NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Chemically altered nucleosides derived from canonical ribo-or deoxyribonucleoside-derivatives of adenosine, cytosine, guanosine, and uridine or thymidine are found in all types of nucleic acids, DNA and RNA. They are particularly abundant in noncoding RNAs, such as transfer RNAs and ribosomal RNA of higher organisms. By increasing the structural diversity of nucleic acids, modified nucleosides play important roles in gene expression and in regulating many aspects of RNA functions. They also contribute to nucleic acid stability and to protection of genetic materials against virus aggression. In this chapter we present a historical overview of the discovery, occurrence, and diversity of the many naturally occurring modified nucleosides that are present in both DNA and RNA of diverse organisms. We also briefly describe the different enzymes that accomplish these nucleic acid ‘decorations’. More information about the structure, function, biosynthesis and evolutionary aspects of selected modified nucleosides in DNA and RNA and their corresponding modification enzymes can be found elsewhere in this volume.
Origin of Nucleic Acids Research
Discovery of Deoxyribonucleic Acid (DNA)
Friedrich Miescher discovered an unknown compound, later identified as chromatin, in 1869. He extracted a gelatinous material from various cells (initially human pus), and discovered it contained much inorganic phosphorus. This newly identified biochemical material was named ‘nuclein’ because it was always associated with what the histologists designated nuclei. During the period 1885-1900, it was discovered that beside phosphorus, ‘nuclein’ was also rich in a carbohydrate (later identified as a deoxypentose) and in the organic bases adenine, thymine, guanine, and cytosine. The linear structure of the purified organo-phosphate polymer was finally solved by Phoebus Levene (period 1909-1929). At that time, the DNA polymer was thought to be the scaffold of some important elements within the chromatin. No connection was made between this ‘boring long polymer with only four types of nucleotides’ and the molecular basis of transmission of hereditary characteristics that geneticists were eagerly seeking.
Detailed study of polymeric DNA began in 1928 when Fred Griffith suspected that a “genetic transforming principle” was associated with the ‘nuclein’. However, it was only in 1944 that Oswald Avery and his research group,1 using almost-pure DNA from Streptococcus cell extracts, inferred that DNA contains genetic information. It took another year before Avery demonstrated that the transforming activity disappeared after DNAse treatment.2 Then the race to identify the detailed chemical structure of the ‘genetic’ DNA really started. First, Rollin Hotchkiss3 confirmed the genetic nature of DNA, while Erwin Chargaff4 discovered that adenine with thymine and guanine with cytosine always exist in a 1:1 ratio, although the ratio of G+C/A+T varies from species to species. Based on these crucial observations, together with the very first crystallographic data of DNA fibers obtained by Rosalind Franklin working in the laboratory of Maurice Wilkins, and based on competitor Linus Pauling’s suggestion that DNA could have an helical shape, Francis Crick and James Watson proposed in 1953 the double helix structure of DNA,5,6 which revolutionized our concept of the transmission of genetic characters. Next came the identification and purification of the first DNA restriction enzymes7,8 that recognize a defined sequence in DNA and cut it specifically. Together with the invention of techniques for DNA sequencing,9,10 these advances allowed the development of recombinant DNA technology11 and opened the field of modern molecular biology.12
Discovery of Ribonucleic Acids (RNAs)
It was not until later in the 20th century that scientists realized there are two types of nucleic acids, DNA and RNA, the latter involving ribose instead of deoxyribose and uridine (or pseudouridine, the ‘fifth ribonucleoside’—see below) instead of thymidine. The reason was that little attention was given to the presence of RNases, and any ‘RNA’ identified in cell extracts was just a mix of degradation products a few nucleotides long.13,14 Degradation of DNA by metal-dependent DNases was easier to avoid. Thus while DNA research was progressing well, the chemistry of the second type of nucleic acid (RNA) remained obscure until the 1950s. Only after introducing detergent (as for DNA preparation15), associated with phenol for purification, were the first long RNA polymers finally identified in 1956-58 (ribosomal RNA16 and ‘soluble’ RNA—now called transfer RNA17).
Wide interest in these new types of nucleic acids emerged only after Crick hypoth esized in 1955 (but published only in 1958) that an RNA molecule should be the inter mediate between DNA and proteins (known as the ‘RNA adaptor hypothesis18), and later on advanced the ‘Wobble hypothesis’ for decoding mRNA.19 Initially, Crick thought the adaptor molecules might be the small RNA molecules that were known to be present in cell extracts, until the ‘soluble’ RNAs (tRNAs), able to be specifically aminoacylated,20 were identified and characterized in 1958. The concept of messenger RNA and regulatory mechanisms in the synthesis of proteins was formulated in 1961 (refs. 21, 22). The genetic code was finally solved and officially presented during a Cold Spring Harbor Symposium23 in 1966. In the meantime (1965-67) the first fully sequenced tRNAs specific for alanine,24 tyrosine,25 serine26 and phenylalanine,27 all from yeast were fully sequenced. These sequences included the identification and location of no less than 17 different noncanonical nucleosides, among them two hypermodified nucleosides N6-isopentenyladenosine (i6A) and wyosine (yW). The first crystals of tRNAs were produced and the first three dimensional structure of one of them28 was finally solved in 1974. This was the birth of structural biology of the nucleic acids.
