NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Major intrinsic proteins (MIPs) form a large superfamily of proteins that can be divided into different subfamilies and groups according to phylogenetic analyses. Plants encode more MIPs than other organisms and seven subfamilies have been defined, whereof the Nodulin26-like major intrinsic proteins (NIPs) have been shown to permeate metalloids. In this chapter we review the phylogeny of MIPs in general and especially of the plant MIPs. We also identify bacterial NIP-like MIPs and discuss the evolutionary implications of this finding regarding the origin and ancestral transport specificity of the NIPs.
Introduction
Major intrinsic proteins (MIPs) form channels in membranes that facilitate the permeation of water and other small uncharged polar molecules. The function of MIPs seems fundamental to life as we know it today since MIPs are found in virtually every organism. All studied MIPs share a common structure which consists of two transmembrane helices (TMHs), half a TMH followed by a third TMH. This topology is repeated once and due to the odd number of TMH in the first half, the second half is inserted in the opposite orientation in the membrane where the two half TMHs meet at a conserved NPA (Asn-Pro-Ala) motif situated at the N-terminus of each half TMH. A tetrad of amino acid residues situated in helix 2 and 5 and in loop E (H2, H5, LE1, LE2) referred to as the ar/R (aromatic/Arginine) selectivity region, appears to be the major determinant for substrate specificity. Water specific channels or orthodox AQPs (aquaporins) typically have the amino acid residues FHTR (one letter code) at these positions. Phylogenetic analyses of MIP sequences can shed light on how this super family has evolved and will potentially increase our understanding of the functions of the many different subfamilies that have been identified in these studies. In this chapter we review the phylogenetic analyses that have been reported regarding the whole super family irrespectively of species and in particular the studies of the abundant plant MIPs which are about three times as numerous as compared to MIPs in mammalian species. In addition we analyse the phylogeny of the plant NIP subfamily and discuss their evolutionary origin and ancestral selectivity region.
A Historical Account of the MIP Phylogeny
A prerequisite for phylogenetic analyses and recognition of protein families is of course that a sufficient number of homologous sequences are available. In hindsight it is surprising how quickly the MIPs were recognized as belonging to the same family, taking into account the few sequences available at the time and the differences in their proposed physiological roles. The nomenclature for the different isoforms has changed several times over the years and can sometimes be confusing. In this chapter we are using the original protein name when first citing a publication but the accepted systematic names preceded by the initials of the species name are mentioned in brackets and subsequently used to allow the reader to easily classify the MIP and refer to the current literature and the phylogenetic trees presented in this chapter.
The first cDNA sequence of a MIP appeared in the 1984 publication on bovine MIP-26 (Bos taurus; BtAQP0).1 BtAQP0 was believed to function as a gap junction protein in lens fiber tissue. Surprisingly though, comparisons of protein properties showed that the similarity to other known junctional proteins, such as connexins, was low. Further analyses indicated that BtAQP0 had six transmembrane helices similar to many of the transport proteins known at the time. In 1987, an integral membrane protein from soybean, NOD26 (Glycine max NOD26-like intrinsic protein; GmNIP1;1), was identified in a search for proteins associated with the peribacteroid membrane surrounding nitrogen fixating symbiotic bacteria in the root nodule and became the second MIP to have its cDNA sequenced.2 In the following year two groups, independently of each other, noticed a surprisingly high degree of sequence similarity between the lens fiber protein BtAQP0 and the peribacteroidal membrane protein GmNIP1;1.3,4 This was the very start of what would become the large and diverse MIP family of proteins. In 1989, sequencing of the Escherichia coli glpF gene encoding a glycerol transporter (EcGlpF) was finished5 and in 1990 Baker et al noticed that EcGlpF, BtAQP0 and GmNIP1;1 were all similar and members of the same family of proteins sharing a common ancestor.6 1990 also saw three publications reporting five new MIP sequences, the Drosophila melanogaster (DmBIB),7 two closely related bean tonoplast intrinsic proteins (Phaseolus vulgaris; PvTIP3s)8 and two TIPs expressed in the root of Arabidopsis thaliana (AtTIP1;1) and tobacco (Nicotiana tabacum; NtTIP2) respectively.9
With a growing number of proteins recognized as MIPs, the first real phylogenetic analysis of the family was conducted in 1991.10 Six MIPs were included in the analysis (EcGlpF, GmNIP1;1, BtAQP0, PvTIP3, DmBIB and a partial sequence of a bacterial GlpF from Streptomyces coelicolor) and although this was too few sequences to be able to draw any general conclusions, one test result still stands out as very interesting. By splitting each protein sequence into the two repeated halves, Pao et al actually got 11 sequences from the five full length MIPs and the partial MIP for their analysis. The result showed that all of the first halves form one cluster and all of the second halves form another. This is indicating that the corresponding halves of all MIPs are closer related to each other than to the other half of any of them. The authors thus concluded that the internal gene duplication responsible for the repeat must have happened before the split of eukaryotes and prokaryotes.
