NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The majority of the human genome is found to be transcribed and generates mostly noncoding (nc) RNAs that do not possess protein information. MicroRNAs are one of the well-identified small ncRNAs, but occupy merely a fraction of ncRNAs. Long (large) ncRNAs are emerging as a novel class of ncRNAs, but knowledge of these ncRNAs is far less accumulated. Long ncRNAs are tentatively classified as an ncRNA species containing more than 200 nucleotides. Recently, a long promoter-associated ncRNA (pncRNA) has been identified to be transcribed from the cyclin D1 promoter upon induction by genotoxic factors like ionizing-irradiation. The cyclin D1 pncRNA is specifically bound with an RNA-binding protein TLS (Translocated in liposarcoma) and exerts transcriptional repression through histone acetyltransferase (HAT) inhibitory activity. Analysis of TLS and the pncRNAs could provide a model for elucidating their roles in regulation of mammalian transcriptional programs. The pncRNA binding to TLS turns out to be an essential event for the HAT inhibitory activity. A key consensus sequence of the pncRNA is composed of GGUG, while not every RNA sequence bearing GGUG is targeted by TLS, suggesting that a secondary structure of the GGUG-bearing RNAs is also involved in recognition by TLS. Taken together, TLS is a unique mediator between signals of the long ncRNAs and transcription, suggesting that RNA networking functions in living cells.1-3
INTRODUCTION
Ninety percent of the human genome appears to be expressed to transcripts mainly consisting of noncoding (nc) RNAs.4-7 Lists of ncRNAs have been rapidly growing especially regarding micro (mi) RNAs and their related small ncRNAs including siRNAs of length 20 to 25 nucleotides.8,9 These small ncRNAs share core machinery composed of the RNA induced silencing complex (RISC) for exerting their function.5,6 miRNAs repress translation by blocking activity of target mRNAs with the RISC. Translational repression by miRNAs has been well analyzed, while long ncRNAs (also with their specific RNA binding proteins) have been emerging to play a role in the regulation of transcription. Long ncRNAs are tentatively defined as the ncRNAs that have a length of more than two hundred nucleotides. There have been reported divergent species of long ncRNAs. Some long ncRNAs function as coactivator and corepressor of transcription. Indeed, the ncRNAs, SRA and Evf-2 work as a coactivator,10,11 while HOTAIR and P15AS work as a corepressor,12,13 Furthermore, we found that long ncRNAs transcribed from the cyclin D1 promoter with the 200- and 330-nucleotides (nt) length represses transcription of the cyclin D1 gene.14 The cyclin D1 promoter-associated ncRNAs (pncRNAs) bind an RNA binding protein TLS (Translocated in Liposarcoma) and exert inhibition of histone acetyltransferase (HAT) activity to execute the transcriptional repression.14 We have performed extensive biochemical analysis on the pncRNA-TLS transcriptional repression and obtained a suitable model for studying long ncRNA functions.
TLS was initially identified as TLS-CHOP, a fusion protein arising from a chromosomal translocation presumed to be causative of myxoid liposarcoma.15 It has been shown to play roles in RNA processing, homologous DNA pairing and transcription.8,9 Deletion of the TLS gene in mice resulted in male infertility, chromosomal instability and increased sensitivity to DNA damaging agents,9 with the data from analysis of TLS null mice showing that TLS plays a pivotal role in maintaining genomic integrity.16,17
TLS has been reported to possess homologs referred to as the TET (TLS, EWS and TAF15) protein family.18,19 Intriguingly, EWS and TAF15 also have been identified as partners in fusion genes that are linked to sarcomas. EWS was initially identified as EWS-FLI1 in Ewing's sarcoma.18 TAF15 was initially described as a component of core transcriptional machinery19 but was also found to be a partner in a fusion gene (TAFII68-CHN) in a chondrosarcoma patient20,21 in which the fusion partner is an orphan nuclear hormone receptor. In each case, the relevant fusion proteins generated through the translocation event combine the N-terminus of the TET family member with the DNA binding domain of its corresponding partner. These fusion proteins cause mostly sarcomas.21-23
