NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Wyosine derivatives are tricyclic ribonucleosides present exclusively at position 37 of tRNA specific for phenylalanine in most Eukarya (cytoplasmic only) and Archaea, but not Bacteria. They occur by posttranscriptional modification of encoded guanosine in the tRNA precursor. Depending on the organism examined, eight wyosine derivatives have been identified in naturally occurring tRNAPhe, eleven if one also includes biosynthetic intermediates. The large diversity of wyosine derivatives attests to the existence of complex biosynthetic routes that differ from one organism to another. In this review, we describe the chemical structures of these hypermodified guanosine derivatives and the biosynthetic pathway of the wyosine derivative found in Saccharomyces cerevisiae. Not surprisingly given their location at position 37, 3′adjacent to the GAA anticodon, wyosine derivatives have been demonstrated to play an essential role in translation, particularly in the regulation of frameshifting.
Introduction
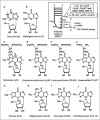
All transfer RNA genes sequenced so far harbor a purine (A or G) at position 37, adjacent to the 3′side of the anticodon.1,2 After transcription this purine is often enzymatically modified into more complex derivatives, the nature of which depends on the sequence of the anticodon and the organism,3,4 (recently reviewed in refs 5 and 6) recently reviewed in refs. 5 and 6. In the case of tRNAPhe (anticodon GAA, 31 sequences deposited in the tRNA databank—http://trnadb.bioinf.uni-leipzig.de/), two sets of unrelated compounds are found: i) isopentenyladenylate derivatives, such as isopentenyladenosine (i6A) in the cytoplasmic tRNAPhe of some eukaryotes and methylthiolated i6A and/or hydroxylated derivatives (io6Ams2i6Ams2io6A) in tRNAPhe of Bacteria and eukaryotic organelles; ii) a simple N1-methylguanine (m1G) in tRNAPhe of some Bacteria, Archaea and Eukaryotes and more complex derivatives of the hypermodified nucleoside wyosine (in fact of demethylwyosine, the minimalist derivative in this series, see below) in the cytoplasmic tRNAPhe of other Eukarya7 and Archaea (Fig. 1A and below). Recently, Bujnicki and coworkers have analyzed in great detail the occurrence and biosynthetic pathway of the isopentenylated adenosine derivatives in 63 organisms.8 Here, we focus on the structure and the biosynthetic pathway of the other family of purine-37 derivatives, wyosines.
Discovery of the So-Called ‘Y’ Base
In the flurry of excitement over the purification and sequencing of tRNAs, scientists in the nineteen sixties observed some peculiarities in one fraction of nucleic acids containing a tRNA specific for phenylalanine and isolated from baker's yeast or animal liver. This eukaryotic tRNAPhe was found to contain a highly fluorescent nucleoside.9-11 The presence of this peculiar base accounts for the remarkable hydrophobic behavior of tRNAPhe during column chromatography12 and has been shown to be essential for proper codon-anticodon interaction.11,13 Also, due to the lability of the glycosidic bond this compound was easily excised from tRNA by incubation under mildly acidic conditions (pH 2-4, 37°C for a few hours) without breaking the tRNA phosphodiester backbone or affecting any other bases of the tRNAPhe.14 The structure of the acid liberated ‘Y’ base of baker's yeast was subsequently determined to be a tricyclic purine (imidazopurine) now designated wybutine.14,15 Since the acid treatment might have altered the chemistry of the isolated Y base, its structure was independently confirmed by total synthesis.16,17 The position of the glycosidic linkage was established by chemical analysis of the nucleoside obtained after mild enzymatic hydrolysis of purified S. cerevisiae tRNAPhe 18 and much later verified by chemical synthesis.19 Wybutosine (yW, Fig. 1C and Table 1), the nucleoside of wybutine, has a UV light absorption maxima at 234, 263 and 310 nm and a fluorescence emission maximum at 443 nm with excitation maxima at 239 and 318 nm at pH 7.5.20 In view of the steric hindrance between the N4-methyl group and the ribose moiety, it is likely that yW exists in tRNA exclusively in the anti conformation (not in syn conformation as it is drawn in Fig. 1C), a conformation that has been confirmed in solution for the nucleoside by NMR21 and in the solid state by crystallography.22 Wyosine derivatives are highly photoreactive, 23 a characteristic that has to be taken into account when testing the function of tRNAPhe. In this chapter, we use the numbering of the atoms as defined by Blobstein and coworkers24 for the tricyclic compounds when the imidazole is fused to the purine. This numbering differs from the IUPAC convention used for purines (compare the numberings in the Figs. 1A and 1C).
Table 1
Wybutine and wyosine derivatives found at position 37, 3′adjacent to anticodon GAA of tRNAPhe in Eukarya (E) and Archaea (A).
Other Members of the Wyosine Families in Eukaryota
Experiments aimed at identifying the structures of the modified purine-37 in tRNAPhe were carried out on a large panel of eukaryotes, including the yeast Torulopsis utilis, animal liver (bovine, rat, chicken, rabbit), several plants (wheat germ, yellow lupine and maize seeds), insects (Drosophila melanogaster and Bombix mori), firmicute Mycoplasma kid and the aquatic fungus Geotrichum candidum. While a fluorescent tricyclic imidazopurine was detected in most cases, it was clear that the precise chemical structure was dependent on the organism and the method of isolation.
Interestingly, the structure of wyosine derivatives isolated from plants or animal liver were first reported to be β-hydroperoxywybutosine (o2yW, Fig. 1D and Table 1), the only peroxy group within the lateral amino acid chain of wybutine.24-29 The same structure was later reported in tRNAPhe isolated from the aquatic fungus Geotrichum candidum.30 The presence of a hydroperoxide in tRNAPhe from L. luteus and wheat germ was supported by a positive Fe(SCN)2 coloring test.28,29 However, based on a negative Fe(SCN)2 coloring test and mass spectrometry data, Itaya and coworkers suggested that a hydroxyl group (OHyW, Fig. 1E and Table 1) instead of a hydroperoxide group was present in wybutosine of rat liver tRNAPhe 31, and later synthesized OHyW.32,33 However, because hydroperoxywybutosine (o2yW) can be formed during storage or manipulation of hydroxywybutosine (OHyW)33 and decomposition of o2yW leads to the formation of hydroxywybutosine (OHyW) and wybutosine (yW),29 the ultimate resolution of the relevant structure(s) may require the identification of the corresponding metabolic enzyme(s) forming one or another of these two experimentally identified wyosine derivatives.