There was a rapid increase in the discovery rate of new MIPs following the publication of the first cDNA sequence of a human aquaporin CHIP28 (HsAQP1)11 and in 1993 Reizer and colleagues made a new phylogenetic analysis of the 18 sequences known to be members of the MIP family at the time.12 Even though this is also a very limited number of sequences, it is still possible to recognize five of the subfamilies as we group them today and a similar analysis of the sequence repeats as that conducted by Pao et al10 confirmed the previous finding. By 1996 the number of known MIPs had increased to an amount that allowed a more comprehensive phylogenetic analysis. Of the 84 MIP sequences available at this time, Park and Saier analyzed the 52 most divergent and complete and found them to belong to 12 subfamilies (4 bacterial, 3 yeast, 3 plant and 2 animal subfamilies).13 They concluded that all present MIPs originate from two divergent bacterial MIPs that gave rise to the aquaglyceroporin- (Glycerol facilitator-like protein; GLP) and the AQP-cluster respectively. The former cluster consists of 5 subfamilies as defined in this study but corresponds to one subfamily in phylogenetic analysis as of today where the different groups of GLPs in general reflect the phylogeny of the species. Animal water-specific AQPs as well as the plant specific subfamilies PIPs (plasma membrane intrinsic proteins), TIPs and NIPs are all very clearly resolved in the phylogenetic analysis. It is interesting to see that the water specific AQP from E. coli (EcAqpZ) clusters together with NIPs and that both these subfamilies are next to the GLPs in the phylogenetic tree.13 Froger and colleagues used a set of 142 MIPs in a sequence alignment to, among other things, identify positions potentially important for substrate specificity that could discriminate between GLPs and other MIPs.14 The following year Heymann and Engel included a phylogenetic analysis in their review on aquaporins.15 It was based on 46 MIPs, selected to reflect the diversity of the 160 MIP protein sequences known at the time and resulted in 16 subfamilies divided into the two clusters of AQPs and GLPs with a Archaeoglobus fulgidus aquaporin (the only archaeal MIP in the analysis) being the closest relative to the GLPs.
Between 2001 and 2005, Rafael Zardoya and colleagues published three articles forming the most comprehensive phylogenetic analysis of the MIP family of proteins available as of today. In the first of these papers all 153 full-length, nonredundant, MIP sequences available at the time were analyzed together.16 Another very important improvement in the analysis was the use of bootstrapping to test the validity of the different nodes in the phylogenetic tree. Based on this analysis Zardoya and Villalba concluded that MIPs could reliably be grouped into six major paralogous groups (subfamilies) GLPs, animal AQPs, PIPs, TIPs, NODs (NIPs) and AQP8s. However, the exact phylogenetic relationship between these subfamilies was not resolved even though there was some support for PIPs, TIPs and animal AQPs being more closely related. Phylogenetic analysis and variability profiles also showed that the PIP group is highly conserved, likely due to stringent evolutionary constraints.16,17 In an analysis published in 2002, Zardoya and coworkers saw a close phylogenetic relationship between NIPs and bacterial AQPs and concluded that plant NIPs likely arose by horizontal gene transfer 1190 million years ago.18 This could however not be confirmed in a more thorough analysis published in 2005.17 For the latter analysis, 463 nonredundant and complete or almost complete MIP sequences were aligned and subsets of these were analyzed to get an overall phylogeny of the complete MIP family as well as a more detailed analysis for each subfamily.
As the available MIP sequences have gone from scarce to plentiful, the problem has shifted from how to retrieve enough sequences to how to make an appropriate selection of sequences for your analysis. Since many of the early sequences of MIPs were picked up as cDNA sequences, there was a large bias toward highly expressed MIPs in the early sequence alignments, often resulting in having many sequences of close homologs in the same dataset. With more and more genomes being sequenced, it has become possible to instead retrieve all MIP sequences encoded in the genome of a species and in that way avoid the bias to highly expressed genes. This strategy has proven to be successful for identifying new subfamilies and also provides a way to find out whether MIPs from different species are separated by a gene duplication event (paralogs) or a speciation event (orthologs).19,20 However, this approach has also introduced the problem of accidentally identifying pseudogenes as functional MIPs since it does not discriminate between expressed and non expressed sequences.
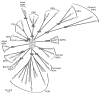
The general phylogeny of the MIP superfamily as of today can be seen in Figure 1. The dataset used for this analysis was constructed to contain a few representative sequences from all of the major groups including the subfamilies identified by Zardoya.17 In total 13 subfamilies are defined whereof five appear to be specific for plants. All of the central nodes, indicated by grey shading in Figure 1, have very low bootstrap values (≤52%) and hence the relationship between the subfamilies is not resolved. We would like to point out that what is often referred to as the AQP cluster or AQP group of MIPs is actually not a clear monophyletic clade, but rather constitutes a heterogeneous group, that can only be defined as all MIPs except the GLPs. The GLP cluster/group, on the other hand, is a monophyletic group with high bootstrap support and contains eukaryotic as well as bacterial sequences suggesting an ancient origin of the aquaglyceroporins. It is worth noting that higher plants are lacking GLPs whereas mosses contain a GLP (PpGIP1;1) suggesting that the GLPs were lost during the evolution of higher plants.21
Plant MIPs
Plant MIPs were early identified and subsequent studies have shown that plants have more isoforms than other organisms. In 1992, a total of six MIP sequences from plants were known in addition to GmNIP1;1 allowing a phylogenetic analysis.22 Two paralogous TIP groups was discerned, the seed specific αTIPs (TIP3s) and the vegetatively expressed γTIPs (TIP1s). Although the two other MIPs included in the study were annotated as TIPs they were quite different and are now recognized as PIPs. It became evident from studies of the model plant Arabidopsis thaliana that plants express several isoforms of PIPs, classified as PIP1s or PIP2s based on their sequence.23-25 By 1997 the rapidly growing sequence databases allowed the identification of MIP homologs in at least 15 different plant species. More importantly the large number of deposited EST sequences from Arabidopsis thaliana allowed a systematic search of expressed MIP genes in a single plant species.26 In total 23 MIP-encoding genes were identified in Arabidopsis, representing an overwhelming multitude of genes compared to the five human AQP genes known at the time. The AtMIPs formed three distinct subfamilies, consisting of 1 NIP, 11 PIP and 11 TIP sequences. Although 11 of the sequences were only partial, 20 of these genes were later confirmed by comprehensive analyses of the MIPs encoded in the genomic sequence (see below). However, due to the many partial sequences the exact relationship especially within the TIP subfamily remained speculative. This was resolved in a phylogenetic analysis of 38 TIPs from different plants.27 In this study high bootstrap support was provided for three different groups that had emerged within the TIP subfamily, γTIPs (TIP1s), δTIPs (TIP2s) and the αTIPs (TIP3s). The analysis showed that at least the TIP1 and the TIP2 groups formed before the split of the monocots and the dicots. In addition, PIPs were clearly divided in PIP1s and PIP2s with a similar high support. Preliminary analyses of the nearly completed Arabidopsis genome identified about 30 MIP members and suggested that the TIP subfamily consisted of at least four groups. Most significant was the expansion of the NLM subfamily (NIPs) since six MIPs were classified as NIPs based on sequence comparisons.28 At this time several groups of plant MIPs were well established phylogenetically and could be expected to be found in virtually every higher plant. However, there was not a common standard for how new MIPs should be named and some MIPs had several alternative names that could cause confusion. To facilitate identification of orthologs or co-orthologs, a uniform and systematic nomenclature based on phylogenetic analysis was informally agreed upon the third international aquaporin meeting, MIP2000, Gothenburg, Sweden, 2000.