Here, I review recent publications regarding the RNA binding of TLS and focus on transcriptional regulation by newly identified long ncRNAs transcribed from the cyclin D1 promoter.14 This chapter reviews long ncRNA-dependent transcriptional repression and presents an overview of biological consequences of the long ncRNAs, suggesting that the transcriptional repression is one of examples of RNA network in mammalian cells.1-3
THE TARGET RNA SEQUENCES RECOGNIZED BY TLS
This section details our experiments to investigate the molecular mechanism of TLS and its target RNA sequences. We use this as a model of how long ncRNA-based mechanisms can be examined. Our quest for RNA molecules that specifically bind TLS began just after the identification of TLS as a HAT inhibitory molecule, because of the possibility of the regulatory function of RNA specifically binding to TLS.14 These following experiments indicated the specific RNA sequences recognized by TLS.24 The SELEX (systematic evolution of ligands by exponential enrichment) experiments used twenty-five nucleotides with randomized sequences.24 Four rounds of the SELEX through binding of bacterially expressed GST-TLS protein captured on GST-agarose gel beads with a 32P-labeled randomized RNA pool were performed and a subsequent three rounds of a selected RNA pool with a gel-shift assay were processed. After selection, the three clones (9, 60 and 75) turned out to possess the best bindings to TLS and the GGUG consensus TLS binding site (Table 1).24 Clone 9 is referred to as the GGUG-RNA and was selected for further analysis. The SELEX system was assembled artificially with synthetic RNA oligos and bacterially expressed GST-TLS. However, the selected consensus sequence GGUG possesses a robust binding to TLS and also to other TET family proteins in biochemical assay systems.24 The GGUG RNA is bound with TLS to enhance the inhibitory activity of TLS,14 and is a useful tool to assess the RNA dependency of the TLS HAT inhibitory activity.
Table 1
RNA sequences related to the binding of TLS.
After identification of the GGUG RNA as a regulatory RNA sequence, we explored naturally occurring RNA sequences with the ability to enhance the HAT inhibition by TLS,14 especially since the expression of cyclin D could be repressed by ionizing irradiation (IR).14 The cyclin D1 promoter DNA sequence region was found to contain the GGUG sequence at six regions (A, B, C, D, E and F). The RT-PCR experiments showed enhancement of the transcripts from regions A, B, D and E, while they showed no increment of transcript from the regions C and F, upon IR. Sequences of region A and B have the GGUG sequence (Table 1) and bind TLS at gel-shift assay. The expression of the region A and B is induced by IR. Region C is not induced by IR, but contains the GGUG sequence (Table 1). The oligos containing GGUG sequence designed from the region C, however, have no binding to TLS and no HAT inhibitory effect. These results showed that not all GGUG sequences are a determinant for their binding specificity to TLS and that sequences outside the GGUG and also a secondary structure of the GGUG RNAs might be involved in their binding specificity to TLS. Actually, the sequence outside of the GGUG consensus has been proved to be important. I discuss this issue in detail later.
Extensive biochemical experiments demonstrated that the cyclin D1 pncRNAs are transcribed through RNA polymerase II and have a poly A tail, but no CAP structure.14 The transcription is induced by IR and other DNA-damaging reagent treatments, although the mechanism of the induction of the cyclin D1 pncRNAs remains unclear.25 The transcripts appeared at distinct bands of 200 and 330 nucleotides, but always exhibited diffused electrophoresis patterns around these two major bands, suggesting that the transcription might be started from multiple sites or terminated as premature products. Elucidation of the transcriptional mechanism of the long ncRNAs would pose a crucial clue to understanding of the biology of these molecules. More than 1000 long ncRNAs have been identified, but transcriptional mechanisms of most of the long ncRNAs have not been well investigated.26-28 Understanding the transcriptional mechanisms of the ncRNAs will provide us what kind of cellular signals induces the ncRNA transcription, suggesting biological functions of the ncRNAs. So far our knowledge of the cyclin D1 pncRNAs make this of the best models to execute the analysis of the ncRNA transcription.14