A simpler wyosine derivative, designated wyosine (imG), has been isolated and characterized from tRNAPhe of the yeast Torulopsis utilis (alternative names Candida utilis, Torula utilis, Pichia jadinii).34,35 Its structure (Fig. 1G and Table 1) has been unambiguously verified by several methods, including comparison with the chemically synthesized nucleoside.19 Like the wyosine derivatives described above, imG is characterized by the susceptibility of the glycosidic bond to acid-catalyzed hydrolysis34 and remarkable fluorescence when illuminated with a UV-lamp.35
Finally, intermediates of wybutosine biosynthesis have been isolated from a variety of cell types. For example, when Vero cells (a cell line derived from African green monkey kidney) are grown in medium deprived of methionine (Met starvation), the yW (normally present in an important fraction of the cellular tRNAPhe) is absent; addition of methionine to the growth medium leads to reappearance of tRNAPhe fully modified with wybutosine.36 The tRNAPhe isolated from various tumor cells (e.g., Ehrlich ascites, neuroblastoma and Novikoff hepatoma) has been shown to be hypo-modified at the lateral chain (OHyW*, Fig. 1F and Table 1) and contain a fraction of tRNAPhe with 1-methylguanosine at position 37 (m1G, Fig. 1B).37-39 The presence of m1G in tRNAPhe has also been observed in a number of other cell types and organisms, including rat liver hepatomas,40 the previtellogenic oocyte of amphibian Tinca tinca,41 the posterior silkgland of Bombyx mori,42 Mycoplasma kid43 and from Drosophila melanogaster.44,45
Wyosine Derivatives also Exist in Archaea
To date, the only tRNAPhe that has been sequenced from an archaeal organism originates from the euryarchaeon H. volcanii: it harbors an m1G37.46 However, wyosine derivatives have been identified in the tRNAPhe of other Archaea by analysis of tRNA enzymatic digests with combined liquid chromatography-mass spectrometry (LC-MS). The first wyosine derivative was found in digests of the hyperthermophilic crenoarchaeota Sulfolobus solfataricus, Thermoproteus neutrophilus and Pyrodictium occultum. Based on the comparison of the UV absorption spectrum, fluorescence properties and mass spectrometry with those of eukaryotic wyosine derivatives and of synthetic bases, a new wyosine derivative was identified47 as 7-methylwyosine (mimG, Fig. 1H). However, further analysis of eleven additional thermophilic Archaea, including phylogenetically diverse representatives of thermophilic methanogenes and sulfur metabolizing hyperthermophiles of the euryarchaeota group (e.g., Methanobacterium thermoautotrophicum, Thermoplasma acidophylum and Archaeoglobus fulgidus), revealed that only a few Archaea (mainly the crenoarchaeota) contain mimG in their tRNAs,48 albeit demethylwyosine, which lacks the methyl group at N4, can also be detected. In enzymatic tRNA extracts of Sulfolobus solfataricus P2, Methanococcus thermolithotrophicum and Stetteria hydrogenophila, two new compounds have been identified: one corresponds to an isomer of wyosine (imG) and is designated isowyosine (imG2, Fig. 1I and Table 1), while the other one is a minimalist form of imG/imG2 with a molecular mass of 321, designated imG-14 because its molecular mass is 14 Da less than that of wyosine imG (Mr = 335, Fig. 1J and Table 1).49 Pyrolobus fumarii, a submarine crenarchaeote which grows optimally at 106°C, has been shown to contain several wyosine derivatives, including mimG, imG2, imG-14 and imG (the same as in T. utilis—Fig. 1G).50 The imG-14 (not mimG and imG/imG2) has also been detected in the psychrotolerant archaeon Methanococoides burtonii, which grows at 4-23°C,51 demonstrating that imG-14 derivatives in archaea are not exclusively synthesized in thermophilic or hyperthermophilic organisms. A compound of yet unknown structure with Mr = 422, exhibiting UV absorption spectrum characteristic of wyosine derivatives (designated N422), has been found in M. maripaludis and M. vannielii52 but not in the other archaea analyzed so far.
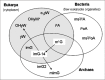
Thus, depending on the archaeon, at least four imG-14 derivatives can be identified in hydrolysates of bulk tRNA (imG, imG2, mimG (see Fig. 1, G-J and possibly N422). Their structures appear to be unique to Archaea because of the presence of a simple methyl group (or none) at C7 of the imidazopurine (Figs.1H and 1I) instead of α-amino-α-carboxypropyl side chain as in eukaryal wybutosines (Fig. 2, C-F). Unlike all eukaryal wyosine derivatives identified so far, archaeal derivatives do not always harbor a methyl group at N4.
