing protein and promotes rapid turnover of actin structures in vivo. J Cell Sci. 2006;119(Pt 8):1547–1557. [PubMed: 16569665]
- 47.
- Brieher WM, Kueh HY, Ballif BA. et al. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin and Aip1. J Cell Biol. 2006;175(2):315–324. [PMC free article: PMC2064572] [PubMed: 17060499]
- 48.
- David V, Gouin E, Troys MV. et al. Identification of cofilin, coronin, Rac and capZ in actin tails using a Listeria affinity approach. J Cell Sci. 1998;111(Pt 19):2877–2884. [PubMed: 9730980]
- 49.
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145(5):1009–1026. [PMC free article: PMC2133125] [PubMed: 10352018]
- 50.
- Suzuki K, Nishihata J, Arai Y. et al. Molecular cloning of a novel actin-binding protein, p57, with a WD repeat and a leucine zipper motif. FEBS Lett. 1995;364(3):283–288. [PubMed: 7758584]
- 51.
- Oku T, Itoh S, Okano M. et al. Two regions responsible for the actin binding of p57, a mammalian coronin family actin-binding protein. Biol Pharm Bull. 2003;26(4):409–416. [PubMed: 12673016]
- 52.
- Nakamura T, Takeuchi K, Muraoka S. et al. A neurally enriched coronin-like protein, ClipinC, is a novel candidate for an actin cytoskeleton-cortical membrane-linking protein. J Biol Chem. 1999;274(19):13322–13327. [PubMed: 10224093]
- 53.
- Kammerer RA, Kostrewa D, Progias P. et al. A conserved trimerization motif controls the topology of short coiled coils. Proc Natl Acad Sci USA. 2005;102(39):13891–13896. [PMC free article: PMC1236532] [PubMed: 16172398]
- 54.
- Rosentreter A, Hofmann A, Xavier CP. et al. Coronin 3 involvement in F-actin-dependent processes at the cell cortex. Exp Cell Res. 2007;313(5):878–895. [PubMed: 17274980]
- 55.
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274(22):15538–15546. [PubMed: 10336448]
- 56.
- Rybakin V, Stumpf M, Schulze A. et al. Coronin 7, the mammalian POD-1 homologue, localizes to the Golgi apparatus. FEBS Lett. 2004;573(1-3):161–167. [PubMed: 15327992]
- Introduction
- Deconstructing Coronin: Isoforms, Domains and Interactions
- Life Without Coronin
- Coronin-Arp2/3 Complex: Conditional Inhibition and Recruitment Drive Front-End Assembly
- Coronin Influence on Cofilin Activity: Protect the Front, Dismantle the Rear?
- An Integrated Working Model for Coronin Mechanism and Function
- Conclusions and Perspective
- Acknowledgements
- References
- Coronin: The Double-Edged Sword of Actin Dynamics - Madame Curie Bioscience Data...Coronin: The Double-Edged Sword of Actin Dynamics - Madame Curie Bioscience Database
- PARP and the Release of Apoptosis-Inducing Factor from Mitochondria - Madame Cur...PARP and the Release of Apoptosis-Inducing Factor from Mitochondria - Madame Curie Bioscience Database
- THE RNA INFRASTRUCTURE: An Introduction to ncRNA Networks - Madame Curie Bioscie...THE RNA INFRASTRUCTURE: An Introduction to ncRNA Networks - Madame Curie Bioscience Database
- The Tn4371 ICE Family of Bacterial Mobile Genetic Elements - Madame Curie Biosci...The Tn4371 ICE Family of Bacterial Mobile Genetic Elements - Madame Curie Bioscience Database
- Idiopathic Parkinson's Disease: Staging an α-Synucleinopathy with a Predictable ...Idiopathic Parkinson's Disease: Staging an α-Synucleinopathy with a Predictable Pathoanatomy - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...