It has been reported based upon analyses of the RNA binding regions, that TLS binds single stranded RNA molecules and their possession of a RNA recognition motif (RRM) and the zinc finger domain, indicates that they can bind single stranded RNAs.14,24,29 Indeed, the single stranded CCND1-pncRNA bound TLS and had inhibitory effect on HAT activity.14 Treatment of HeLa cells with IR induced bi-directional transcription of the pncRNAs from the cyclin D1 promoter.14 Preliminary experiments showed that both sense and antisense strands of pncRNAs are active on the HAT inhibition (Kurokawa, unpublished data). In case that the double stranded pncRNAs are not bound with TLS, transcription of both strands of the pncRNAs might be regulatory mechanism of the inhibitory activity of the pncRNAs, although the formation of double stranded pncRNA with sense-antisense strand has not been examined.14 The sense-antisense pairing ncRNAs have been found also in Alu repeats and the untranslated regions of protein coding genes,30,31 although biological meaning of formation of these double stranded RNAs has not been well understood. Another example of antisense ncRNA is a tumor suppressor gene p15.13 The antisense RNA of the p15 gene was reported to exert epigenetic silencing of this gene itself, but not by formation of the double stranded RNAs.13 Formation of the double stranded long ncRNAs is possible mechanism for regulation of the function of these ncRNAs, although specific examples are not widely characterized. Furthermore, subcellular fractionation studies revealed that the long pncRNAs and TLS are mainly localized to nuclear fraction.14 The nuclear localization of the long pncRNAs suggests that the pncRNAs and their binding partner TLS play a role in transcriptional regulation.
DOMAIN STRUCTURE OF TLS
TLS is a key molecule to execute the signal from the cyclin D1 ncRNAs. Dissection of the TLS domain structure presents the molecular mechanism of its binding to RNA sequences. TLS forms a modular structure consisting of several functional domains between an acidic N-terminus and a basic C-terminal region. Analysis of the amino acid composition of TLS indicates that the N-terminus of TLS (1-211) is an acidic peptide (pI: 2.6) and that the C-terminus (211-526) is a basic one (pI: 10.45) (Fig. 1A,B). The protein binding assay showed that the N-terminus of TLS interacts with the C-terminus and that this interaction is blocked by binding of the GGUG-RNA to the RNA binding regions of the C-terminus.14
The N-terminus region of TLS has high content of three amino acids, serine (S), glutamine (Q) and tyrosine (Y) and has been reported as a putative transcriptional activation domain (Fig. 1A).19,32 Pasting the N-terminus to a heterogenic DNA binding domain of the yeast transcription factor GAL4 results in transcriptional activation measured with a GAL4 reporter assay, indicating that the N-terminus is a transcriptional activation domain.32 The N-terminal region of the TET proteins also contains a glutamine-rich domain that is a common structure of transcriptional activation domains, as described above (Fig. 2).14 These data suggest that the fusion of the transcriptional activation domain of a TET protein to the DNA-binding domain of CHOP, Fli or CHN results in aberrant transcriptional activation that promotes the development of their related sarcomas. The N-terminus is bound by CREB-binding protein CBP and adenovirus p300 that are potent transcriptional coactivators with the potent HAT activity.14 The binding of CBP/p300 to the N-terminus at least partly contributes to its transcription activity.
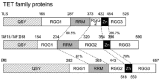
Figure 2.
Alignment of TLS homologues. Alignment of the TET family protein with homology. The homology percentages are calculated compared to the TLS sequence. The sequences are from human genes. Abbreviation: "Zn" represents the zinc finger domain.