Biosynthesis of Wyosine Derivatives in Eukarya
Origin of Various Carbon Atoms in Wybutosine
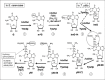
The biosynthetic pathway of imG-14 derivatives remained largely unknown for many years. Early work with S. cerevisiae demonstrated that wybutosine (yW) is derived from posttrancriptional modification of the encoded guanosine of the tRNAPhe precursor,53,54 a conclusion that appears evident today with the many sequences of tRNAPhe genes available. Subsequent work demonstrated incorporation of the α-amino-α-carboxypropyl group (symbol acp) from methionine into wybutosine. 55 NMR analysis of tRNAPhe isolated from yeast grown in the presence of (methyl-13C)-S-adenosylmethionine (AdoMet) demonstrated that each of the two methyl groups of the acp side chain, one of the two carbons at position 6 or 7 of the imidazo ring and the methyl group at position N4 of the guanine moiety are all derived from methionine, but the methyl group attached to the C6 atom is not.56 Identical results were observed in the biosynthesis of methylwyosine (mimG) in Sulfolobus acidocaldarius grown at 65° in the presence of (methyl-13C)-AdoMet.57
Understanding how the imidazo ring is formed has been (and still is) a challenge. Early studies demonstrated incorporation of radioactivity into tRNAPhe of Vero cells (a monkey kidney cell line) incubated with labeled lysine.58 This observation was interpreted in the context of formation of the third ring and it was proposed that lysine is converted into α-aminodipic acid semialdehyde (a C3 precursor), which then reacted with the guanosine. Demonstration that the C7 atom from the imidazo ring in fact originates from N1-methylation of guanosine-37 came from experiments in which a chimeric yeast tRNAPhe harboring an unmodified G37 in place of yW37 was microinjected into the cytoplasm of Xenopus laevis oocyte. After microinjection rapid formation of N1-methylguanosine was observed (Fig. 1B), followed by its successive transformation into two wyosine derivatives of unknown structures.59 Later, the gene encoding AdoMet-dependent tRNA:m1G37 methyltransferase Trm5p was identified in S. cerevisiae (see below) and deletion of TRM5 gene was shown to abolish not only the formation of m1G37 in many cellular tRNAs and m1I37 in tRNAAla, but also the formation of yW37 in tRNAPhe, consistent with the first step of wybutine biosynthesis being the formation of m1G37.60 While the hypothesis of Pergolizzi concerning the origin of the atoms C6, C7 and the attached methyl group of the imidazo ring is therefore not valid, the possibility remains that the C2 precursor used for the imidazo ring formation is connected to lysine metabolism. Identification of the next steps of wyosine metabolism came only recently from a comparative genomics approach followed by genetic/biochemical verifications.
Identification of Genes Coding for Enzymes of the yW Metabolism
The first gene encoding a protein involved in wybutosine biosynthesis, trmD, which encodes tRNA:m1G37 methyltransferase, was identified in Archaea (M. jannaschii and M. vannielii) by Björk and coworkers during the course of long-time studies on m1G37 modification in tRNA.60 The yeast ortholog TRM5 was identified by sequence similarity to the archaeal genes. Interestingly, bacterial TrmD is evolutionarily unrelated to the archaeal/eukaryal enzymes.61,62 These enzymes belong to the large Rossmann fold-containing superfamily of methyltransferases, while TrmD and all bacterial homologues belong to the SPOUT fold-containing superfamily of methyltransferases,63 (see also chapter by Czerwoniec et al in this volume).
The discovery of additional genes in wyosine biosynthesis has been achieved in recent years through the application of various genome mining techniques. For example, analysis of orphan genes in the genomes of organisms that synthesize wyosine derivatives (e.g., M. jannashii, S. cerevisiae, S. pombe, H. sapiens and A. thaliana) with those that don't (D. melanogaster, E. coli and B. subtilis) led to the identification of a single gene family belonging to the Cluster of Orthologous Genes COG073. Deletion of the S. cerevisiae gene belonging to this COG (YPL207w) and nucleotide analysis of the tRNAPhe isolated from the mutant strain demonstrated the presence of m1G37 instead of wyosine, leading to the conclusion that YPL207w catalyzes the second step of wyosine biosynthesis64 (Fig. 2).
In another approach, yeast genes encoding putative AdoMet-dependent methyltransferases harboring the characteristic seven ß-strand structures were systematically deleted and the nucleotide content of the tRNA isolated from the gene-deleted strains was analyzed. One of the candidate mutant strains (ΔYML005w) was shown to lack the yW derivative while accumulating the minimalist demethylwyosine (imG-14),65 (Fig. 1J), demonstrating that imG-14, initially discovered in archaeal tRNAs (see above), is a genuine intermediate of wybutine biosynthesis in S. cerevisiae. The newly discovered enzyme (YML005w, renamed Trm12p) was proposed to be involved in one of the steps following the ring cyclization, possibly the addition of the α-amino-α-carboxypropyl group (acp) on C7 atom of imG-14 or the methylation of N4 atom of the guanine ring.
Finally, in an exhaustive, systematic analysis of bulk RNAs isolated from 351 different S. cerevisiae deletion mutants by high performance liquid chromatography coupled to mass spectrometry (LC-MS), termed ‘ribonucleome analysis’,66 only four strains were found to lack the yW modification. In addition to the YPL207w and YML005w genes, previously shown to be involved in the formation of wybutosine, deletion of the YGL050w and YOL141w genes were also shown to disrupt the pathway leading to yW formation. In the corresponding mutant strains additional wyosine derivatives were observed; yW-86 in the tRNA from the ΔYGL050w strain and yW-72 in the ΔYOL141w strain (see explanation below and Fig. 2, also ref. 67). Using the corresponding recombinant proteins Yml005w, Ygl050w and Yol141w (renamed Tyw2p, Tyw3p and Tyw4p respectively), 3 steps of the wybutosine (yW) biosynthetic pathway were reconstituted in vitro.67
The Biosynthetic Route of Wybutosine Biosynthesis in S. cerevisiae