Discovery of Noncanonical Nucleosides
Modified Nucleosides in Genomic DNAs
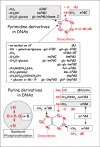
During the period 1920-45, naturally occurring nucleic acid polymers (DNA and RNA) were thought to contain only four canonical nucleosides (ribo-or deoxy-derivatives): adenosine, cytosine, guanosine, and uridine or thymidine. However, after analyzing a picrate precipitate from a hydrolysate of DNA of avian tubercule bacilli, Johnson and Coghill29 detected a minor amount of a methylated cytosine derivative (m5dC, Fig. 1). This report was later disputed by Vischer et al30 because they could not reproduce the result, but Johnson and Coghill were in fact correct. Only in 1948 was the presence of m5dC in DNA from calf thymus31,32 firmly established using the new technique of paper chromatography of DNA hydrolysates.31,32 This was followed in 1958 by the detection of N6-methyl adenine (m6dA) in microbial DNA.33 It was not until much later—1964—that the methylation of cytosines and adenosines within DNA molecules was shown to occur by enzymatic post-replicative modification (see below the section concerning enzymes). A surprise discovery during the period 1953-63 was that the DNA of some bacterial viruses lacks deoxycytosine (dC) or deoxythymidine (dT) and instead contains 5-hydroxymethyldeoxycytosine34 (hm5dC), 5-hydroxymethyldeoxyuridine35 (hm5dU) or simply deoxyuridine36 (dU). These modified cytosines or thymidines (100% replacing of the standard base dC or dT completely), unlike the m5dC and m6dA in bacterial and mammalian DNA, are generated at the precursor level (prereplicative modification) and subsequently incorporated into phage DNA by the bacteriophage polymerase.37 However, the hm5dC in phage DNA can be further glucosylated at the polymer level38,39 by direct transfer of glucose from UDP-glucose to form hexosylated derivatives glc-hm5dC / glc-hm5dU and even di-glucosylhydroxy methyl deoxy uridine glc-glc-hm5dC. Note that a minor amount of hm5dU and glc-hm5dU (also designated Base J) have been found recently in genomic DNA of flagellated protozoa of the order Kinetoplastida (Trypanosoma brucei for example) and in the closely related unicellular alga Euglena gracilis (see chapter by Sabatini et al). In this case hydroxylation of deoxyribothymine and the subsequent glycosylation step occur at the polymer level. Later (1972-81) came the discovery of new uridine derivatives containing putrescinyl-, glutamyl-or dihydroxpentyl groups linked to C5 of the uracil ring (symbolized by Put-m5dU, Glu-m5dU and Dhp5dU respectively40,41). In the case of Dhp5dU, glucose or gluconolactone-1-phosphate can be further attached on one of the two free hydroxyl groups leading to hypermodified Glc-Dhp5dU and GlcP-Dhp5dU, respectively. In these latter cases, depending on the type of chemical alteration, the extent of replacement of the canonical dT by modified dU derivatives was estimated to be 15-60%. In E. coli phage Mu, a substantial number of adenines were found modified to N6-carbamoylmethyl adenine (ncm6A, 15% of dA), while in S. elongatus phage S-2L 100% of adenines are methylated to 2-aminoadenine42 (m2A) or N2-N6-dimethyladenine43 (m2,6A). Also, in the phage DDV1 infecting Shigella sonnei, a trace amount of 7-methylguanine (m7dG, about 1% of dG) was found. In contrast, other types of noncanonical modified deoxynucleosides would most probably be identified were more bacterial and phage DNAs to be explored—an endeavor that unfortunately has been much neglected in the past decade (discussed in the chapter by Forterre and Grosjean). Quite recently (1983-87), N4-methylcytosine (m4dC) and also deoxyinosine (dl) were identified in some bacterial DNA,44-46 especially from thermophiles. The selective advantage of m4dC over m5dC at high temperatures is thought to be to avoid production of mutagenic m5dU resulting from heat-induced deamination of m5dC, and m4dC is indeed more resistant to deamination at high temperature than m5dC (discussed in ref. 47). A surprising recent discovery (2005) is that the phosphoryl group in bacterial DNA can be thiolated to form a phosphoro thionate linkage of the Sp chiral configuration;48,49 the mechanism remains to be elucidated (commented by Eckstein50).
In conclusion, so far relatively few naturally occurring modified deoxynucleosides have been identified in genomic DNAs (summarized in Fig. 1). The most common modifications are simple methylation of either the C5 atom of the cytidine ring (m5dC in almost all kinds of organisms) or the exocyclic amine groups of adenine (m6dA mainly in bacteria and archaea) or cytidine (m4dC mainly in thermophilic bacteria and Archaea). Unusual, deoxynucleosides (sometimes hypermodified) are confined to bacteriophages and viruses (reviewed in refs. 40-41).
Modified Nucleosides in Coding and Noncoding RNAs
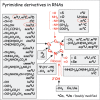
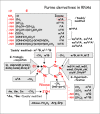
In the case of RNA, the story is very different and far more complex. In contrast to DNA, we now know that every position of a pyrimidine or a pyrimidine ring (Figs. 2 and 3 respectively) can be posttranscript ionally modified, not only by methylation or hydroxymethylation, but also by deamination, transglycosyl ation, acetyl ation, reduction, thiolation, oxidation, ribosylation, formylation, isomerization, selenat ion, or multiple group additions or transfer …. singly or sequentially (Fig. 2). Moreover, the 2’-hydroxyl group of the ribose moiety can be methylated (alone or in combin ation with base modific ations) or ribosylated with a bulky adenosine-5’-phosphate group. To date 110 - 119 (depending on how certain ‘hypermodified’ modified nucleosides are considered) naturally occurring modified nucleosides have been identified in different types of RNAs, not only tRNAs and rRNAs, but also mRNAs and snRNAs like sn/snoRNAs, miRNAs, and chromosomal RNAs. The most widespread RNA modifications are base or ribose methylations (symbolized by mX or Xm respectively) and isomerization of uridine into pseudouridine (Psi). The majority of hypermodified ribonucleosides occur in transfer RNAs; these modifications include long lateral chains or multiple substituents on two or more atoms of the same purine or pyrimidine ring (see below).
How was this vast body of information on the identity and location of the many modified nucleosides in RNA acquired? The story starts only in 1951, after the discovery of m5dC in DNA, when W. Cohn and his colleagues51 used paper chromatography of an acid hydrolyzate of enriched ‘soluble RNA’ of yeast to identify a new compound in addition to the four expected ribonucleosides. This compound, initially designated by a question mark ‘?’, was shown later in 1957-58 to be 5-ribosyluridine, also called the ‘fifth nucleoside in RNA’ (now designated pseudouridine52,53). Pseudo uridine accounts for about 4% of the molecular weight of the total constituent nucleosides in yeast tRNAs and is the most abundant modified nucleoside identified so far in all kinds of tRNA as well as rRNA. The next modifications to be identified (about one year later) were 2’-O-methylribose derivatives (Xm = Cm, Um, Gm, Am; these are the second most abundant class.54,55 Also identified in the same period were 5-methylribouridine (m5U, ribothymine or ribo-T, one of which occurs in almost every tRNA), 5-methylribocytosine (m5C, also abundant in mammal ian tRNAs, up to 2-3 per tRNA) and a few other simple methylated adenine and guanine derivatives (m1A, m2G, m1G, m7G…; refs. 56-59). Mainly from sequence information of tRNAs from yeast, E. coli and mammals, many other modified nucleosides were identified, including N6-isopentenyladenosine (i6A and its variant ms2i6A), wybutosine (yW—see chapter by Urbonavicius et al), and N6-carbamoylthreonineadenosine (t6A). By 1970, thus just 20 years after the discovery of Psi in RNAs, 35 well-characterized modified nucleosides had been identified compared to only five in DNA (reviewed in the book by RH Hall,60 the only one available to date dealing with RNA and DNA modifications. For details concerning chemical structures, occurrence and classification of modified ribonucleosides, as well as on metabolic pathways, and enzymes catalyzing RNA modification reactions, consult the MODOMICS database at http://modomics.genesilico.pl; see also Appendix 1 by Rother et al and additional Web links in Appendix 6 in this volume. Other useful sources of information are in references 61-64.