The C-terminus of TLS and also the TET protein is markedly a basic region that fits a criterion for nucleic acid binding domain (Fig. 1B and Fig. 2). Actually, the C-terminus has several putative RNA interacting regions: RRM; the RGG-repeats that have multiple tandem repeats of arginine, glycine and glycine: RGG1, RGG2, RGG3; a zinc finger domain (Fig. 1A,B).32,33 Lerga et al showed that the GGUG-RNA bound to the RRM (271-392; amino acid numbers shown), activation domain (AD)-RGG1 (1-271) and RGG2-3 containing zinc finger (392-526). Extensive experiments with gel-shift assays using mutants of the GGUG-RNA (Clone 9, Table 1) indicated that RRM and RGG2-3 with the zinc finger (RGG2-3/zinc finger) are able to bind the mutants at the outside of core GGUG sequence (CUC-GGUG RNA, ΔUAG-RNA), while AD-RGG1 is not able to bind these mutants (Table 1). These data imply that AD-RGG1, RRM and RGG2-3/zinc finger play distinctive roles in recognition of the GGUG-RNA and that sequences outside the GGUG motif support the binding of full-length TLS to the GGUG-RNA. Contrast to the data, NMR analysis showed that the RRM fragment (266-375) failed to bind the RNA oligos (5'-UAGUUUGGUGAU-3') that contain the core GGUG motif trimmed down to 12 nucleotides, while the zinc finger fragment (398-468) bound to the RNA oligos.29 There is a remarkable discrepancy regarding the binding of the RRM to the GGUG RNA between these two data. The discrepancy could be explained by the difference in the RRM fragments used at these two experiments.24,29 Iko et al employed the RRM fragment (266-375) that does not contain the RGG repeat at the C-terminus,29 while Lerga et al utilized the RRM (271-392) that contains the C-terminal RGG repeat.24 Addition to this explanation, shorter RNA oligos used by Iko et al might lose their ability to bind to the RRM. The binding of the zinc finger domain of TLS to the GGUG RNA has been confirmed by the data from both groups.24,29 Moreover, the NMR analysis showed that the RRM (266-375) and the zinc finger have no interaction with or without the GGUG RNA. These data have indicated that TLS has multiple regions of interaction to the GGUG-RNA and each region could have a distinctive role to support the RNA binding.
The zinc finger domain of TLS plays a crucial role in its binding to the GGUG-RNA and belongs to the RanBP2-type zinc finger family.34 The RanBP2-type zinc finger has been analyzed regarding the RNA-binding protein ZRANB2 showing that ZRANB2 possesses two zinc finger domains. These zinc fingers, F1 (9-41) and F2 (65-95) bind an AGGUAA sequence with dissociation constants (Kds) of 3 μM and 2 μM, respectively.34 The AGGUAA sequence has been identified by a SELEX experiment using gel-shift assays and randomized 25 mer RNA oligonucleotides.34 Both F1 and F2 can bind the AGGUAA RNA sequence. These two zinc fingers in a fragment F12 (1-95) synergistically bind the tandem repeat of AGGUAA with higher Kd of 0.19mM, indicating that bipartite fragments of the zinc fingers coordinately recognize the target RNA.34 NMR analysis of the structure of the F2 of ZRANB2 dictated that the RanBP2-type zinc finger domain recognizes the AGGUAA sequence and also revealed a core sequence of AGGUAA as the GGU of the target mRNA for splicing.34 A protein sequence alignment of the zinc fingers of ZRANB2 and TET family proteins like TLS, EWS and TAF15 indicates significant homology.34 The sequence alignment between ZRANB2-F2 and TET proteins also indicates exact matching at the amino acids (D425, N435, F438, R440, N445: amino acid numbers of TLS are shown) contacting the core sequence, GGU and confirms that the zinc finger of the TET family proteins binds the GGU tract of the GGUG-RNA. Therefore, TET family proteins need to recognize the additional "G" residue at the end of GGU"G". This indicates that more residues in the TET proteins than the homologous amino acids to ZRANB2-F2 are involved in recognition of the GGUG-RNA.
Gel-shift binding assay using 32P-GGUG-RNA indicated that Kd of full-length TLS is 250 nM,24 while the NMR analysis of the zinc finger fragment (398-468) of TLS with the truncated GGUG-RNA (5'-UAGUUUGGUGAU-3') showed that Kd is 10000 nM.29 The zinc finger domain alone displays 40 times lower affinity to the GGUG consensus although affinity to the zinc finger domain might be underestimated because of possible loss of the affinity to shorter RNA oligos used. This suggests that the zinc finger of TLS cooperates with other domains to achieve the higher affinity to grasp the GGUG-RNA. These data also imply that TLS is able to bind multiple RNA molecules simultaneously because of its having multiple binding regions, to have synergistic and allosteric effect on its protein binding ability. Therefore, the multiple binding regions to RNA sequences give TLS versatility in its regulatory functions.