In the following year three articles conforming to the new nomenclature were published. The first study was a systematic analysis and classification of ESTs from maize (Zea mays),29 followed by two articles reporting the annotation and classification of AtMIP genes based on the recently published Arabidopsis genomic sequence.19,30 In total 33 ZmMIP cDNAs and 35 full-length AtMIP genes were found. In addition three and seven pseudogenes encoding partial AtMIP sequences were identified by Quigley et al30 and Johanson et al,19 respectively, representing partial gene duplications and deletion events. Although bootstrap analysis was only employed by Johanson and colleagues,19 the very similar results presented in all three articles demonstrated the high reliability of the phylogenetic classifications. Four different subfamilies in higher plants were now recognized, the PIPs, TIPs, NIPs and the small basic intrinsic proteins (SIPs). The latter subfamily is formed by rather divergent and atypical MIPs that are lacking a fully conserved first NPA motif (Fig. 1).31 The gene structure in each subfamily was found to be conserved and furthermore the two PIP-, five TIP- and two SIP-groups were preserved in both dicots and monocots, suggesting that the last common ancestor had at least these groups of MIPs and the gene structure characteristic for each subfamily. Hence, these genes and gene structures could be expected in all monocot and dicot plants.19 Regarding the NIP groups, the situation was more complicated. In Arabidopsis a fixed criterion of maximum distance of 30% within a group were applied in all subfamilies. The high diversity among AtNIPs resulted in seven NIP groups, whereas the phylogenetic classification of ZmNIPs and other plant NIPs without a specific criterion defined only three groups, where the NIP1 group included AtNIP1s to AtNIP4s. Interestingly, there were no orthologs in Arabidopsis to the ZmNIP2 group. The early diversification of plant MIPs was confirmed by an analysis of ESTs from the moss, Physcomitrella patens.32 P. patens was found to have at least 12 different MIPs that could be classified into the four plant subfamilies. It was also concluded that the PIP1 and PIP2 groups formed before the divergence of mosses and the lineage leading to vascular plants. In contrast the five TIP groups found in higher plants were proposed to have evolved later in the lineage leading to vascular plants. It has been suggested that the NIPs evolved from a water-specific AQP to fill the functional role of GLPs, which are hitherto not found in higher plants.18 In this context it was unexpected that further studies of P. patens ESTs identified a GLP homolog, PpGIP1;1 (GlpF-like intrinsic protein).21 Interestingly, this MIP is closely related to the type II GLPs generally found in Gram positive bacteria and it was concluded that a GLP was most likely acquired from this group of bacteria by a horizontal gene transfer event about 1040 million years ago, i.e., 100 to 150 million years after the NIPs were suggested to have been acquired in plants.
Analysis of the rice genome provided the first comprehensive list of MIPs in a monocot. In an early study 33 full length MIP genes were identified and the encoded proteins were analyzed phylogenetically and classified according to the maize MIPs.29,33 All the groups within the four subfamilies in maize were confirmed and in addition two MIPs, OsNIP4;1 and OsPIP2;8 were identified that could not be classified into any of the predefined groups. Although not supported by the phylogenetic analysis, OsPIP2;8 was assigned to the PIP2 group based on sequence similarity.
The other protein, OsNIP4;1, was considered as a founder of a new and fourth NIP group in rice. In a later study focusing on homology modeling of AtMIPs, ZmMIPs and OsMIPs the updated rice genome was reanalyzed regarding encoded MIPs.34 The result of Sakurai et al33 was confirmed and six additional MIP genes were included although two of these (OsPIP1;4 and OsPIP1;5) originated from a different cultivar and might therefore represent allelic variation. All six new OsMIPs could be added to the existing groups as defined by Sakurai et al.33
Although comparisons of nucleotide sequences are potentially more informative than comparisons of protein sequences all phylogenetic analyses reviewed here except one are based on the protein sequences. Forrest and Bhave compared nucleotide sequences encoding PIPs and TIPs in wheat, rice and Arabidopsis.35 Surprisingly, the PIP1s and PIP2s cluster together in an Arabidopsis clade and a monocot clade in this study, although it is well established that the PIP1 and PIP2 subfamilies separated long before the divergence of monocots and dicots. This result could be due to that much of the nucleotide variation is at synonymous sites since the amino acid sequence is highly conserved among the PIPs. Thus the two clades could reflect a difference in codon usage between the species and it might be more reliable to use protein sequences when PIPs are compared between monocots and dicots.