Degree, Extent, and Pattern of Nucleoside Modifications
A modified nucleoside at a given position within a population of RNA molecules may not be present in all of them so that the molar ratio (or % proportion) of a given nucleoside in a population of RNA molecules (referred to as the degree of modification) can be less than 1/1 (less than 100% modified) at a given site. The degree of modification may vary according to the physiological conditions (oxygen concentration, temperature, availability of metabolic intermediates or cofactors, metabolic stress, malignancy…) of the cell from which the RNA came, thus creating a micro-heterogeneity in the RNA population (‘modivariants’; see chapter by Giegé and Lapointe). In some cases, modivariants can be separated by simple chromatographic procedures. For example, the molar ratio of ribothymine (m5U, ribo-T) at position 54 in the T-Psi loop of all types of tRNAs is usually 1/1 (100% U-54 methylated), while the molar ratio of thiolation on C2 of the ring of the same uridine-54 in the tRNA of thermophiles (harboring m5s2U instead of m5U, as in Thermus thermophilus—see chapter by Noma et al) can be less than 1/1 or even zero, especially when the organism is grown at temperatures below that optimal for growth. It is important to remember that in the RNA modification data banks (tRNA, rRNA, snRNA) the presence of a given modified nucleotide at a given position of an RNA molecule is indicated (m5s2U as in the example above), but never the degree of modification of that particular base or ribose. This caveat is particularly relevant for ribosomal RNA, where for instance the degree of modification of a particular Psi or 2’-O-methylribose (Xm) can be very low. Since the DNA genome in principle exists in only one copy per cell, the notion of degree of modification does not apply to DNA. However in some microorganisms this is not the case: Synechococcus for example has about 10 copies of the chromosome, while in certain hyperthermophilic and halophilic archaea, this number can be as high as 20 copies of the chromosome. Then the notion of degree of methylation should apply.
The extent of nucleic acid modification concerns the relative amount of a given modified nucleoside that exists at several positions within a given RNA or a DNA molecule, usually expressed as % replacement of total nucleosides (or total of a particular canonical one) in the whole nucleic acid molecule. For example, the extent of post-replicative modification of dC into m5dC for bacterial, archaeal, and eukaryal genomic DNA is generally 1-8 % of the total dC, except for mammalian and plant DNAs where m5dC can reach 30% of total dC. In phage DNAs, where modifications arise by a prereplicative event, the extent of modification can reach 100%. The extent of total modifications in tRNA molecules from plant and mammals is also high (up to 25%), whereas that in homologous tRNAs from bacteria is lower (2-15%—reviewed in refs. 40, 60).
The pattern of modifications in RNA/DNA is a more complex, qualitative concept. Here, comparison of different nucleic acids is made by taking into the account type, location and diversity of modifications which of course differ greatly from one type of nucleic acid to another (for example DNA versus RNA, or rRNA versus tRNA or mRNA). More interesting is that distinct and characteristic patterns of modification exist between homologous nucleic acids from phylogenetically distant organisms (see Fig. 1), as well as between tRNAs of the same organism (see below). The pattern of modification is the ‘fingerprint’ or ‘identity card’ of a RNA molecule, in the same way that a restriction pattern is the ‘fingerprint’ or ‘identity card’ of a DNA molecule. As more sequences of RNAs themselves (not the sequence of their genes or RT-PCR products) become available, this important feature of nucleic acids will become more evident.
Distribution of Modified Nucleosides in the Three Domains of Life
Nucleosides Found in Coding and Noncoding RNAs
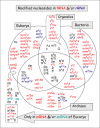
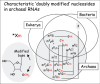
Figure 4, shows the symbols of 107 structurally distinct modified ribo nucleo sides identified so far in different RNAs from various Eukarya, Bacteria, or Archaea.65 The information comes primarily from RNA sequence data and from analysis of RNA nucleoside composition by thin-layer chromatography, high performance liquid chromatography and/or mass spectrometry (for examples see refs. 66-70). Symbols indicated in normal characters (in red in the version on the Web Site of this chapter), initalics, bold (in blue) or in normal charaters, underlined (in black) correspond to modified nucleosides found in tRNAs, rRNAs, or in both t+rRNAs, respectively. Organelle (mitochondrial and chloroplastic) tRNAs and rRNAs contain their own set of modified nucleosides, some of which (like cmnm5U, k2C, τm5U, τm5s2U, f5C, f5Cm) are not present in cytoplasmic RNAs of the eukaryotic host cell. The corresponding modification enzymes, now encoded in the host genome, are believed to have originated from ancient bacterial endosymbionts. Therefore, while present in the Eukaryal domain, mitochondrial-modified nucleosides should be considered as ‘bacterial by origin’ or at least belonging to both Eukarya and Bacteria (they are boxed in the intersector E-B in Fig. 4). Symbols of modified nucleosides outside the circles correspond to those found in eukaryal mRNAs (normal characters, in red) and snRNAs (italics, bold, in green) or in both mRNAs and snRNAs (italics, underlined, in black). Five members of this eukaryal group are unique to mRNAs and/or snRNAs, while others are also present in eukaryal tRNAs and/or rRNAs (indicate by an arrow).
Figure 4 shows that half of the modified ribonucleosides are domain specific. These presumably arose later during more than evolution, after the separation of organisms into the three domains. About one fifth of the other modified nucleosides are located within overlapping sectors of the circles and thus found in two or more domains: either between Eukarya and Bacteria (E+B), or Bacteria and Archaea (B+A) or Archaea and Eukarya (E+A). The remaining fifth of modified nucleosides are present in all kinds of organisms (E+B+A). They are the simplest types of modification, several are found in all types of RNAs. From this observation, it has been inferred71 that they correspond to relics of modified nucleosides that were present in primordial organisms existing before the three biological domains separated. However, the reality may not be so simple. Symbols like m1G or m5U within the central common sector E+B+A correspond to modifications that are located in different positions and in different types of RNA molecules, each of them being produced by site-specific as well as RNA-specific enzymes that do not necessarily belong to the same protein family. Some cases most probably represent convergent rather than divergent evolution, so that the evolutionary history of the emergence of RNA modification machinery is complex (see for examples refs. 72,73, also chapters by Czerwoniec et al, by Myllykallio et al and by Forterre and Grosjean).
Concerning doubly modified nucleosides of the type xNm (like m2Gm or ac4Cm), a majority of them were found so far in archaeal RNAs. They correspond in fact to combinations of simple methylation of the ribose (Gm or Cm) and of enzymatic alteration of the base (m2G or ac4C), each of the ‘independent’ modific at ions being found within the three overlapping E+B+A sectors, or in the E+A sectors (Fig. 5). Thus, while modified nucleosides like xNm’s are indeed found mainly in archaeal RNAs, the corresponding modification enzymes may not necessarily be unique to archaea.
Lastly, modifications like imG, imG2, mimG, yW, OHyW, o2yW and OHyW*, or preQo, preQ1, Q, oQ, gluQ, manQ, GalQ and G+, or nm5U, or cmnm5U, mnm5U are merely intermediates of the same phylogenetically related stepwise metabolic reaction chain (see chapters by Urbonavicius et al for wyosine derivatives, by Iwata-Reuyl and de Crécy-Lagard for queuosine derivatives, and by Bessho and Yokoyama for the modified uridines series). Consequently, the real diversity of naturally occurring modified nucleosides as it appears in Figure 4 could probably be reduced from 107 to about half truly distinct, biosynthetically unrelated types of chemical structures in RNAs. However, exploring more RNAs, especially from extremophiles, may uncover new types of modified nucleoside.