TRANSCRIPTION-REGULATORY LONG ncRNA, CYCLIN D1 pncRNA
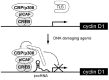
After characterising the HAT-inhibitory effect of the GGUG RNA, we can then explore the naturally occurring RNA sequence bound by TLS and the cyclin D1 pncRNA can be identified. The cyclin D1 pncRNA is a regulatory long ncRNA for transcription and requires TLS as a compulsory binding partner to execute its inhibitory effect on the HAT activity to repress the transcription (Fig. 3). There have been a few other ncRNAs transcribed from 5'-untranslated regulatory regions, promoters and enhancers.35,36 One example of a long ncRNA transcribed from a promoter is that of dehydrofolate reductase (DHFR).35 In quiescent mammalian cells, expression of DHFR is repressed. It has been reported that a transcript of a minor promoter located upstream of a major promoter is involved in the repression of DHFR.35 In the quiescent cells, the transcript of this minor promoter inhibits transcriptional initiation from the major promoter through direct binding to TFIIB of the pre-initiation complex. There are various genes that possess major and minor promoters like DHFR gene.37,38 Therefore, this could be considered a general mechanism in that the transcripts from a minor promoter have a regulatory role in transcription of the major promoter. The ncRNAs transcribed from this kind of alternative promoters could have a significant impact on the regulation of the genes in these promoter contexts. The DHFR pncRNA does not need to bind any RNA binding protein, but is directly interacted with the target molecule of the basic core transcription factor TFIIB.
Recently, novel ncRNAs (eRNA) transcribed from enhancers have been reported in primary culture of mouse cortical neurons.36 Genome-wide sequencing analysis with a KCl-dependent enhancer function in mouse cortical neurons, indicated 12,000 neuronal activity-regulated enhancers that are bound by CBP in a neuronal activity-dependent manner. A function of CBP is to recruit RNA polymerase II. Actually, activity-regulated binding of RNA polymerase II with thousands of enhancers was observed. Intriguingly, RNA polymerase II at enhancers transcribes bi-directionally eRNA within enhancer regions defined by the presence of histone H3 monomethylated at lysine 4 (H3K4me1). The level of eRNA expression at neuronal enhancers positively correlates with the level of mRNA synthesis at nearby genes. This suggests that eRNA expression is engaged in inducing mRNA synthesis. Experiments with the ARC gene, which regulates synaptic functions, indicate that truncation of the ARC promoter resulted in no eRNA synthesis although the ARC promoter was bound with transcription factor SRF and RNA polymerase II. Thus, the recruitment of RNA polymerase II to the ARC enhancer is not sufficient for eRNA synthesis and suggests that eRNA synthesis might require an interaction of the enhancer with a promoter. Given that the enhancers are interacted with the promoter to tether them to each other, eRNA might be bound with an RNA-binding protein like TLS to exert transcriptional regulations. Contrast to these two ncRNAs, the cyclin D1 pncRNA has been proved to require TLS as a binding partner to exert transcriptional regulation, although the DHFR ncRNAs and the eRNAs may have unidentified specific RNA-binding protein. Thus far, TLS and the cyclin D1 pncRNA are characteristic of a unique machinery to exert transcriptional repression in eukaryotic transcriptional programs (Fig. 3).
TLS is the distinctive enzyme inhibitor with specific target genes. Molecular mechanisms that enable TLS to target specific sets of genes have been explored. Our biochemical experiment showed that TLS inhibits the HAT activity of CBP/p300. This data prompted us to examine if TLS could repress the CREB-target genes that have been reported for more than 240 genes, because CBP/p300 is a major component of the CREB coactivator complex.39 Unexpectedly, TLS only repressed two genes, cyclin D1 and E1. Further analysis has been performed just on the cyclin D1 promoter indicating that TLS could specifically recognize the CBP just on the cyclin D1 gene promoter. The mechanism of the specific recognition of TLS to the CBP located on the cyclin D1 promoter remains unsolved. One possible explanation is that the cyclin D1-pncRNA specifically localizes TLS to the promoter sites at which the cyclin D1-pncRNAs are transcribed and also CBP/p300 is bound. In this case, transcription of the cyclin D1-pncRNA itself should function as a signal for recruiting TLS to the cyclin D1 promoter site. Another possible mechanism is that TLS could recognize specific histone-modifications near by the binding sites of the cyclin D1 promoter. Targeting only cyclin D1 and E2 by TLS indicates that every CREB binding site is not equivalent regarding specific binding of TLS. Upon binding of the ncRNAs, TLS specifically recognizes the cyclin D1 promoter to repress its expression. This might be another regulatory mechanism for selective repression of a gene during embryonic development or processes requiring differential expression of genes.