Integration of all the above information allows the following scheme to be proposed for the multistep enzymatic formation of wybutosine derivatives (Fig. 2). The complete biosynthetic route encompasses 7 steps (most taken from ref. 67): (1) Formation of m1G37 catalyzed by tRNA:m1G methyltransferase (Trm5p). The crystal structure of only the archaeal homolog (Methanocaldococcus jannaschii62) is known and raises interesting questions about the mechanism of action of this AdoMet-dependent methyltransferase (for the human homologue Trm5p, see also;68 (2) Addition of a C2-unit followed by cyclization of the 3rd ring catalyzed by Tyw1p. This enzyme contains a flavodoxin domain in its N-terminus and a radical-AdoMet domain including one (or possibly two) (4Fe-4S) cluster(s) in its C-terminus. The FMN cofactor in the flavodoxin domain probably serves as an electron storage unit. Mutations in one of the (4Fe-4S) cluster motifs abolished the yW biosynthesis in vivo, demonstrating that it is essential for catalytic activity.67 Crystal structures of the archaeal Tyw1p homologues lacking the N-terminal flavodoxin domain have recently been solved from M. jannaschii and Pyrococcus horikoshii.69,70 Their overall structures are similar to other radical-AdoMet enzymes71,72 and are consistent with the predicted enzymatic activity for imidazo ring formation (for details see chapter by Atta et al in this volume). The identity of the two-carbon donor in this reaction remains to be identified; (3) Transfer of the α-amino-α-carboxypropyl group (acp) from the methionine moiety of AdoMet to imG-14 at C7 atom catalyzed by Tyw2p. This is consistent with the earlier observation that the acp moiety of yW originates from methionine;55 (4) AdoMet-dependent methylation of the yW-86 (7-aminocarboxypropyl-demethylwyosine) to give yW-72 (7- aminocarboxypropylwyosine) by Tyw3p. In S. cerevisiae it appears that the Tyw2p activity occurs before Tyw3p and not vice versus, while in T. utilis imG is the apparent final modified nucleoside, demonstrating that in some organisms Tyw3p can catalyze the AdoMet-dependent conversion of imG-14 to imG (Fig. 2, dashed arrow); (5,6) The AdoMet-dependent conversion of yW-72 (aminocarboxypropylwyosine) to yW-58 (aminocarboxypropylwyosine methyl ester) and further to yW (methoxycarbonyl-aminocarboxypropylwyosine methyl ester), catalyzed by Tyw4p. Only scanty amount of yW-58 intermediate is formed in vitro. Still, it demonstrates that recombinant TYW4p has an apparent Ado-Met-dependent methylation activity. The fact that yW is a major product of TYW4p-catalyzed reaction together with the absence of detectable yW-14 or other possible intermediates indicate that multistep enzymatic formation of yW from yW-72 is most probably triggered by Ado-Met-dependent methylation. Since small quantity of other protein involved in methoxycarbonylation of yW-58 might copurify with the his-tagged TYW4p protein by a Ni-chelating column, TYW4p was further purified by anion exchange chromatography. Such purified protein directly converts yW-72 to yW with production of only low amount of yW-58 intermediate, suggesting that TYW4p is a bifunctional enzyme catalyzing both methylation of α-carboxyl group and methylcarbonylation of the α-amine group in the lateral chain of wyosine. (7) Hydro(pero)xylation of wybutosines. The enzymes catalyzing such reactions in plants and higher eukaryotes are unknown. Altogether, at least five molecules of AdoMet are consumed in the biosynthesis, making wybutosine perhaps the most costly modified nucleotide formed in a single cellular tRNA.73 To conclude this part of the chapter, Figure 3 summarizes the present knowledge about the origin of various ‘building blocks’ of wyosine derivatives in Eukarya and Archaea.
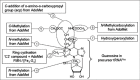
Figure 3
Origins of ‘building blocks’ in wybutosine. This figure complements Figure 2 concerning the biosynthetic pathway. Various ‘building blocks’ of wyosine derivatives in Eukarya and Archaea are presented.
Role of Wyosine Derivatives During Translation Process
The high-energetic cost and the requirement of many specific enzymes for its biogenesis beg the question of the cellular function of wybutosine in tRNAPhe. Its location exclusively at position 37, 3′-adjacent to the GAA anticodon, suggests a function in the decoding process on the ribosome. However, since there is significant structural diversity in the wyosine deriviatives found in Eukarya and Archaea (and Bacteria completely lack wyosine derivatives), how these nucleosides might participate in decoding is not immediately obvious. It appears from many studies that, in general, the modification status of tRNAPhe, especially at purine-37, plays a role in modulating the stability of the codon-anticodon interaction by dangling end-type base stacking (stabilization function74). Also, because most modified purines at position 37 cannot base pair in a Watson-Crick mode, their presence 3′-adjacent to the anticodon restricts the tRNA to base pair with the in-frame codon-anticodon triplet, thus limiting (but not completely avoiding, see below) the risk of frameshifting during translation (‘antislip’ function, reviewed in refs. 6,75,76). In the special case of tRNAPhe (anticodon GAA), which has to read ‘potentially weakly binding’ UUU/C codons, sometimes in a run of ‘slippy U-rich’ codon contexts, there may be a particular need for the ‘stabilization and antislip’ functions of highly hydrophobic bases like the wyosine derivatives. Notably, the level of modification at purine-37 and especially of the wybutosine derivatives in eukaryal tRNAPhe, depends on cell growth or stress conditions and the availability of the cofactor(s) needed for their enzymatic formation (see references above). Thus, the presence or absence of wybutosine derivatives in tRNAPhe may affect and possibly regulate, the expression level of certain proteins. For example, in several retroviruses the production of gag-pol or pro-pol fusion proteins requires -1 frameshifting at the so-called ‘programmed frameshifting’ site U-UUU-UUA (the slippery sequence read by tRNAPhe is underlined).77 Interestingly, the tRNAPhe isolated from HIV infected H9 cells lacks the wyosine derivatives.78 Also, using a rabbit reticulocyte in vitro translation system and the ‘shifty’ A-AAU-UUU sequence, Hatfield and collaborators demonstrated that the rabbit reticulocyte tRNAPhe bearing a m1G37 instead of the yW37 stimulates -1 frameshifting fourfold.79,80 However, when tRNAPhe from the S. cerevisiae trm5 mutant, which contains G37, was tested in vivo with another slippery sequence U-UUU-UUA, no significant difference in the level of frameshifting was observed when compared with the wild type tRNAPhe containing yW37.81 These contradictory observations indicate that either the consequences of a lack of yW37 depend on the position of the U-UUU sequence in the heptameric shifty sequence, or the two biological systems (in vitro or in vivo) are not comparable. Recently, Schimmel and coworkers demonstrated a clear correlation between the degree of modification of the G37 in yeast tRNAPhe and the efficiency of -1 frameshifting.82 Using an in vivo system based on mutants of S. cerevisiae ΔTYW1 and ΔTYW2 that accumulate tRNAPhe bearing m1G or imG-14, respectively (see Fig. 2), they probed all six naturally occurring shifty sequences in SCV-LA virus: (G-G(G,A,U)U-UU(U,C)), followed by a characteristic pseudo-knot that was shown to favor the frameshift event. The results were very compelling; tRNAPhe bearing m1G37 was more prone to frameshifting (up to 35% in some cases) than the one bearing imG-14 at position 37 (about 25%), which itself was more prone to frameshift than the wild type tRNAPhe containing yW37 (around 15%). It now remains to explain these data at the molecular level and importantly, to understand why some types of cells do not contain wyosine derivatives and have m1G37 instead.