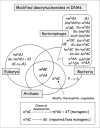
Nucleosides Found in Genomic DNAs
Figure 6 summarizes the types of modified deoxyribonucleosides found in DNA from different origins. Symbols for modified deoxynucleosides found in genomic DNA of a cell, or in DNA of bacteriophages and eukaryal viruses, are indicated in different types of boxes. In the cases of cellular genomic DNA, almost all (if not all) modified deoxynucleosides are formed by post-replicative enzymatic modification processes; their extent ranges from 1% to 30% (refs. 40,60). In viruses, on the other hand, modified deoxynucleosides are derived either by post-replicative modification processes or via incorporation of modified deoxynucleotide precursors directly into DNA by the virus DNA-dependent DNA polymerase (prereplicative process). In this later case the extent of DNA modification can reach 100%. In the case of viruses and bacteriophages, only few modified deoxynucleosides are found to be common (m5dC, m6dA and hm5dU, there are also those found in genomic DNA of bacteria or Eukarya (reviewed in refs. 40,41).
The Case of Transfer and Ribosomal RNAs
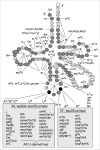
To date (January 2009) more than six hundreds of tRNAs from more than one hundred different organisms of the three domains of life (with strong bias for bacterial tRNAs) have been sequenced, and the type and location of each individual naturally occurring modified nucleoside have been identified (see ref. 74 and http://trnadb.bioinf.uni-leipzig.de). Figure 7 summarizes the available information in one cumulative ‘tRNA modification map’. As can be seen, a large number of nucleotides in tRNAs can be enzymatically altered in many different ways, the most common modification being pseudouridine. Independent maps for Eubacteria, Archaea, protists, animals, plants, mitochondria and chloroplasts, are available in reference 75 (not updated since 1995, but nevertheless still useful). As a rule, tRNAs from eukaryotes (and plants) are more heavily modified than the homologous tRNAs from Eubacteria. Transfer RNAs from organelles and parasitic organisms like Mollicutes76 are those for which the extent of modification is the lowest (1-6 %). Only 60 archaeal tRNAs have been sequenced so far (majority from halophiles), so it is hard to generalize about them. However, analysis of the base composition of bulk tRNAs from several hyperthermophilic organisms indicates that they are heavily modified and are rich in stabilizing 2’-O-methylated nucleosides (reviewed in ref. 77), while for tRNAs of halophiles,78 where there is a compensatory stabilizing effect of high salt concentration in the cytoplasm, the extent of modification is rather low.
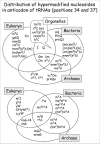
Some modified nucleosides, like m5U (ribo-T) and Psi located at positions 54 and 55 of the so-called T-Psi loop, are almost ubiquitous in all kinds of tRNAs. They usually correspond to modified nucleosides whose function is to stabilize the 3D-core of the nucleic acid. Other modified nucleosides are unique to a given tRNA isoacceptor, like the wyosine derivatives found exclusively at position 37 of eukaryal and archaeal tRNA-Phe (see chapter by Urbonavicius et al) or lysidine (k2C) present in all bacterial and most organelle. They are generally located in the tRNA anticodon loop, whose function is to decode the genetic information in mRNAs. Note that the distribution of the modified nucleosides of the anticodon loop is clearly ‘domain specific’ (Fig. 8). Among them, 5’-substituted hypermodified uridines of the type Xo5(s2)U(m) and Xm5(s2)U(m) involved in decoding the two-codon boxes (discussed in chapters by Bessho and Yokoyama and by Weixelbaumer and Murphy) are the most diversified. These modified nucleosides are genuine ‘signatures’ of the origin of tRNA; this applies also to certain anticodon base modifications (essentially for the wobble base of anticodon) identified in the ‘endosymbiotic’ mitochondrial and chloroplastic tRNAs.
The same kind of analysis can be performed with ribosomal RNAs of the small and large subunits (refs. 62-64 and http://biochem.umass.edu/fournierlab/3dmodmap). Much less information about the types and locations of modified nucleosides is available for rRNAs than for tRNAs. However, from what is known, rRNAs from eukaryotes are much more heavily modified than their homologs in bacteria (commented in ref. 76). Concerning archaeal rRNAs, the only ones that have been carefully investigated are those from the halophile Haloferax volcanii and the closely related Haloarcula marismortui (refs. 63,64,78), which are not representative of the whole domain of Archaea. Only pseudouridine and 2’-O-methylation of various archaeal rRNAs (as well as of tRNAs) are being currently studied because of their special interest to RNA-guide machineries.79,80
RNA and DNA Modification Enzymes
Discovery of RNA Modification Enzymes
The first evidence for existence of enzymes able to modify nucleic acids at the polymer level came in 1962-63. After incubating transfer RNA with E. coli cell extract and S-AdoMet labeled in the methyl group, three groups81-83 demonstrated independently that radioact ivity appeared in methylated bases in RNA. The first identified modification enzyme82 was tRNA:m5U54 methyltransferase, now designated TrmA in Bacteria and Trm2 in Eukarya. Soon after followed the discovery of similar activities for other methyl transfers specific for the formation of m1G, m7G, m2A, m6A, m2,2G and m5C in E. coli transfer RNAs84 and four additional distinct activities for the formation of m6A, m6,6A, m7G and m5C in ribosomal RNA.85 These simple but important experiments illuminated a new feature of RNA metabolism, namely that methyl group incorporation can take place after polymerization and not, as was shown earlier (1958) for bacteriophage DNA, by incorp oration of deoxyribonucleotide triphosphate analogs (such as m5dCTP) during replication.37 Since the pioneering work on RNA methylation, all subsequent modifications identified in RNAs of many different types of cells have been found to occur the same way, i.e., by enzymatic posttranscriptional alteration of a base and/or of the ribose at the RNA precursor level. Many other RNA processing enzymes catalyzing reactions as diverse as 5’-and 3’-trimming, 5’-capping, RNA-splicing, CCA and polyA addition likewise act posttranscriptionally (RNA maturation process, see chapter by Hall and Li). The precise interplay of these various types of RNA alterations allows in fine to produce fully mature RNAs with many new chemical ‘decorations’ as described in preceding paragraphs.
Later in 1975, a completely different type of RNA methyltransferase using 5,10-methylene tetrahydrofolate (CH2-THF) instead of S-AdoMet as methyl donor was discovered in Streptococcus faecalis (ref. 86, and chapter by Myllykallio et al). Thus while S-AdoMet is by far the major cellular source of methyl groups (and is often called the ‘universal methyl donor’), an alternative solution exists for methylating RNAs.
An enzyme catalyzing the insertion of a guanine in tRNA (via a transglycosylation reaction)87 was identified in rabbit erythrocytes 1973-75. It was only few years later that the physiological function of this ‘G’-inserting enzyme was discovered:88-89 the insertion of a deazaguanine derivative in the anticodon of few selected tRNAs. This enzyme, now designated tRNA-guanine-34 insertase (abbreviated tgt), removes the encoded guanine located at the first position of the anticodon of precursor tRNA by cleaving the canonical C1-N1 glycosidic bond and inserting in its place a premodified 7-deazaguanosine derivative precursor (or a guanine as in the original observation of Farkas and coworkers;87 see chapter by Iwata-Reuyl and de Crécy-Lagard). A similar type of enzyme was recently found in Archaea (in 1997). In this case,90 formation of archaeosine (G+, another type of deazaguanine derivative) at position 15 in the D-loop of archaeal tRNAs depends on a similar, phylogenetically related tRNA-guanine-15 insertase designated a-tgt. It should be mentioned that formation of pseudouridine in RNA proceeds by a similar mechanism, except that it is the genetically encoded uracil base that is replaced in RNA after a 180° rotation and reformation of a noncanonical C1-C5 glycosidic bond (cis-transglycosylation or isomerization reaction—see chapter by Mueller and Ferre d’Amare).