A significant percentage of the human genome is transcribed. Many of them are ncRNAs which localize to both nuclei and cytoplasm, although copy numbers of each ncRNA are not abundant. The cyclin D1-pncRNAs are also low copy number ncRNAs. For instance, the cyclin D1-pncRNA-D that is transcribed from the region D of the cyclin D1 promoter was found as low as two copies in a single cell without the treatment of ionizing radiation and induced up to four copies after the treatment. Amounts of other pncRNAs from the cyclin D1 promoter are supposed to be at a similar level based upon observations with quantitative PCR.14 Low abundance of the cyclin D1-pncRNA-D suggests that it functions as a regulatory agent instead of a structural component in living cells. Indeed, the cyclin D1-pncRNAs has a regulatory role in transcription of the cyclin D1 gene.14 The low abundance of the cyclin D1-pncRNAs limits their effective area or forces their function just around a milieu where their transcription occurs. This enforces the cyclin D1-pncRNAs to work as a cis-acting agent, but not as a trans-acting one. The benefit of the cis-acting mode of TLS is that the regulatory effect is limited to the promoter region in which the cyclin D1 pncRNAs are generated. Hence, the localization of the effect enhances the specificity of the cyclin D1 pncRNAs just towards the cyclin D1 expression.
The originally selected GGUG-RNA oligos (25 mers) have a relatively low affinity to TLS. TLS binds the GGUG-RNA oligos with a Kd of 250 nM.24 Gel-shift assays with the GGUG-RNA and the cyclin D1-pncRNAs A and B showed that TLS has similar level of the Kd to the pncRNAs.14 However, the estimated concentration of the cyclin D1 pncRNA is much lower than the level of the Kd. Indeed, a single HeLa cell has only four molecules of the cyclin D1 pncRNA-D even induced by IR. Given that the cell volume is 1pL, concentration of the cyclin D1 pncRNA-D is merely 0.0066 nM. Therefore, TLS needs to enhance its affinity to the pncRNAs in the cells to capture the cyclin D1 pncRNAs. A possible mechanism to enhance the TLS affinity to the cyclin D1-pncRNAs is allosteric effect by complex formation of TLS with other RNA-binding proteins. Protein-protein interactions during formation of the TLS complex may enhance the affinity of TLS to the pncRNAs and it is noteworthy that other TET family proteins, EWS and TAF15 also form distinctive complexes. Considering that EWS and TAF15 have a similar HAT inhibitory effect upon binding with RNAs,14 this complex formation could contribute to enhancement of affinities to each target ncRNA.
Protein modifying enzymes in the complex could modify TLS to induce higher affinity to the pncRNAs. Indeed, our mass spectrometric analysis showed that the protein arginine methyltransferase1 (pRMT1) that has been reported to have a role in transcription40 is a component of the TLS complex and pRMT1 methylates TLS and modifies its function (Kun Du and Kurokawa, unpublished data). Taken together, the formation of the TLS complex could be an essential process to enable TLS to play a pivotal role in mammalian transcription regulatory programs.
CONCLUSION AND FUTURE SCOPE
Long ncRNAs are extremely heterogeneous molecules transcribed from numerous sites of the eukaryotic genome. It seems unlikely that all of these long ncRNAs utilize a common mechanism to exert their biological functions. A potential clue to unravel the functions of long ncRNAs is to explore their binding partners, RNA-binding proteins like TLS.
I have presented here a detailed description of the structures and functions of TLS as a model system for understanding biological functions of long ncRNAs and corresponding RNA binding proteins in transcription regulatory programs. This discussion might assist analysis of the biological consequence of other classes of long ncRNAs. Characterisation of the function of RNA-binding proteins specific to their target long ncRNAs should include efficient strategies to explore their biological consequence in the human genome.