Analysis of MIPs in plant lineages that diverged from higher plants a long time ago can provide a better understanding of early evolution of the MIP family in terrestrial plants. We therefore identified and classified the MIPs encoded in the genome of P. patens.20 The result shows that the bryophyte has an unexpected multitude and variation in the MIP family. In total 23 encoded MIPs were found that could be divided into seven subfamilies. One of the new subfamilies was the HIPs (hybrid intrinsic proteins). This class of protein was also found in spikemoss (Selaginella moellendorffii) and as indicated by the name has similarities to both TIPs and PIPs. The ar/R constriction was predicted to have a histidine both at H2 like the TIPs and at H5 like the PIPs. The other new subfamily was the XIPs (X intrinsic proteins) that are present also in many dicots. The physiological role of both HIPs and XIPs awaits further studies. The origin and early evolution of plant MIPs is still unresolved. None of the seven subfamilies are close homologs to the MIPs found in unicellular green algae, e.g., CrMIP1 from Chlamydomonas reinhartii (Fig. 1). Although the GIPs and the NIPs are suggested to have been acquired by horizontal gene transfer from bacteria, there is no evidence to suggest that any of the other five subfamilies originates from a similar event.
Phylogenetic Analysis of NIPs
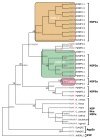
As previously mentioned one of the first MIPs identified was a plant NIP from soybean, GmNIP1;1. This NIP is localized to the peribacteroid membrane of root nodules, but it was soon apparent that NIPs were not restricted to this highly specific membrane of leguminous plants. Today, NIPs are known to be one of the largest families of plant MIPs and also one of the most divergent, both in regard to substrate specificities as well as amino acid sequences. Phylogenetic analyses show that the NIP subfamily can be divided into several well defined subgroups which are remarkably well conserved across species (Fig. 2).17,20,29 The NIP1, NIP2 and NIP3 subgroups are all present in higher plants, although NIP2s seem to be present only in some plants including-but not restricted to-monocots. Arabidopsis only possesses the NIP1 (the AtNIP1 to AtNIP4 groups) and NIP3 group (AtNIP5;1 and AtNIP6;1). The bryophyte P. patens has one NIP3 and three NIPs belonging to another distinct group, the NIP5s, so far only detected in primitive plants. There are also divergent NIPs that do not belong to any of these four groups, such as OsNIP4;1, PpNIP6;1 and AtNIP7;1. As seen in Figure 2 these diverse NIPs tend to group together due to the phenomenon known as long-branch attraction. The low levels of support in Figure 2 and previous analyses indicate that it is not clear how the different groups are related to each other.20,36 It is interesting to note that at least the NIP3 group had already evolved in a common ancestor to bryophytes and higher plants. Thus it is possible that this conserved group of NIPs represents and has retained, the original function of NIPs in early terrestrial plants.
Solute Transport
The substrate specificity of MIPs is governed by the tetrad of amino acid residues at the most narrow region of the pore referred to as the ar/R region. In Figure 2 the tetrad of each isoform is shown as well as the ancestral state of each node according to parsimony analysis. Based on the structure of the ar/R constriction region, AtNIPs have been divided into two functional groups, NIP-I and NIP-II.37 Whereas NIP-Is are able to transport both formamide, glycerol and to a moderate extend also water, NIP-IIs show very low water permeability but can instead transport urea.38,39 This have been accredited to the very wide pore of NIP-IIs, as it has been shown that decreasing the aperture of the ar/R decreasing, by replacing an alanine at the H2 position with a tryptophan, abolishes urea transport.39 In 2006, Ma et al identified a rice NIP in a mutant screen for silicon transporters.40 This NIP was found to be OsNIP2;1 and since then several other NIP2s have been shown to function as silicon transporters.41,42 Although, originally classified as NIP-II,43 further analysis has revealed that these NIPs have an ar/R filter distinctly different from that of the NIP-I and NIP-II groups and they have therefore been suggested to form a separate functional group, NIP-III in Figure 2.34,44-46 However, as seen in Figure 2, the functional grouping is at large covered by the phylogenetic groups and it would therefore be recommended that the phylogenetic grouping is used when referring to the NIP groups. This will avoid the confusion resulting from the discrepancy between the numbering of the functional and phylogenetic groups.
Apart from silicon transport, NIPs have also been shown to be capable of transporting other metalloids (boron, arsenite and antimony) both in vivo47-51 and in vitro.44,36 It has been speculated whether metalloid transport is the original function of NIPs.52 For example boron is essential in higher plants and needed in high amounts as it is used in the cross linking of the pectin Rhamnogalacturonan II in the cell walls. However, the low level of borate found in bryophytes53 and the relatively complex NIP group found in the bryophyte P. patens suggest that at least boron might not be the original physiological NIP substrate in mosses. This would favor the idea of another solute as the original substrate of NIPs, see further discussion below.