Another remarkable recent discovery (1996-97) is that some RNA modification enzymes are ‘guided by RNA’. This was first demonstrated in the case of enzymatic form ation of 2’-O-methyl ribose in yeast and mammalian rRNAs,91,92 immediately followed by the same discovery in the case of Psi formation also in rRNAs.93,94 This observation has since been extended to the formation of 2’-O-methylribose and Psi in many other RNAs (tRNAs, snrRNAs, snoRNAs) of Eukarya and/or Archaea; however, neither bacteria, nor organelles examined so far use this ‘RNA-assisted’ type of enzyme, in fact ‘RNA-assisted’ multiprotein enzymatic complex (see chapters by Gagnon et al, by Grozdanov and Meier and by Karijolich et al). Note that a given ribose methylation or uridine isomerization in RNA can be mediated by a ‘classical’ all–protein enzyme in one organism, while in another organism the same modification is catalyzed by the RNA-assisted multiprotein machinery (see for examples refs. 95-99). This observation raises interesting questions about the evolutionary pressures that favour one type of RNA modification system over the other. Perhaps the main advantage for a cell using an ‘RNA-assisted’ enzyme machinery instead of an ‘only protein’ enzyme is that, with only few proteins (besides the enzyme) required for elaborating the RNA-guided RNA machineries, and with a huge array of guide RNAs (of which the sequence is more versatile than that of proteins) many more nucleosides in RNAs can be targeted. However, this might not be the sole advantage (discussed in chapters by Gagnon et al, by Grozdanov and Meier and by Karijolich et al).
Discovery of DNA Modification Enzymes
At almost the same time as tRNA:m5U54 methyltransferase was discovered (1963), however before the first sequence-specific restriction enzyme was identified8,12 (and the importance of restriction/modification self-defence mechanism in bacteria was recognized), enzymatic activities for ‘post-replicative’ methylation in polymeric DNA were beginning to be identified.100 Partially purified S-AdoMet-dependent methyltransferases of E. coli were shown to catalyze the formation of m5dC and m6dA in double-stranded DNA.101 Similar enzymes were subsequently identified in many other types of bacterial and eukaryotic cells, as well as in certain bacteriophages (reviewed in refs. 40,41; and chapters by Coffin et al; by Cheng and Blumenthal and by Jeltsch and Jurkowski). Enzymatic post-replicative DNA glucosylation (in fact formation of hyper-modified glucopyranosyloxymethyluracil, base J) was discovered with DNA of bacteriophages38,39,102 before it was found in eukaryotic DNA103,104 (see chapter by Sabatini et al). In the 1980s, enzymes catalyzing formation of m4dC in bacteria were discovered,44 and also a new family of demethyl/dealkyl-methylases acting on both RNA and DNA (AlkB family of enzymes—see chapter by Falnes et al). Another family of dual-enzymes exists, catalyzing the conversion of C-to-U in single-stranded DNA or RNAs and cellular mRNAs (Apobec deaminases—see chapter by Smith). These deaminases play an essential role in cellular defense against viruses and allow new opportunities for variability in gene expression
Conclusion and Future Prospects
DNA and RNA are key cellular polymers in all organisms. To fulfil their multiple functions, these molecules need more than just four canonical nucleosides. To date more than one hundred of chemically distinct noncanonical modified nucleosides have been identified in nucleic acids of many different organisms of the three domains of life (although mesophilic free-living bacteria and viruses have received most attention). The majority of these modified nucleosides occur in RNAs, especially tRNAs. However, the organisms that have been explored represent only a tiny fraction of extant terrestrial taxa. The analysis of nucleic acids of more organisms, especially of the many types of extremophiles (often Archaea) is consequently very likely to reveal additional peculiar ‘decorations’ of nucleic acids. Another limitation is the type of RNA species that can be examined. Some, such as mRNA, sn(o)RNA, microRNA, and viral RNA, are hard to isolate in sufficient amounts for unambiguous identification of their modified nucleoside content (see however refs. 105-109). Hopefully, technical developments, including a new generation of very sensitive mass spectrometers, will help the identification of new modified (deoxy)ribonucleosides, their fine structures, and most importantly their distributions (pattern of modification/ identity card) among many different nucleic acids (RNA and DNA) of the three domains of life.
To account for the many different modified (deoxy)ribonucleosides identified so far in different types of nucleic acids, a correspondingly large number of different enzymes with distinct specificities must exist. Already 130 RNA-modification enzymes are catalogued in MODOMICS (end 2008). They correspond to more than one hundred distinct types of chemical reactions, most of which are S-AdoMet-dependent methylations of a base or a base already modified, or the 2’-hydroxyl of ribose (see Appendix 1 by Rother et al). In the case of DNA-modification enzymes, due to their considerable interests (and commercial values) in relation to restriction/modification process, the few DNA-methyltransferases from many different organisms have been characterized, purified and studied (see Appendix 1 by Rother et al). The number of identified RNA or DNA modification enzymes is increasing very fast, and within the next decade we might reasonably expect it to double or triple. How many different DNA/RNA modification enzymes exist in a given cell is still difficult to estimate, and of course, how many such enzymes exist in all types of living organisms is impossible to predict.
Nowadays, we have techniques that allow identification and characterization of both genes and corresponding modification enzymes. The enzymes can be produced in recombinant form and studied in vitro to identify their mechanism and specificity, as well as their crystal structure. The next challenges will be to understand how all these enzymatic activities are coordinated/regulated in the cell, where each individual reaction occurs within the cellular milieu, how enzymes are organized in complexes with other proteins of the nucleic acid maturation process, how post-replicative and post-transcriptional processes emerged and diversified within each of the three domains of life, and—most importantly—what are the functions of these entire ‘dam’ modified nucleosides in RNA and DNA. Nucleic acids are emphatically not ‘boring long polymers of only four nucleotides’. The purpose of this book is precisely to respond, at least in part, to the important questions that they raise.
Acknowledgements
HG is Emeritus Scientist at University of Paris-XI in Orsay, working in the laboratory of Professor Jean-Pierre Rousset who is acknowledged for his kind hospitality. I deeply acknowledge critical reading of this manuscript by Prof. Andrew Hanson (University Florida, Gainesville) and by Kristian Rother (Laboratory of Bioinformatics, Warsaw).
References
- 1.
- Avery OT, MacLeod CM, McCarthy M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med. 1944;79:137–158. [PMC free article: PMC2135445] [PubMed: 19871359]
- 2.
- McCarthy M, Avery OT. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: II. Effect of deoxyribonuclease on the biological activity of the transforming substance. J Exp Med. 1946;83:89–96. [PubMed: 21010046]
- 3.