Conclusion and Future Prospects
Wyosine and related derivatives are found exclusively at position 37, 3′-adjacent to anticodon GAA in tRNAPhe of many Eukarya and Archaea. They are absent in tRNAPhe of Bacteria, which instead harbor m1G37. In Figure 4, arrows indicate the direction of chemical complexity of different derivatives found in the three phylogenetic domains, from simple m1G to the more elaborate imG-14, 7-methylwyosine (mimG) in Archaea and wybutosine derivatives (yW and o2yW/OHyW) in Eukarya. Biosynthesis of these guanosine derivatives, especially in higher eukaryotes and plants, involves many enzymatic steps and is energetically costly in terms of AdoMet consumption. Its function is clearly to facilitate and regulate the production of proteins at the translation level (stabilization of codon-anticodon interaction and avoidance of slippage out of reading frame). However, there is also evidence that wyosine biosynthetic enzymes might be associated with other functions. For example, overexpression of the TRM12 gene, which encodes the human homologue of Tyw2p (step 3 in Fig. 2), has been observed in human breast cancer cells.83 Since it has been previously observed that in some tumors a hypomodified tRNAPhe (bearing m1G37) is utilized in translation instead of the fully modified isoacceptor,84 overproduction of this tRNA modification enzyme may trap a fraction of the imG-14 harbouring tRNAPhe, leaving free the m1G37-bearing variant. Thus, exploring the origin of the overexpression of TRM12 in cancer cells may reveal novel pathways of tumorogenesis.
While considerable progress has been made in recent years elucidating the wyosine biosynthetic pathway, much remains to be done. For example, the two-carbon donor for the radical-AdoMet enzyme Tyw1p (step 2 in Fig. 2) has yet to be identified, as well as the donor of methylcarbonyl (step 6 in Fig. 2) and the enzyme(s) involved in the hydro(peroxy)lation of wybutosine (step 7 in Fig. 2) in plants and higher eukaryotes are still unknown. Moreover, the potential exists for the organization of wyosine biosynthetic enzymes into a multiprotein complex (metabolon).67 At least in plants, genes coding for homologues of Tyw2p, Tyw3p and the C-terminal domain of Tyw4p are fused and code for a large ‘TYW3-4C-2’ protein.67 The close proximity of enzymes active-sites and substrates within multi-enzyme complexes generally enables the reactions to proceed more efficiently and without accumulation of intermediates. Crystallization of such type of complex should shed light about how the successive reactions are performed in a sequential manner. The three-dimensional structures of several wyosine and wybutosine biosynthetic enzymes have been solved, but so far only for archaeal homologues.62,69,70
Presently, the set of genes encoding enzymes of the wybutosine biosynthetic pathway is known only in S. cerevisiae. There are several different imG-14 derivatives in eukaryal and archaeal tRNA and searching for homologous genes in fully sequenced genomes (53 archaeal and 21 eukaryal ones are available at http://img.jgi.doe.gov, August 2008) and correlating them with the presence of imG-14 derivatives in each of these organisms may reveal the existence of alternative metabolic routes and provide insight on the emergence and evolution of this huge family of genes.
Acknowledgements
We thank Jef Rozenski (Katholieke Universiteit Leuven, Belgium) for suggestions regarding the nomenclature of wybutosine derivatives and Dirk Iwata-Reuyl (Portland State University, Portland, Oregon, USA) for advices and considerable improvements of the text. Current work on wyosine derivatives in LD laboratory is financed by the Fonds pour la Recherche Fondamentale Collective (FRFC), Fonds Jean Brachet Recherche, Fonds E. Defay and Fonds D. et A. Van Buuren. JA thanks the Commissariat à l′Energie Atomique for financial supports. JU was supported by a postdoctoral fellowship from the FRFC and by a FEBS Distinguished Young Investigator Award. HG (emeritus scientist) thanks Prof. Jean-Pierre Rousset from Université Paris-Sud for providing facilities to continue working in his laboratory.
References
- 1.
- Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8(10):1189–1232. [PMC free article: PMC1370332] [PubMed: 12403461]
- 2.
- Juhling F, Morl M, Hartmann RK. et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes Nucleic Acids Res 200937Database issueD159–62. Epub 2008 Oct 28 . [PMC free article: PMC2686557] [PubMed: 18957446]
- 3.
- Nishimura S. Minor components in transfer RNA: their characterization, location and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed: 4557059]
- 4.
- Grosjean H, Cedergren RJ, McKay W. Structure in tRNA data. Biochimie. 1982;64(6):387–397. [PubMed: 6180777]
- 5.
- Bjork GR, Hagervall T. Transfer RNA Modification. In: Curtis R. I, ed. Escherichia coli and Salmonella: Cellular and molecular biology. Washington, D.C.: ASM Press 2005. Module 4.6.2.
- 6.
- Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32(1):223–238. [PMC free article: PMC384350] [PubMed: 14715921]
- 7.
- Fairfield SA, Barnett WE. On the similarity between the tRNAs of organelles and prokaryotes. Proc Natl Acad Sci USA. 1971;68(12):2972–2976. [PMC free article: PMC389572] [PubMed: 4257127]
- 8.
- Kaminska KH, Baraniak U, Boniecki M. et al. Structural bioinformatics analysis of enzymes involved in the biosynthesis pathway of the hypermodified nucleoside ms(2)io(6)A37 in tRNA. Proteins. 2008;70(1):1–18. [PubMed: 17910062]
- 9.