NIP–Like Bacterial MIPs and Ancestral State of ar/R Filter
The origin of NIPs is still unclear. In a very recent article the green algae Ostreococcus lucimarinus was claimed to encode a NIP, although no evidence for this statement was presented.54 Furthermore, we were not able to find any support for this classification neither in phylogenetic analyses, in motif comparisons, nor in BLAST searches. In an attempt to resolve the ancestry of the NIPs we made TBLASTN searches for the closest prokaryotic homologs to PpNIP6;1, one of the basal NIPs according to our earlier analysis.20 Some of the hits are included in the phylogenetic analysis and as seen in Figure 2 they neither appear to form a stable monophyletic group, nor do they correspond to a clear bacterial taxonomic subgroup. However, the relatively high bootstrap value for the common node with the NIPs shows that these bacterial sequences, representing a group of widely distributed but previously unrecognized NIP-like MIPs, are likely to be more closely related to NIPs than AqpZs are. A common origin of NIPs and the bacterial NIP-like MIPs (bNIPs) would exclude the suggested horizontal gene transfer of a bacterial AqpZ encoding gene as the origin of plant NIPs. However, a direct evolutionary link between AqpZs and an ancestral bNIP might still exist.
Interestingly, some bNIPs have an ar/R filter identical to that of PpNIP5 group (FAAR) and the parsimony analysis suggests that this was the ancestral state of NIPs and bNIPs (Fig. 2). Whether this ar/R tetrad would be specific for transport of metalloids can only be speculated since no MIP with this filter has yet been tested. The ar/R tetrad of HsAQP9, which transports the metalloid arsenite, is similar (FACR) taking into account that it is mainly the backbone carbonyl oxygen of the Cys at LE1 that is likely to interact with a substrate. However, this extrapolation might not be valid since HsAQP9 belongs to the GLP subfamily and therefore differs from NIPs at many other positions.
Conclusion
A coherent picture of the MIP superfamily divided into many different subfamilies has emerged. However, the relationship between the subfamilies and the earliest events that formed these subfamilies still remain unclear. Several of the different groups within the plant MIP subfamilies were already present in the last common ancestor of bryophytes and vascular plants suggesting an early evolution of e.g., the NIP3 group in terrestrial plants. However, none of the algal MIPs as identified today is closely related to MIPs in terrestrial plants. Here we identify bacterial NIP-like MIPs as close relatives of the plant NIPs, suggesting that NIP-like MIPs were already present in a common ancestor. Interestingly, the bryophyte NIP5 group has the same ar/R constriction region as these bacterial NIPs leading us to speculate that an original function is retained. Further studies are required to elucidate the substrate specificity of these MIPs and identification of MIPs in an algal sister clade of terrestrial plants is likely to contribute to our understanding of the early evolution of plant MIPs.
Acknowledgements
This work was supported by the Swedish Research Council (VR).
References
- 1.
- Gorin MB, Yancey SB, Cline J, et al. The major intrinsic protein (MIP) of the bovine lens fiber membrane: Characterization and structure based on cDNA cloning. Cell . 1984;39(1):49–59. [PubMed: 6207938]
- 2.
- Fortin MG, Morrison NA, Verma DP. Nodulin-26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res . 1987;15(2):813–824. [PMC free article: PMC340469] [PubMed: 3822816]
- 3.
- Shiels A, Kent NA, McHale M, et al. Homology of MIP26 to NOD26. Nucleic Acids Res . 1988;16(19) [PMC free article: PMC338721] [PubMed: 3174458]
- 4.
- Sandal NN, Marcker KA. Soybean nodulin 26 is homologous to the major intrinsic protein of the bovine lens fiber membrane. Nucleic Acids Res . 1988;16(19) [PMC free article: PMC338720] [PubMed: 3174457]
- 5.
- Muramatsu S, Mizuno T. Nucleotide sequence of the region encompassing the GlpKF operon and its upstream region containing a bent DNA sequence of Escherichia coli. Nucleic Acids Res . 1989;17(11) [PMC free article: PMC317952] [PubMed: 2544860]
- 6.
- Baker ME, Saier MH Jr. A common ancestor for bovine lens fiber major intrinsic protein, soybean nodulin-26 protein and E. coli glycerol facilitator. Cell . 1990;60(2):185–186. [PubMed: 2404610]
- 7.
- Rao Y, Jan LY, Jan YN. Similarity of the product of the Drosophila neurogenic gene big brain to transmembrane channel proteins. Nature . 1990;345(6271):163–167. [PubMed: 1692392]
- 8.
- Johnson KD, Hofte H, Chrispeels MJ. An intrinsic tonoplast protein of protein storage vacuoles in seeds is structurally related to a bacterial solute transporter (GIpF). Plant Cell . 1990;2(6):525–532. [PMC free article: PMC159908] [PubMed: 2152174]
- 9.
- Yamamoto YT, Cheng CL, Conkling MA. Root-specific genes from tobacco and Arabidopsis homologous to an evolutionarily conserved gene family of membrane channel proteins. Nucleic Acids Res . 1990;18(24) [PMC free article: PMC332893] [PubMed: 2129561]
- 10.
- Pao GM, Wu LF, Johnson KD, et al. Evolution of the MIP family of integral membrane transport proteins. Mol Microbiol . 1991;5(1):33–37. [PubMed: 2014003]
- 11.
- Preston GM, Agre P. Isolation of the cDNA for erythrocyte integral membrane protein of 28 kilodaltons: Member of an ancient channel family. Proc Natl Acad Sci USA . 1991;88(24):11110–11114. [PMC free article: PMC53083] [PubMed: 1722319]
- 12.
- Reizer J, Reizer A, Saier MH Jr. The MIP family of integral membrane channel proteins: Sequence comparisons, evolutionary relationships, reconstructed pathway of evolution and proposed functional differentiation of the two repeated halves of the proteins. Crit Rev Biochem Mol Biol . 1993;28(3):235–257. [PubMed: 8325040]
- 13.
- Park JH, Saier MH Jr. Phylogenetic characterization of the MIP family of transmembrane channel proteins. J Membr Biol . 1996;153(3):171–180. [PubMed: 8849412]
- 14.