- Hotchkiss RD, Marmur J. Double marker transformation as evidence of linked factors in deoxyribonucleate transforming agents. Proc. Natl Acad Sci USA. 1954;40:55–60. [PMC free article: PMC527940] [PubMed: 16589434]
- 4.
- Chargaff E. Structure and function of nucleic acids as cell constituents. Fed Proc. 1951;10:654–659. [PubMed: 14887699]
- 5.
- Watson JD, Crick FHC. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. [PubMed: 13054692]
- 6.
- Watson JD, Crick FHC. General implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. [PubMed: 13063483]
- 7.
- Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. [PubMed: 4868368]
- 8.
- Smith HO, Wilcox KW. A restriction enzyme from hemophilus-influenza. I. Purification and general properties J Mol Biol 197051379–391. See also the paper by Danna K, Nathans D. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. Proc Natl Acad Sci 1971; 68:2913-2917. [PMC free article: PMC389558] [PubMed: 4332003]
- 9.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;4:5463–5467. [PMC free article: PMC431765] [PubMed: 271968]
- 10.
- Maxam A, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. [PMC free article: PMC392330] [PubMed: 265521]
- 11.
- Berg P. Dissections and reconstructions of genes and chromosomes. Science. 1981;213:296–303. [PubMed: 6264595]
- 12.
- Roberts RJ. How restriction enzymes became the workhorses of molecular biology. Proc Natl Sci USA. 2005;102:5905–5908. [PMC free article: PMC1087929] [PubMed: 15840723]
- 13.
- Singh H, Lane BG. The separation, estimation and characterization of alkali-stable derived from commercial ribonucleate preparation. Can J Biochem. 1964;42:87–93.
- 14.
- Holley RW, Apgar J, Merrill SH. Evidence for the liberation of a nuclease from human fingers. J Biol Chem. 1961;236:PC42. [PubMed: 13715349]
- 15.
- Marko AM, Butler GC. The isolation of sodium deoxyribonucleate with sodium dodecyl sulphate. J Biol Chem. 1951;190:165–176. [PubMed: 14841162]
- 16.
- Colter JS, Brown RA. Preparation of nucleic acids from Ehrlich ascites tumor cells. Science. 1956;123:1077–1078. [PubMed: 13380422]
- 17.
- Hoagland MB, Stephenson ML, Scott JF. et al. A soluble RNA intermediate in Protein Synthesis. J Biol Chem. 1958;231:241–257. [PubMed: 13538965]
- 18.
- Crick FHC. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed: 13580867]
- 19.
- Crick FHC. Codon-Anticodon pairing: The Wobble hypothesis. J Mol Bio. 1966;19:184–191. [PubMed: 5969078]
- 20.
- Hoagland MB, Stephenson ML, Scott JF. et al. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958;231:241–257. [PubMed: 13538965]
- 21.
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. [PubMed: 13718526]
- 22.
- Brenner S, Jacob F, Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961;190:576–581. [PubMed: 20446365]
- 23.
- The Genetic Code. Vol 31. Cold Spring Harbor Symp Quant Biol. 1966 [PubMed: 5237191]
- 24.
- Holley RW, Apgar J, Everett GA. et al. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. [PubMed: 14263761]
- 25.
- Madison JT, Everett GA, Kung H. Nucleotide sequence of a yeast tyrosine tRNA. Science. 1966;153:531–534. [PubMed: 5938777]
- 26.
- Zachau HG, Dutting D, Feldman H. Nucleotidsequenzen zweier serin spezifischer tRNA. Angew Chem. 1966;78:392–393.
- 27.
- RajBhandary UL, Chang SH, Stuart A. et al. Studies on polynucleotide: The primary structure of yeast phenylalanine tRNA. Proc. Natl Acad Sci. 1967;57:751–758. [PMC free article: PMC335572] [PubMed: 16591527]
- 28a.
- Kim SH, Suddath FL, Quigley GJ. et al. Three-dimensional tertiary structure of yeast tRNA. Science. 1974;185:435–440. [PubMed: 4601792]
- 28b.
- Robertus JD, Ladner JE, Finch JT. et al. Structure of yeast phenylalanine tRNA at 3 angstroms resolution. Nature. 1974;250:546–551. [PubMed: 4602655]
- 29.
- Johnson TB, Coghill RD. The discovery of 5-methyl-cytosine in tuberculinic acid, the nucleic acid of the Tubercle bacillus. J Am Chem Soc. 1925;47:2838–2844.
- 30.
- Vischer E, Zamenhof S, Chargaff E. Microbial nucleic acids: the desoxypentose nucleic acids of avian tubercle bacilli and yeast. J Biol Chem. 1949;177:429–438. [PubMed: 18107446]
- 31.
- Hotchkiss RD. The quantitative separation of purines, pyrimidines and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–332. [PubMed: 18873306]
- 32.
- Wyatt GR. Occurrence of 5-methylcytosine in nucleic acids. Nature. 1950;166:237–238. [PubMed: 15439258]
- 33.
- Dunn DB, Smith JD. The occurrence of 6-methylaminopurine in microbial deoxyribonucleic acids Nature London 1955175336–339. and Biochem J 1958; 68:627-636. [PubMed: 14389251]
- 34.
- Wyatt GR, Cohen SS. The base of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953;55:774–782. [PMC free article: PMC1269533] [PubMed: 13115372]
- 35.
- Kallen RG, Simon M, Marmur J. The occurrence of a new pyrimidine base replacing thymine in a bacteriophage DNA: 5-hydroxymethyluracil. J Mol Biol. 1962;5:248–250. [PubMed: 13961966]
- 36.
- Takahashi I, Marmur J. Replacement of thymidylic acid by deoxyurydilic acid in the DNA of a transducing phage for B. subtilis. Nature. 1963;197:794–795. [PubMed: 13980287]
- 37.
- Bessman MJ, Lehman IR, Adler J. et al. Enzymatic synthesis of DNA. 3. The incorporation of pyrimidine and purine analogues into DNA. Proc Natl Acad Sci USA. 1958;44:633–640. [PMC free article: PMC528637] [PubMed: 16590253]
- 38.
- Lehman IR, Pratt EA. On the structure of the glucosylated hydroymethylcytosine nucleotides of coliphages T2, T4 and T6. J Biol Chem. 1960;235:3254–3259. [PubMed: 13760441]
- 39.
- Takahashi I, Marmur J. Glucosylated DNA from a transducing phage for B. subtilis. Biochem Biophys Res Commun. 1963;10:289–292. [PubMed: 13980286]
- 40.
- Warren RAJ. Modified bases in bacteriophage DNAs. Ann. Rev. Microbiol. 1980;34:137–158 (review). [PubMed: 7002022]
- 41.
- Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. FASEB J. 1995;9:1034–1042 (review). [PubMed: 7649402]
- 42.
- Kirnos MD, Khudyakov IY, Alexandruschkina NI. et al. 2-aminoadenine in an adenine substituting for a base in S-2L cyanophage DNA. Nature. 1977:369–370. [PubMed: 413053]
- 43.
- Khudyakov IY, Kirnos MD, Alexandrushkina NI. et al. Cyanophage S-2L contains DNA with 2,6-diaminopurine substituted for adenine. Virology. 1978;88:8–18. [PubMed: 676082]
- 44.