- Rajbhandary UL, Chang SH, Stuart A. et al. Studies on Polynucleotides, Lxviii the Primary Structure of Yeast Phenylalanine Transfer RNA. Proc Natl Acad Sci USA. 1967;57(3):751–758. [PMC free article: PMC335572] [PubMed: 16591527]
- 10.
- Yoshikami D, Katz G, Keller EB. et al. A fluorescence assay for phenylalanine transfer RNA. Biochim Biophys Acta. 1968;166(3):714–717. [PubMed: 5722700]
- 11.
- Thiebe R, Zachau HG. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur J Biochem. 1968;5(4):546–555. [PubMed: 5698615]
- 12.
- Wimmer E, Maxwell IH, Tener GM. A simple method for isolating highly purified yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1968;7(7):2623–2628. [PubMed: 4298225]
- 13.
- Ghosh K, Ghosh HP. Role of modified nucleoside adjacent to 3′-end of anticodon in codon-anticodon interaction. Biochem Biophys Res Commun. 1970;40(1):135–143. [PubMed: 5456949]
- 14.
- Thiebe R, Zachau HG, Baczynskyj L. et al. Study on the properties and structure of the modified base Y+ of yeast tRNAPhe. Biochim Biophys Acta. 1971;240(2):163–169. [PubMed: 4327043]
- 15.
- Nakanishi K, Furutachi N, Funamizu M. et al. Structure of the fluorescent Y base from yeast phenylalanine transfer ribonucleic acid. J Am Chem Soc. 1970;92(26):7617–7619. [PubMed: 5490722]
- 16.
- Funamizu M, Terahara A, Feinberg AM. et al. Total synthesis of dl-Y base from yeast phenylalanine transfer ribonucleic acid and determination of its absolute configuration. J Am Chem Soc. 1971;93(24):6706–6708. [PubMed: 5122783]
- 17.
- Itaya T, Mizutani A. Studies on the synthesis of the fluorescent bases from phenylalanine transfer ribonucleic acids. Nucleic Acids Symp Ser. 1984;(15):13–15. [PubMed: 6441153]
- 18.
- Blobstein SH, Gebert R, Grunberger D. et al. Structure of the fluorescent nucleoside of yeast phenylalanine transfer ribonucleic acid. Arch Biochem Biophys. 1975;167(2):668–673. [PubMed: 1092270]
- 19.
- Itaya T, Kanai T, Iida T. Practical synthesis of wybutosine, the hypermodified nucleoside of yeast phenylalanine transfer ribonucleic acid. Chem Pharm Bull (Tokyo). 2002;50(4):530–533. [PubMed: 11964003]
- 20.
- Maelicke A, von der Haar F, Sprinzl M. et al. The structure of the anticodon loop of tRNAPhe from yeast as deduced from spectroscopic studies on oligonucleotides. Biopolymers. 1975;14(1):155–171. [PubMed: 1100138]
- 21.
- Kan LS, Ts'o PO, von der Haar F. et al. Proton magnetic resonance studies on the conformation of the hexanucleotide, GmpApApYpApsiP and Related fragments from the anticodong loop of baker's yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1975;14(14):3278–3291. [PubMed: 1096939]
- 22.
- Suddath FL, Quigley GJ, McPherson A. et al. Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0 angstroms resolution. Nature. 1974;248(443):20–24. [PubMed: 4594440]
- 23.
- Paszyc S, Rafalska M. Photochemical properties of Yt base in aqueous solution. Nucleic Acids Res. 1979;6(1):385–397. [PMC free article: PMC327695] [PubMed: 424298]
- 24.
- Blobstein SH, Grunberger D, Weinstein IB. et al. Isolation and structure determination of the fluorescent base from bovine liver phenylalanine transfer ribonucleic acid. Biochemistry. 1973;12(2):188–193. [PubMed: 4566585]
- 25.
- Dudock BS, Katz G, Taylor EK. et al. Primary structure of wheat germ phenylalanine transfer RNA. Proc Natl Acad Sci USA. 1969;62(3):941–945. [PMC free article: PMC223689] [PubMed: 5257014]
- 26.
- Nakanishi K, Blobstein S, Funamizu M. et al. Structure of the “peroxy-Y base” from liver tRNAPhe. Nat New Biol. 1971;234(47):107–109. [PubMed: 5289220]
- 27.
- Fink LM, Lanks KW, Goto T. et al. Comparative studies on mammalian and yeast phenylalanine transfer ribonucleic acids. Biochemistry. 1971;10(10):1873–1878. [PubMed: 4327495]
- 28.
- Feinberg AM, Nakanishi K, Barciszewski J. et al. Isolation and characterization of peroxy-Y base from phenylalanine transfer ribonucleic acid of the plant, Lupinus luteus. J Am Chem Soc. 1974;96(25):7797–7780. [PubMed: 4464312]
- 29.
- Barciszewska M, Kaminek M, Barciszewski J. et al. Lack of cytokinin activity of Y-type bases isolated from phenylalanine specific tRNAs. Plant Science Letters. 1981;20:387–392.
- 30.
- Moshizuki A, Omata Y, Miyazawa Y. Structure determination of the fluorescent base from Geotrichum candidum phenylalanine tRNA. Bull Chem Soc Jpn. 1980;53:813–814.
- 31.
- Kasai H, Yamaizumi Z, Kuchino Y. et al. Isolation of hydroxy-Y base from rat liver tRNAPhe. Nucleic Acids Res. 1979;6(3):993–999. [PMC free article: PMC327747] [PubMed: 440976]
- 32.
- Itaya T, Watanabe N, Mizutani A. Studies on the synthesis of the hypermodified base isolated from rat liver phenylalanine transfer ribonucleic acid. Nucleic Acids Symp Ser. 1986(17):49–51. [PubMed: 3645548]
- 33.
- Itaya T, Kanai T. Synthesis and structure of the hypermodified nucleoside of rat liver phenylalanine transfer ribonucleic Acid. Chem Pharm Bull (Tokyo). 2002;50(10):1318–1326. [PubMed: 12372857]
- 34.