However, biological function of long ncRNAs still remains largely unanalyzed. The cyclin D1 pncRNAs and TLS is a model system for establishing a biological significance of long ncRNAs.41 Indeed, TLS has been discovered as a new causative gene of amyotrophic lateral sclerosis (ALS).42,43 TLS has been reported to have multiple mutants to cause ALS. ALS and frontotemporal degeneration (FTLD) are neurodegenerative diseases with clinical and pathological similarity.44 Remarkable discoveries of more than 20 mutations including H517Q and R521G in TLS as causatives of ALS, combined with the abnormal aggregation of the protein, have initiated a shifting paradigm for the underlying pathogenesis of multiple neurodegenerative diseases.42,43,45 This TLS association with ALS and other neurodegenerative diseases is redirecting research efforts toward understanding the role of RNA metabolism in neurodegenerative diseases and provide a biological significance of TLS and related ncRNAs.
ACKNOWLEDGEMENTS
The author thanks Ms. R. Tanji for preparation of the manuscript and Dr. C.K.Glass for critical discussion. This work was supported by Takeda Science Foundation, the Naito foundations, Astellas Foundation for Research on Metabolic Disorders Foundation and also by Grant-in-Aid for Scientific Research (B: nos22390057) and Grant-in-aid for "Support Project of Strategic Research Center in Private Universities" from the Ministry of Education, Culture, Sports, Science and Technology to Saitama Medical University Research Center for Genomic Medicine.
REFERENCES
- 1.
- Pawlicki JM, Steitz JA. Nuclear networking fashions premessenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2009;20:52–61. [PMC free article: PMC2821161] [PubMed: 20004579]
- 2.
- Collins LJ, Penny D. The RNA infrastructure: dark matter of the eukaryotic cell? Trends Genet. 2009;25:120–128. [PubMed: 19171405]
- 3.
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. [PubMed: 19158785]
- 4.
- ENCODE-consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. [PubMed: 15499007]
- 5.
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. [PubMed: 16141072]
- 6.
- Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. [PubMed: 15539566]
- 7.
- Willingham AT, Orth AP, Batalov S, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. [PubMed: 16141075]
- 8.
- Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. [PubMed: 9486653]
- 9.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of posttranscriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. [PubMed: 18197166]
- 10.
- Lanz RB, McKenna NJ, Onate SA, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. [PubMed: 10199399]
- 11.
- Feng J, Bi C, Clark BS, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. [PMC free article: PMC1475760] [PubMed: 16705037]
- 12.
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. [PMC free article: PMC2084369] [PubMed: 17604720]
- 13.
- Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. [PMC free article: PMC2743558] [PubMed: 18185590]
- 14.
- Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. [PMC free article: PMC2823488] [PubMed: 18509338]
- 15.
- Crozat A, Aman P, Mandahl N, et al. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. [PubMed: 8510758]
- 16.
- Kuroda M, Sok J, Webb L, et al. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. EMBO J. 2000;19:453–462. [PMC free article: PMC305582] [PubMed: 10654943]
- 17.
- Hicks GG, Singh N, Nashabi A, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–179. [PubMed: 10655065]
- 18.
- Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. [PubMed: 1522903]
- 19.
- Bertolotti A, Lutz Y, Heard DJ, et al. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article: PMC452240] [PubMed: 8890175]
- 20.
- Bertolotti A, Bell B, Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. [PubMed: 10637511]
- 21.
- Attwooll C, Tariq M, Harris M, et al. Identification of a novel fusion gene involving hTAFII68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene. 1999;18:7599–7601. [PubMed: 10602520]
- 22.
- Ladanyi M. The emerging molecular genetics of sarcoma translocations. Diagn Mol Pathol. 1995;4:162–173. [PubMed: 7493135]
- 23.
- Riggi N, Cironi L, Provero P, et al. Expression of the FUS-CHOP fusion protein in primary mesenchymal progenitor cells gives rise to a model of myxoid liposarcoma. Cancer Res. 2006;66:7016–7023. [PubMed: 16849546]
- 24.
- Lerga A, Hallier M, Delva L, et al. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem. 2001;276:6807–6816. [PubMed: 11098054]
- 25.