- Janulaitis A, Klimasauskas S, Petrusyte M. et al. Cytosine modification in DNA by BcnI methylase yields N4-methylcytosine. FEBS Lett. 1983;161:131–134. [PubMed: 6884523]
- 45.
- Ehrlich M, Gama-Sosa MA, Carreira LH. et al. DNA methylation in thermophilic bacteria: N4-methylcytosine and N6-methyladenine. Nucl Acids Res. 1985;13:1399–1412. [PMC free article: PMC341080] [PubMed: 4000939]
- 46.
- Ehrlich M, Wilson GG, Kuo KC. et al. N4-methylcytosine as a minor base in bacterial DNA. J Bact. 1987;169:939–943. [PMC free article: PMC211883] [PubMed: 3029036]
- 47.
- Grosjean H, Oshima T. 2007. How nucleic acids cope with high temperature. In: Gerday C, Glansdorff N, eds. Physiology and Biochemistry of Extremophiles. Washington, DC: ASM Press; pp. 39–56.
- 48.
- Zhou X, He X, Liang J. et al. A novel DNA modification by sulphur. Mol Microbiol. 2005;57:1428–1438. [PubMed: 16102010]
- 49.
- Wang L, Chen S, Xu T. et al. Phosphothioation of DNA In bacteria by dnd genes. Nature Chem Biol. 2007;3:709–710. [PubMed: 17934475]
- 50.
- Eckstein F. News and views: Phosphorothioation of DNA in bacteria. Nature Chem Biol. 2007;3:689–670. [PubMed: 17948013]
- 51.
- Cohn WE, Volkin E. Nucleoside-5’-phosphates from ribonucleic acid. Nature. 1951;167:483–484.
- 52.
- Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem. 1957;227:907–915. [PubMed: 13463012]
- 53.
- Cohn WE. 5-Ribosyl uracil, ribofuranyl nucleoside in RNA. Biochim. Biophys Acta. 1959;32:569–571. [PubMed: 13811055]
- 54.
- Smith JD, Dunn DB. An additional sugar component of RNA. Biochim Biophys Acta. 1959;31:573–575. [PubMed: 13628698]
- 55.
- Lane BG, Butler GC. The isolation, identification and properties of dinucleotides from alkali hydrolyzates of RNA. Can J Biochem Physiol. 1959;37:1329–1350. [PubMed: 14413833]
- 56.
- Littlefield JW, Dunn DB. The occurrence and distribution of thymine and three methylated adenine bases in RNA from several sources. Biochem J. 1958;70:642–651. [PMC free article: PMC1196721] [PubMed: 13607422]
- 57.
- Adler M, Weissmann B, Gutman AB. Occurrence of methylated purine bases in RNA. J Biol Chem. 1958;230:717–723. [PubMed: 13525389]
- 58.
- Smith JD, Dunn DB. The occurrence of methylated guanines in ribonucleic acids from several sources. Biochem J. 1959;72:294–301. [PMC free article: PMC1196922] [PubMed: 13662298]
- 59.
- Dunn DB. Additional components in RNA of rat liver fractions. Biochim Biophys Acta. 1959;34:286–288. [PubMed: 13818674]
- 60.
- Hall RH. The Modified Nucleosides in Nucleic Acids. New York/London: Columbia University Press. 1971.
- 61.
- Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucl Acids Res. 1994;22:2183–2196. [PMC free article: PMC523672] [PubMed: 7518580]
- 62.
- McCloskey JA, Rozenski J. The small subunit rRNA modification database. Nucleic Acids Res. 2005;33:D135–138. [PMC free article: PMC539969] [PubMed: 15608163]
- 63.
- Piekna-Przybylska D, Decatur WA, Fournier MJ. New bioinformatics tool for analysis of nucleotide modifications in eukaryotic rRNA. RNA. 2007;13:1–8. [PMC free article: PMC1800513] [PubMed: 17283215]
- 64.
- Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucl Acids Res. 2008;36:D178–183. [PMC free article: PMC2238946] [PubMed: 17947322]
- 65.
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eukarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. [PMC free article: PMC54159] [PubMed: 2112744]
- 66.
- Gehrke CW, McCune RA, Gama-Sosa MA. et al. Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J Chromatogr. 1984;301:199–219. [PubMed: 6209294]
- 67.
- Gehrke CW, Kuo KC. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J Chromatogr. 1989;471:3–36. [PubMed: 2670985]
- 68.
- Grosjean H, Keith G, Droogmans L. Vol. 265. 2004. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. In: Gott JM, ed. RNA Interference, Editing and Modification - Methods in Molecular Biology. Totowan: Humana Press; pp. 357–392. [PubMed: 15103084]
- 69.
- Wagner TM, Nair V, Guymon R. et al. A novel method for sequence placement of modified nucleotides in mixtures of tRNA. Nucleic Acids Symp Series. 2004;48:263–264. [PubMed: 17150579]
- 70.
- Gott JM. 2007. Methods in Enzymology. Vols. 424 and 425. Academic Press-Elsevier.
- 71.
- Cermakian N, Cedegren R. 1998. Modified nucleosides always were: an evolutionary model. In: Grosjean H, Benne R, eds. Modification and Editing of RNA. Washington DC: ASM Press; pp. 535–541.
- 72.
- Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucl Acids Res. 2002;30:1427–1464. [PMC free article: PMC101826] [PubMed: 11917006]
- 73.
- Uurbonavicius J, Auxilien S, Walbott E. et al. Acquisition of a bacterial RumA-type tRNA(uracil-54,C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol Microbiol. 2008;67:323–333. [PubMed: 18069966]
- 74.
- Jühling J, Mörl M. Hartmann V et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37, Database issue:D159–D162. [PMC free article: PMC2686557] [PubMed: 18957446]
- 75.
- Grosjean H, Sprinzl M, Steinberg S. Posttrancriptionally modified nucleosides in tRNA: their locations and frequencies. Biochimie. 1995;77:139–141. [PubMed: 7541252]
- 76.
- de Crécy-Lagard V, Marck C, Grosjean H. Comparative RNomics and modomics in mollicutes: prediction of gene function and evolutionary implications. IUBMB Life. 2007;59:634–658. [PubMed: 17852564]
- 77.
- Grosjean H, Gupta H, Maxwell S. 2008. Modified nucleotides in archaeal RNAs. In: Blum P, ed. Archaea, New Models for Prokaryotic Biology. Norwich: Horizon Press; pp. 164–196.www
.caister.com. - 78.
- Grosjean H, Gaspin C, Marck C. et al. RNomics and Modomics in the halophile Haloferax volcanii: identification of RNA modification genes. BMC Genomics. 2008;9:470–496. [PMC free article: PMC2584109] [PubMed: 18844986]
- 79.
- Omer AD, Ziesche S, Decatur WA. et al. RNA-modifying machines in archaea. Mol Microbiol. 2003;48:617–629. [PubMed: 12694609]
- 80.
- Muller S, Charpentier B, Branlant C. et al. A dedicated computational approach for the identification of archaeal H/ACA sRNAs. Methods Enzymol. 2007;425:355–387. [PubMed: 17673091]
- 81.