- Takemura S, Kasai H, Goto M. Nucleotide sequence of the anticodon region of Torulopsis phenylalanine transfer RNA. J Biochem. 1974;75(5):1169–1172. [PubMed: 4472218]
- 35.
- Kasai H, Goto M, Ikeda K. et al. Structure of wye (Yt base) and wyosine (Yt) from Torulopsis utilis phenylalanine transfer ribonucleic acid. Biochemistry. 1976;15(4):898–904. [PubMed: 942858]
- 36.
- Pergolizzi RG, Engelhardt DL, Grunberger D. Formation of phenylalanine transfer RNA lacking the wye base in Vero cells during methionine starvation. J Biol Chem. 1978;253(18):6341–6343. [PubMed: 681356]
- 37.
- Salomon R, Giveon D, Kimhi Y. et al. Abundance of tRNAPhe lacking the peroxy Y-base in mouse neuroblastoma. Biochemistry. 1976;15(24):5258–5262. [PubMed: 999805]
- 38.
- Kuchino Y, Kasai H, Yamaizumi Z. et al. Under-modified Y base in a tRHAPhe isoacceptor observed in tumor cells. Biochim Biophys Acta. 1979;565(1):215–218. [PubMed: 259424]
- 39.
- Kuchino Y, Borek E, Grunberger D. et al. Changes of posttranscriptional modification of wye base in tumor-specific tRNAPhe. Nucleic Acids Res. 1982;10(20):6421–6432. [PMC free article: PMC326932] [PubMed: 6924749]
- 40.
- Grunberger D, Weinstein IB, Mushinski JF. Deficiency of the Y base in a hepatoma phenylalanine tRNA. Nature. 1975;253(5486):66–67. [PubMed: 163007]
- 41.
- Mazabraud A. Deficiency of the peroxy-Y base in oocyte phenylalanine tRNA. FEBS Lett. 1979;100(2):235–240. [PubMed: 37112]
- 42.
- Keith G, Dirheimer G. Primary structure of Bombyx mori posterior silkgland tRNAPhe. Biochem Biophys Res Commun. 1980;92(1):109–115. [PubMed: 6766719]
- 43.
- Kimball ME, Szeto KS, Soll D. The nucleotide sequence of phenylalanine tRNA from Mycoplasma sp. (Kid). Nucleic Acids Res. 1974;1(12):1721–1732. [PMC free article: PMC343451] [PubMed: 4375278]
- 44.
- White BN, Tener GM. Properties of tRNAPhe from Drosophila. Biochim Biophys Acta. 1973;312(2):267–275. [PubMed: 4198761]
- 45.
- Altwegg M, Kubli E. The nucleotide sequence of phenylalanine tRNA2 of Drosophila melanogaster: four isoacceptors with one basic sequence. Nucleic Acids Res. 1979;7(1):93–105. [PMC free article: PMC327998] [PubMed: 114987]
- 46.
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids and the sequences of 33 class I tRNAs. J Biol Chem. 1984;259(15):9461–9471. [PubMed: 6746655]
- 47.
- McCloskey JA, Crain PF, Edmonds CG. et al. Structure determination of a new fluorescent tricyclic nucleoside from archaebacterial tRNA. Nucleic Acids Res. 1987;15(2):683–693. [PMC free article: PMC340459] [PubMed: 3103099]
- 48.
- Edmonds CG, Crain PF, Gupta R. et al. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J Bacteriol. 1991;173(10):3138–3148. [PMC free article: PMC207908] [PubMed: 1708763]
- 49.
- Zhou S, Sitaramaiah D, Noon KR. et al. Structures of two new “minimalist” modified nucleosides from archaeal tRNA. Bioorg Chem. 2004;32(2):82–91. [PubMed: 14990307]
- 50.
- McCloskey JA, Liu XH, Crain PF. et al. Posttranscriptional modification of transfer RNA in the submarine hyperthermophile Pyrolobus fumarii. Nucleic Acids Symp Ser. 2000(44):267–268. [PubMed: 12903371]
- 51.
- Noon KR, Guymon R, Crain PF. et al. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature (Topt), 23 degrees C) and Stetteria hydrogenophila (Topt, 95 degrees C). J Bacteriol. 2003;185(18):5483–5490. [PMC free article: PMC193749] [PubMed: 12949100]
- 52.
- McCloskey JA, Graham DE, Zhou S. et al. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29(22):4699–4706. [PMC free article: PMC92529] [PubMed: 11713320]
- 53.
- Li HJ, Nakanishi K, Grunberger D. et al. Biosynthetic studies of the Y base in yeast phenylalanine tRNA. Incorporation of guanine. Biochem Biophys Res Commun. 1973;55(3):818–823. [PubMed: 4586620]
- 54.
- Thiebe R, Poralla K. Origin of the nucleoside Y in yeast tRNAPhe. FEBS Lett. 1973;38(1):27–28. [PubMed: 4589554]
- 55.
- Munch HJ, Thiebe R. Biosynthesis of the nucleoside Y in yeast tRNAPhe: incorporation of the 3-amino-3-carboxypropyl-group from methionine. FEBS Lett. 1975;51(1):257–258. [PubMed: 1091511]
- 56.
- Smith C, Schmidt PG, Petsch J. et al. Nuclear magnetic resonance signal assignments of purified (13C) methyl-enriched yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1985;24(6):1434–1440. [PubMed: 3886007]
- 57.
- McCloskey JA BG, Lindstrom EB, Peltier JM. Methylation of tRNA by S-adenosylmethionine in archaeal hyperthermophiles. Nucleic Acids Symposium Series. 1996;35:277–278.
- 58.
- Pergolizzi RG, Engelhardt DL, Grunberger D. Incorporation of lysine into Y base of phenylalanine tRNA in Vero cells. Nucleic Acids Res. 1979;6(6):2209–2216. [PMC free article: PMC327846] [PubMed: 111226]
- 59.
- Droogmans L, Grosjean H. Enzymatic conversion of guanosine 3′ adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: dependence on the anticodon sequence. EMBO J. 1987;6(2):477–483. [PMC free article: PMC553419] [PubMed: 3556165]
- 60.