- Froger A, Tallur B, Thomas D, et al. Prediction of functional residues in water channels and related proteins. Protein Sci . 1998;7(6):1458–1468. [PMC free article: PMC2144022] [PubMed: 9655351]
- 15.
- Heymann JB, Engel A. Aquaporins: Phylogeny, structure and physiology of water channels. News Physiol Sci. 1999;14:187–193. [PubMed: 11390849]
- 16.
- Zardoya R, Villalba S. A phylogenetic framework for the aquaporin family in eukaryotes. J Mol Evol . 2001;52(5):391–404. [PubMed: 11443343]
- 17.
- Zardoya R. Phylogeny and evolution of the major intrinsic protein family. Biol Cell . 2005;97(6):397–414. [PubMed: 15850454]
- 18.
- Zardoya R, Ding X, Kitagawa Y, et al. Origin of plant glycerol transporters by horizontal gene transfer and functional recruitment. Proc Natl Acad Sci USA . 2002;99(23):14893–14896. [PMC free article: PMC137515] [PubMed: 12397183]
- 19.
- Johanson U, Karlsson M, Johansson I, et al. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol . 2001;126(4):1358–1369. [PMC free article: PMC117137] [PubMed: 11500536]
- 20.
- Danielson JÅ, Johanson U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 2008;8(45) [PMC free article: PMC2386804] [PubMed: 18430224]
- 21.
- Gustavsson S, Lebrun AS, Norden K, et al. A novel plant major intrinsic protein in Physcomitrella patens most similar to bacterial glycerol channels. Plant Physiol . 2005;139(1):287–295. [PMC free article: PMC1203378] [PubMed: 16113222]
- 22.
- Höfte H, Hubbard L, Reizer J, et al. Vegetative and seed-specific forms of tonoplast intrinsic protein in the vacuolar membrane of Arabidopsis thaliana. Plant Physiol . 1992;99(2):561–570. [PMC free article: PMC1080500] [PubMed: 16668923]
- 23.
- Kaldenhoff R, Kolling A, Richter G. A novel blue light- and abscisic acid-inducible gene of Arabidopsis thaliana encoding an intrinsic membrane protein. Plant Mol Biol . 1993;23(6):1187–1198. [PubMed: 8292783]
- 24.
- Kammerloher W, Fischer U, Piechottka GP, et al. Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J . 1994;6(2):187–199. [PubMed: 7920711]
- 25.
- Yamaguchi-Shinozaki K, Koizumi M, Urao S, et al. Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: Sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol. 1992;33:217–224.
- 26.
- Weig A, Deswarte C, Chrispeels MJ. The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol . 1997;114(4):1347–1357. [PMC free article: PMC158427] [PubMed: 9276952]
- 27.
- Karlsson M, Johansson I, Bush M, et al. An abundant TIP expressed in mature highly vacuolated cells. Plant J . 2000;21(1):83–90. [PubMed: 10652153]
- 28.
- Johansson I, Karlsson M, Johanson U, et al. The role of aquaporins in cellular and whole plant water balance. Biochim Biophys Acta. 2000;1465(1–2):324–342. [PubMed: 10748263]
- 29.
- Chaumont F, Barrieu F, Wojcik E, et al. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol . 2001;125(3):1206–1215. [PMC free article: PMC65601] [PubMed: 11244102]
- 30.
- Quigley F, Rosenberg JM, Shachar-Hill Y, et al. From genome to function: The Arabidopsis aquaporins. Genome Biol . 2001;3(1) RESEARCH0001. [PMC free article: PMC150448] [PubMed: 11806824]
- 31.
- Johanson U, Gustavsson S. A new subfamily of major intrinsic proteins in plants. Mol Biol Evol . 2002;19(4):456–461. [PubMed: 11919287]
- 32.
- Borstlap AC. Early diversification of plant aquaporins. Trends Plant Sci . 2002;7(12):529–530. [PubMed: 12475491]
- 33.
- Sakurai J, Ishikawa F, Yamaguchi T, et al. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol . 2005;46(9):1568–1577. [PubMed: 16033806]
- 34.
- Bansal A, Sankararamakrishnan R. Homology modeling of major intrinsic proteins in rice, maize and Arabidopsis: Comparative analysis of transmembrane helix association and aromatic/arginine selectivity filters. BMC Struct Biol. 2007;7(27) [PMC free article: PMC1866351] [PubMed: 17445256]
- 35.
- Forrest KL, Bhave M. The PIP and TIP aquaporins in wheat form a large and diverse family with unique gene structures and functionally important features. Funct Integr Genomics . 2008;8(2):115–133. [PubMed: 18030508]
- 36.
- Bienert GP, Thorsen M, Schüssler MD, et al. A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biol. 2008;6(26) [PMC free article: PMC2442057] [PubMed: 18544156]
- 37.
- Wallace IS, Roberts DM. Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol . 2004;135(2):1059–1068. [PMC free article: PMC514140] [PubMed: 15181215]
- 38.
- Wallace IS, Wills DM, Guenther JF, et al. Functional selectivity for glycerol of the nodulin 26 subfamily of plant membrane intrinsic proteins. FEBS Lett. 2002;523(1–3):109–112. [PubMed: 12123814]
- 39.
- Wallace IS, Roberts DM. Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry . 2005;44(51):16826–16834. [PubMed: 16363796]
- 40.
- Ma JF, Tamai K, Yamaji N, et al. A silicon transporter in rice. Nature . 2006;440(7084):688–691. [PubMed: 16572174]
- 41.
- Mitani N, Yamaji N, Ma JF. Identification of maize silicon influx transporters. Plant Cell Physiol . 2009;50(1):5–12. [PMC free article: PMC2638714] [PubMed: 18676379]
- 42.