- Svensson I, Boman HG, Eriksson KG. et al. Studies on microbial RNA: Transfer of methyl groups from methionine to soluble RNA from E. coli. J Mol Biol. 1963;7:254–271. [PubMed: 14065310]
- 82.
- Fleissner E, Borek E. A new enzyme of RNA synthesis: RNA methylase. Proc Natl Acad Sci USA. 1962;48:1199–1203. [PMC free article: PMC220932] [PubMed: 13893516]
- 83.
- Starr JL. The incorporation of methyl groups into amino acid transfer ribonucleic acid. Biochem Biophys Res Comm. 1963;10:175–180. [PubMed: 13983497]
- 84.
- Hurwitz J, Gold M, Anders M. The enzymatic methylation of RNA and DNA. 3. Purification of soluble RNA-methylating enzymes. J Biol Chem. 1964;239:3462–3473. [PubMed: 14245404]
- 85.
- Hurwitz J, Anders M, Gold M. et al. The enzymatic methylation of RNA and DNA. 7. The methylation of ribosomal RNA. J Biol Chem. 1965;240:1256–1266. [PubMed: 14284734]
- 86.
- Delk AS, Rabinowitz JC. Biosynthesis of ribosylthymine in the tRNA of S. faecalis: a folate-dependent methylation not involving S-adenosylmethionine. Proc Natl Acad Sci. 1975;72:528–530. [PMC free article: PMC432345] [PubMed: 804695]
- 87.
- Farkas WR, Hankins WD, Sing R. The guanylation of tRNA: an enzymatic reaction. Biochim Biophys Acta. 1973;294:94–105.
- 88.
- Okada N, Harada F, Nishimura S. Specific replacement of Q-base in the anticodon of tRNA by guanine catalyzed by a cell-free extract of rabbit reticulocytes. Nucl Acids Res. 1976;3:2593–2603. [PMC free article: PMC343115] [PubMed: 792816]
- 89.
- Itoh YH, Itoh T, Haruna I. et al. Substitution of guanine for a specific base in tRNA by extracts of Ehrlich ascites tumor cell. Nature. 1977;267:467. [PubMed: 559945]
- 90.
- Watanabe M, Matsuo M, Tanaka S. et al. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, preceeds via a pathway involving base replacement in the tRNA polynucleotide chain. J Biol Chem. 1997;272:20146–20151. [PubMed: 9242689]
- 91.
- Kiss-Laszlo Z, Henry Y, Bachellerie JP. et al. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. [PubMed: 8674114]
- 92.
- Nicoloso M, Qu LH, Michot B. et al. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their role as guides for 2’-O-ribose methylation of rRNAS. J Mol Biol. 1996;260:178–195. [PubMed: 8764399]
- 93.
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridines in ribosomal RNA. Cell. 1997;89:565–573. [PubMed: 9160748]
- 94.
- Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNA. Cell. 1997;89:799–809. [PubMed: 9182768]
- 95.
- Bonnerot C, Pintard L, Lutfalla G. Functional redundancy of Spb1p and a snR52-dependent mechanism for the 2’-O-ribose methylation of a conserved rRNA position in yeast. Mol Cell. 2003;12:1309–1315. [PubMed: 14636587]
- 96.
- Renalier MH, Joseph N, Gaspin C. et al. The Cm56 tRNA modification in archaea is catalyzed either by a specific 2’-O-methylase, or a C/D sRNP. RNA. 2005;11:1051–1063. [PMC free article: PMC1370790] [PubMed: 15987815]
- 97.
- Ma X, Yang C, Alexandrov A. et al. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 2005;24:2403–2413. [PMC free article: PMC1173158] [PubMed: 15962000]
- 98.
- Gurha P, Joardar A, Chaurasia P. et al. Differential roles of archaeal box H/ACA proteins in guide RNA-dependent and independent pseudouridine formation. RNA Biol. 2007;4:101–109. [PubMed: 17993784]
- 99.
- Decatur WA, Schnare MN. Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Mol Cell Biol. 2008;28:3089–3100. [PMC free article: PMC2423156] [PubMed: 18332121]
- 100.
- Gold M, Hurwitz J, Anders M. The methylation of RNA and DNA. II. On the species specificity of the methylation enzymes. Proc Natl Acad Sci USA. 1963;50:164–169. [PMC free article: PMC300670] [PubMed: 16578536]
- 101.
- Gold M, Hurwitz J. Enzymatic methylation of ribonucleic acid and deoxyribonucleic acid.V. Purification and properties of DNA-methylating activity of E. coli. J Biol Chem. 1964;239:3858–386. [PubMed: 14257620]
- 102.
- Kornberg SR, Zimmerman SB, Kornberg A. Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected E. coli. J Biol Chem. 1961;236:1487–1493. [PubMed: 13753193]
- 103.
- Rae P, Steele R. Modified bases in the DNAs of unicellular eukaryotes. Biosystems. 1978;10:37–53. [PubMed: 566131]
- 104.
- Borst P, Sabatini R. Base J: Discovery, Biosynthesis and possible Functions. Ann Rev. Microbiol. 2008;62:235–251. [PubMed: 18729733]
- 105.
- Yu B, Yang Z, Li J. et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. [PMC free article: PMC5137370] [PubMed: 15705854]
- 106.
- Ebhardt HA, Thi EP, Wang MB. et al. Extensive 3’ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc Natl Acad Sci. 2005;102:13398–13403. [PMC free article: PMC1224661] [PubMed: 16157869]
- 107.
- Ohara T, Sakaguchi Y, Suzuki T. et al. The 3’ termini of mouse Piwi-interacting RNAs are 2’-O-methylated. Nature Struct Biol. 2007;14:349–350. [PubMed: 17384646]
- 108.
- Kawahara Y, Megraw M, Kreider E. et al. Frequency and fate of micro-RNA editing in human brain. Nucl Acids Res. 2008;36:5270–5280. [PMC free article: PMC2532740] [PubMed: 18684997]
- 109.
- Habig J, Taraka D, Bass B. mi-RNA editing, we should have inosine this coming. Molec Cell. 2007;25:712–713. [PMC free article: PMC2583401] [PubMed: 17386255]
- Nucleic Acids Are Not Boring Long Polymers of Only Four Types of Nucleotides: A ...Nucleic Acids Are Not Boring Long Polymers of Only Four Types of Nucleotides: A Guided Tour - Madame Curie Bioscience Database
- Surgical Stress Response and Cancer Metastasis: The Potential Benefit of Periope...Surgical Stress Response and Cancer Metastasis: The Potential Benefit of Perioperative Beta Blockade - Madame Curie Bioscience Database
- Branching Morphogenesis of the Prostate - Madame Curie Bioscience DatabaseBranching Morphogenesis of the Prostate - Madame Curie Bioscience Database
- From Artificial Antibodies to Nanosprings:The Biophysical Properties of Repeat P...From Artificial Antibodies to Nanosprings:The Biophysical Properties of Repeat Proteins - Madame Curie Bioscience Database
- Cell Cycle and Chromosome Segregation Defects in Alzheimer's Disease - Madame Cu...Cell Cycle and Chromosome Segregation Defects in Alzheimer's Disease - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...