- Miyakawa Y, Matsushime H. Rapid downregulation of cyclin D1 mRNA and protein levels by ultraviolet irradiation in murine macrophage cells. Biochem Biophys Res Commun. 2001;284:71–76. [PubMed: 11374872]
- 26.
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature. 2009;458:223–227. [PMC free article: PMC2754849] [PubMed: 19182780]
- 27.
- Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. [PMC free article: PMC2704857] [PubMed: 19571010]
- 28.
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. [PMC free article: PMC2956184] [PubMed: 20673990]
- 29.
- Iko Y, Kodama TS, Kasai N, et al. Domain architectures and characterization of an RNA-binding protein, TLS. J Biol Chem. 2004;279:44834–44840. [PubMed: 15299008]
- 30.
- Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. [PubMed: 17510325]
- 31.
- Wang P, Yin S, Zhang Z, et al. Evidence for common short natural trans sense-antisense pairing between transcripts from protein coding genes. Genome Biol. 2008;9:R169. [PMC free article: PMC2646273] [PubMed: 19055728]
- 32.
- Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. [PubMed: 7958914]
- 33.
- Prasad DD, Ouchida M, Lee L, et al. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9:3717–3729. [PubMed: 7970732]
- 34.
- Loughlin FE, Mansfield RE, Vaz PM, et al. The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5′ splice site-like sequences. Proc Natl Acad Sci USA. 2009;106:5581–5586. [PMC free article: PMC2667063] [PubMed: 19304800]
- 35.
- Martianov I, Ramadass A, Serra Barros A, et al. Repression of the human dihydrofolate reductase gene by a noncoding interfering transcript. Nature. 2007;445:666–670. [PubMed: 17237763]
- 36.
- Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. [PMC free article: PMC3020079] [PubMed: 20393465]
- 37.
- Ayoubi TA, Van De Ven WJ. Regulation of gene expression by alternative promoters. FASEB J. 1996;10:453–460. [PubMed: 8647344]
- 38.
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. [PubMed: 15175754]
- 39.
- Impey S, McCorkle SR, Cha-Molstad H, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. [PubMed: 15620361]
- 40.
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300 and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. [PubMed: 15186775]
- 41.
- Kurokawa R, Rosenfeld MG, Glass CK. Transcriptional regulation through noncoding RNAs and epigenetic modifications. RNA Biol. 2009;6:233–236. [PubMed: 19411842]
- 42.
- Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. [PubMed: 19251627]
- 43.
- Vance C, Rogelj B, Hortobágyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. [PMC free article: PMC4516382] [PubMed: 19251628]
- 44.
- Van Langenhove T, van der Zee J, Sleegers K, et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–371. [PubMed: 20124201]
- 45.
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. [PMC free article: PMC3110083] [PubMed: 19303844]
- PROMOTER-ASSOCIATED LONG NONCODING RNAs REPRESS TRANSCRIPTION THROUGH AN RNA BIN...PROMOTER-ASSOCIATED LONG NONCODING RNAs REPRESS TRANSCRIPTION THROUGH AN RNA BINDING PROTEIN TLS - Madame Curie Bioscience Database
- The Role of Synaptotagmin and Synaptotagmin-Like Protein (Slp) in Regulated Exoc...The Role of Synaptotagmin and Synaptotagmin-Like Protein (Slp) in Regulated Exocytosis - Madame Curie Bioscience Database
- Circadian Organization in the Algal Flagellate Euglena - Madame Curie Bioscience...Circadian Organization in the Algal Flagellate Euglena - Madame Curie Bioscience Database
- Pro-Inflammatory Responses in Macrophages during Toxoplasma gondii Infection - M...Pro-Inflammatory Responses in Macrophages during Toxoplasma gondii Infection - Madame Curie Bioscience Database
- Periodic Selection and Ecological Diversity in Bacteria - Madame Curie Bioscienc...Periodic Selection and Ecological Diversity in Bacteria - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...

 Division of Gene Structure and Function, Research Center for Genomic Medicine, Saitama Medical University, Yamane, Hidaka-shi, Saitama-Ken, Japan
Division of Gene Structure and Function, Research Center for Genomic Medicine, Saitama Medical University, Yamane, Hidaka-shi, Saitama-Ken, Japan