- Chiba Y, Mitani N, Yamaji N, et al. HvLsi1 is a silicon influx transporter in barley. Plant J . 2009;57(5):810–818. [PubMed: 18980663]
- 43.
- Wallace IS, Choi WG, Roberts DM. The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochim Biophys Acta . 2006;1758(8):1165–1175. [PubMed: 16716251]
- 44.
- Mitani N, Yamaji N, Ma JF. Characterization of substrate specificity of a rice silicon transporter, Lsi1. Pflugers Arch . 2008;456(4):679–686. [PubMed: 18214526]
- 45.
- Rouge P, Barre A. A molecular modeling approach defines a new group of nodulin 26-like aquaporins in plants. Biochem Biophys Res Commun . 2008;367(1):60–66. [PubMed: 18155659]
- 46.
- Ali W, Isayenkov SV, Zhao FJ, et al. Arsenite transport in plants. Cell Mol Life Sci . 2009;66(14):2329–2339. [PubMed: 19350206]
- 47.
- Kamiya T, Tanaka M, Mitani N, et al. NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J Biol Chem . 2009;284(4):2114–2120. [PubMed: 19029297]
- 48.
- Takano J, Wada M, Ludewig U, et al. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell . 2006;18(6):1498–1509. [PMC free article: PMC1475503] [PubMed: 16679457]
- 49.
- Isayenkov SV, Maathuis FJ. The Arabidopsis thaliana aquaglyceroporin AtNIP7;1 is a pathway for arsenite uptake. FEBS Lett . 2008;582(11):1625–1628. [PubMed: 18435919]
- 50.
- Ma JF, Yamaji N, Mitani N, et al. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc Natl Acad Sci USA . 2008;105(29):9931–9935. [PMC free article: PMC2481375] [PubMed: 18626020]
- 51.
- Tanaka M, Wallace IS, Takano J, et al. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell . 2008;20(10):2860–2875. [PMC free article: PMC2590723] [PubMed: 18952773]
- 52.
- Bienert GP, Schüssler MD, Jahn TP. Metalloids: Essential, beneficial or toxic? Major intrinsic proteins sort it out. Trends Biochem Sci . 2008;33(1):20–26. [PubMed: 18068370]
- 53.
- Matsunaga T, Ishii T, Matsumoto S, et al. Occurrence of the primary cell wall polysaccharide rhamnogalacturonan II in pteridophytes, lycophytes and bryophytes. Implications for the evolution of vascular plants. Plant Physiol . 2004;134(1):339–351. [PMC free article: PMC316313] [PubMed: 14671014]
- 54.
- Liu Q, Wang H, Zhang Z, et al. Divergence in function and expression of the NOD26-like intrinsic proteins in plants. BMC Genomics. 2009;10(313) [PMC free article: PMC2726226] [PubMed: 19604350]
Appendix
Identifiers for sequences used in the phylogenetic analyses
| Taxa Name | Species | GI Number | CDS-Start | CDS-Stop | Locus Tag |
|---|---|---|---|---|---|
| PpPIP1;1 | Physcomitrella patens | 167997734 | PHYPADRAFT_ | ||
| 62169 | |||||
| PpTIP6;1 | Physcomitrella patens | 168016415 | PHYPADRAFT_ | ||
| 73809 | |||||
| PpNIP3;1 | Physcomitrella patens | -a | |||
| PpSIP1;1 | Physcomitrella patens | 167999246 | PHYPADRAFT_ | ||
| 112053 | |||||
| PpGIP1;1 | Physcomitrella patens | 168057250 | PHYPADRAFT_ | ||
| 171260 | |||||
| PpHIP1;1 | Physcomitrella patens | -a | |||
| PpXIP1;1 | Physcomitrella patens | -a | |||
| AtPIP1;1 | Arabidopsis thaliana | 145339736 | AT3G61430 | ||
| AtTIP1;1 | Arabidopsis thaliana | 145360688 | AT2G36830 | ||
| AtNIP4;1 | Arabidopsis thaliana | 18421689 | AT5G37810 | ||
| AtNIP1;1 | Arabidopsis thaliana | 145340407 | AT4G19030 | ||
| AtNIP1;2 | Arabidopsis thaliana | 42566939 | AT4G18910 | ||
| AtNIP2;1 | Arabidopsis thaliana | 145360604 | AT2G34390 | ||
| AtNIP3;1 | Arabidopsis thaliana | 186479109 | AT1G31885 | ||
| AtNIP4;1 | Arabidopsis thaliana | 18421689 | AT5G37810 | ||
| AtNIP4;2 | Arabidopsis thaliana | 18421690 | AT5G37820 | ||
| AtNIP5;1 | Arabidopsis thaliana | 145340067 | AT4G10380 | ||
| AtNIP6;1 | Arabidopsis thaliana | 42563382 | AT1G80760 | ||
| AtNIP7;1 | Arabidopsis thaliana | 145338170 | AT3G06100 | ||
| AtSIP1;1 | Arabidopsis thaliana | 186509744 | AT3G04090 | ||
| OsPIP1;1 | Oryza sativa | 115447784 | Os02g0666200 | ||
| OsNIP1;1 | Oryza sativa | 115445190 | Os02g0232900 | ||
| OsNIP1;2 | Oryza sativa | 13161410 | Os01g0202800b | ||
| OsNIP1;3 | Oryza sativa | 54144479 | |||
| OsNIP1;4 | Oryza sativa | 58531193 | |||
| OsNIP2;1 | Oryza sativa | 115448656 | Os02g0745100 | ||
| OsNIP2;2 | Oryza sativa | 193811875 | Os06g0228200 | ||