- Bjork GR, Jacobsson K, Nilsson K. et al. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20(1-2):231–239. [PMC free article: PMC140193] [PubMed: 11226173]
- 61.
- Christian T, Hou YM. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol. 2007;373(3):623–632. [PMC free article: PMC2064070] [PubMed: 17868690]
- 62.
- Goto-Ito S, Ito T, Ishii R. et al. Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins. 2008;72(4):1274–1289. [PubMed: 18384044]
- 63.
- Elkins PA, Watts JM, Zalacain M. et al. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J Mol Biol. 2003;333(5):931–949. [PubMed: 14583191]
- 64.
- Waas WF, de Crecy-Lagard V, Schimmel P. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J Biol Chem. 2005;280(45):37616–37622. [PubMed: 16162496]
- 65.
- Kalhor HR, Penjwini M, Clarke S. A novel methyltransferase required for the formation of the hypermodified nucleoside wybutosine in eucaryotic tRNA. Biochem Biophys Res Commun. 2005;334(2):433–440. [PubMed: 16005430]
- 66.
- Noma A, Suzuki T. Ribonucleome analysis identified enzyme genes responsible for wybutosine synthesis. Nucleic Acids Symp Ser (Oxf ). 2006(50):65–66. [PubMed: 17150819]
- 67.
- Noma A, Kirino Y, Ikeuchi Y. et al. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25(10):2142–2154. [PMC free article: PMC1462984] [PubMed: 16642040]
- 68.
- Brule H, Elliott M, Redlak M. et al. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry. 2004;43(28):9243–9255. [PubMed: 15248782]
- 69.
- Suzuki Y, Noma A, Suzuki T. et al. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J Mol Biol. 2007;372(5):1204–1214. [PubMed: 17727881]
- 70.
- Goto-Ito S, Ishii R, Ito T. et al. Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 10):1059–1068. [PubMed: 17881823]
- 71.
- Marsh EN, Patwardhan A, Huhta MS. S-adenosylmethionine radical enzymes. Bioorg Chem. 2004;32(5):326–340. [PubMed: 15381399]
- 72.
- Frey PA, Hegeman AD, Ruzicka FJ. The Radical SAM Superfamily. Crit Rev Biochem Mol Biol. 2008;43(1):63–88. [PubMed: 18307109]
- 73.
- Grosjean H, Marck C, de Crecy-Lagard V. The various strategies of codon decoding in organisms of the three domains of life: evolutionary implications. Nucleic Acids Symp Ser (Oxf ). 2007(51):15–16. [PubMed: 18029563]
- 74.
- Grosjean H, Soll DG, Crothers DM. Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J Mol Biol. 1976;103(3):499–519. [PubMed: 781277]
- 75.
- Grosjean H, Houssier C, Romby P. Modulatory role of modified nucleotides in RNA loop-loop interaction In: Grosjean H, Benne R, ed. Modification and editing of RNA. Washington, DC: ASM press. 1998. pp. 113–133.
- 76.
- Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr Opin Microbiol. 2008;11(2):134–140. [PMC free article: PMC2408636] [PubMed: 18378185]
- 77.
- Jacks T, Madhani HD, Masiarz FR. et al. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55(3):447–458. [PMC free article: PMC7133365] [PubMed: 2846182]
- 78.
- Hatfield D, Feng YX, Lee BJ. et al. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1 and BLV. Virology. 1989;173(2):736–742. [PMC free article: PMC7131661] [PubMed: 2556852]
- 79.
- Carlson BA, Mushinski JF, Henderson DW. et al. 1-Methylguanosine in place of Y base at position 37 in phenylalanine tRNA is responsible for its shiftiness in retroviral ribosomal frameshifting. Virology. 2001;279(1):130–135. [PubMed: 11145896]
- 80.
- Carlson BA, Lee BJ, Hatfield DL. Ribosomal frameshifting in response to hypomodified tRNAs in Xenopus oocytes. Biochem Biophys Res Commun. 2008;375(1):86–90. [PMC free article: PMC2674873] [PubMed: 18675785]
- 81.
- Urbonavicius J, Stahl G, Durand JM. et al. Transfer RNA modifications that alter +1 frameshifting in general fail to affect -1 frameshifting. RNA. 2003;9(6):760–768. [PMC free article: PMC1370442] [PubMed: 12756333]
- 82.
- Waas WF, Druzina Z, Hanan M. et al. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J Biol Chem. 2007;282(36):26026–26034. [PubMed: 17623669]
- 83.
- Rodriguez V, Chen Y, Elkahloun A. Chromosome 8 BAC array comperative genomic hybridization and expression analysis identify amplification and overexpression of TRM12 in breast cancer. Genes, chromosomes and cancer. 2007. pp. 694–707. [PubMed: 17440925]
- 84.
- Smith DW, McNamara AL, Mushinski JF. et al. Tumor-specific, hypomodified phenylalanyl-tRNA is utilized in translation in preference to the fully modified isoacceptor of normal cells. J Biol Chem. 1985;260(1):147–151. [PubMed: 3843839]
- Deciphering the Complex Enzymatic Pathway for Biosynthesis of Wyosine Derivative...Deciphering the Complex Enzymatic Pathway for Biosynthesis of Wyosine Derivatives in Anticodon of tRNAPhe - Madame Curie Bioscience Database
- Tests of a Stereochemical Genetic Code - Madame Curie Bioscience DatabaseTests of a Stereochemical Genetic Code - Madame Curie Bioscience Database
- Origins and Evolution of Cotranslational Transport to the ER - Madame Curie Bios...Origins and Evolution of Cotranslational Transport to the ER - Madame Curie Bioscience Database
- Thermo-Chemo-Radiotherapy Association: Biological Rationale, Preliminary Observa...Thermo-Chemo-Radiotherapy Association: Biological Rationale, Preliminary Observations on Its Use on Malignant Brain Tumors - Madame Curie Bioscience Database
- IgA and Mucosal Homeostasis - Madame Curie Bioscience DatabaseIgA and Mucosal Homeostasis - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...