| OsNIP3;1 | Oryza sativa | 20270142 | Os10g0513200b | ||
| OsNIP3;2 | Oryza sativa | 58531195 | |||
| OsNIP3;3 | Oryza sativa | 37806235 | |||
| OsNIP4;1 | Oryza sativa | 115434109 | Os01g0112400 | ||
| OsTIP1;1 | Oryza sativa | 115450710 | Os03g0146100 | ||
| OsSIP1;1 | Oryza sativa | 115434915 | Os01g0182200 | ||
| HsAQP2 | Homo sapiens | 209180415 | |||
| HsAQP3 | Homo sapiens | 22165421 | |||
| HsAQP5 | Homo sapiens | 186910293 | |||
| HsAQP6 | Homo sapiens | 86792454 | |||
| HsAQP7 | Homo sapiens | 4502186 | |||
| HsAQP8 | Homo sapiens | 45446751 | |||
| HsAQP10 | Homo sapiens | 22538419 | |||
| HsAQP11 | Homo sapiens | 27370564 | |||
| HsAQP12 | Homo sapiens | 156447036 | |||
| CrMIP1 | Chlamydomonas reinhardtii | 159471951 | |||
| VcMIP1 | Volvox carteri | 167172578 + | |||
| 167084018c | |||||
| PfAQP | Plasmodium falciparum | 124804458 | |||
| MmAQPM | Methano thermobacter marburgensis | 54040725d | |||
| ScGLP | Saccharomyces cerevisiae | 51012656 | YFL054C | ||
| ScAQY1 | Saccharomyces cerevisiae | 45270021 | YPR192W | ||
| CgAQY | Candida glabrata | 50285982 | CAGL0D00154g | ||
| CgGLP | Candida glabrata | 50285778 | CAGL0C03267g | ||
| MsAQPM1 | Methanosphaera stadtmanae | 84372150 | 1,146,881 | 1,147,615 | Msp_0998 |
| EcAQPZ | Escherichia coli | 215485161 | 913,630 | 914,325 | |
| EcGLPF | Escherichia coli | 215485161 | 4,432,406 | 4,433,251 | |
| PaAQPZ | Pseudomonas aeruginosa | 115583796 | 1,010,865 | 1,011,554 | aqpZ |
| PaGLPF | Pseudomonas aeruginosa | 115583796 | 1,544,365 | 1,545,204 | glpF |
| SsAQP8 | Sus scrofa | 159461726 | LOC100127152 | ||
| RnAQP8 | Rattus norvegicus | 2358276 | |||
| CtGLP | Clostridium tetani | 28204652 | 2124330 | 2125034 | CTC_01996 |
| BsGLP | Bacillus subtilis | 225184640 | 1002501 | 1003325 | BSU09280 |
| SmHIP | Selaginella moellendorffii | -e | |||
| NbXIP | Nicotiana | 39858292 + | |||
| benthamiana | 39862195c | ||||
| PtXIP | Populus trichocarpa | 224103260 | POPTRDRAFT_ | ||
| 557139 | |||||
| C. parvum | Chlorobaculum parvum | 193085153 | 474251 | 474964 | Cpar_0442 |
| R. baltica | Rhodopirellula baltica | 32397972 | 72504 | 74108 | RB4879 |
| B. cereus | Bacillus cereus | 218540236 | 250503 | 251225 | BCAH820_ B0297 |
| F. bacterium | Flavobacteriales bacterium | 163787877 | 253541 | 254209 | FBALC1_07048 |
| P. irgensii | Polaribacter irgensii | 88803076 | 393143 | 393817 | PI23P_12632 |
| C. flavus | Chthoniobacter flavus | 196229614 | 370933 | 371610 | CfE428DRAFT_ 1912 |
| P. maris | Planctomyces maris | 149176345 | 17718 | 18404 | PM8797T_ 07554 |
- a)
We have described in detail the annotation of this PpMIP in a previous study (Danielson JA, Johanson U. BMC Plant Biol 2008; 8:45);
- b)
This locus only cover part of the MIP sequence used;
- c)
These two EST sequences were combined to get the full length CDS used in the analysis;
- b)
This GI number refers to the AA sequence, the NA sequence can be found in the original article, PMID: 16233136;
- e)
This sequence was retrieved from the genomic sequence available from the Selaginella moellendorffii genome project at the Joint Genome Initiative (http://genome.jgi-psf.org/Selmo1/Selmo1.home.html) and partially corresponds to the gene model "estEXT fgenesh1 pm-C 30069".
- Phylogeny of Major Intrinsic Proteins - Madame Curie Bioscience DatabasePhylogeny of Major Intrinsic Proteins - Madame Curie Bioscience Database
- Molecular Properties of Voltage-Gated Calcium Channels - Madame Curie Bioscience...Molecular Properties of Voltage-Gated Calcium Channels - Madame Curie Bioscience Database
- Matrix Metalloproteinases, Tissue Inhibitors of Metalloproteinase and Matrix Tur...Matrix Metalloproteinases, Tissue Inhibitors of Metalloproteinase and Matrix Turnover and the Fate of Hepatic Stellate Cells - Madame Curie Bioscience Database
- MSK1 and Nuclear Receptors Signaling - Madame Curie Bioscience DatabaseMSK1 and Nuclear Receptors Signaling - Madame Curie Bioscience Database
- Sonic Hedgehog Signaling in the Developing and Regenerating Fins of Zebrafish - ...Sonic Hedgehog Signaling in the Developing and Regenerating Fins of Zebrafish - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...