NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The RNA infrastructure connects RNA-based functions. With transcription- to-translation processing forming the core of the network, we can visualise how RNA-based regulation, cleavage and modification are the backbone of cellular function. The key to interpreting the RNA-infrastructure is in understanding how core RNAs (tRNA, mRNA and rRNA) and other ncRNAs operate in a spatial-temporal manner, moving around the nucleus, cytoplasm and organelles during processing, or in response to environmental cues. This chapter summarises the concept of the RNA-infrastructure, and highlights examples of RNA-based networking within prokaryotes and eukaryotes. It describes how transcription-to-translation processes are tightly connected, and explores some similarities and differences between prokaryotic and eukaryotic RNA networking.
INTRODUCTION
RNA biology in both eukaryotes and prokaryotes exists in a spatiotemporal network of processes we call the RNA-infrastructure. In eukaryotes, there are numerous subtypes of noncoding (nc) RNA genes involved including rRNA, mRNA, tRNA, snRNA, snoRNAs, several classes of regulatory RNAs (RNAi) and many long ncRNAs. In prokaryotes, in addition to tRNAs, mRNAs and rRNAs, we can have small RNAs, CRISPRs and tmRNAs, and even viruses can contain small RNAs. ncRNAs are generally involved in the transcription-to-translation processes surrounding the conversion and regulation of information from DNA to protein, implicated in viral defence mechanisms, or are involved in gene regulation (e.g., RNA interference; RNAi). What we are only beginning to understand is how these processes are integrated, and how RNA plays a previously understated role in the overall regulation of the cell.
There are some key differences in cells that are differentiated (i.e., from multicellular eukaryotes), single celled eukaryotes and prokaryotes, but there are also striking similarities in how RNA processing and regulation works in different types of cells, giving us clues to their evolution. Although we are more familiar with RNA networks from eukaryotes, prokaryotic noncoding RNA research is using concepts developed from eukaryotic work to discover new RNA-based systems in bacteria and archaea. Although finding RNA genes is becoming a standard step in genomic investigations, it is now clear that discovering connections between these genes, and their associated proteins is just as important. Once we add in regulatory and epigenetic elements (such as methylation and histone modification) our regulatory networks can become very complex, but these complex networks have the ability to indicate linkages between cellular machineries not previously observed. The examples in this chapter will show how RNA-based processes within both prokaryotic and eukaryotic cells interact in networks in both a spatial and temporal manner.
RNAs PROCESSING OTHER RNAs
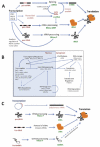
A good example in how RNA-processes are connected comes from examining the transcription-to-translation processes which form the core of the RNA-infrastructure (Fig. 1).1 The processing of the three core-RNAs in eukaryotes (mRNA, tRNA and rRNA) includes the RNA-based mechanisms of RNA cleavage and modification (Fig. 1A). In euaryotes these are: rRNA by RNase MRP and snoRNAs; tRNA by RNaseP; and mRNAs spliced by snRNAs within the spliceosome. We can then expand this idea to include spatial movement and regulation during RNA-processing (Fig. 1B). In prokaryotes we still have tRNAs and rRNAs being processed either directly by RNAs (e.g., RNase P of tRNA and tmRNA) or indirectly (where rRNAs are released by tRNA processing) (Fig. 1C).
Examining the connections between these processes in more detail we see networking between different mRNA machineries. For example, transcription by RNA Polymerase II (Pol II) and mRNA splicing in mammals are carried out in close proximity,2 and this coupling may protect the newly synthesised RNA from degradation3 before the termination of transcription.4,5 Some splicing may occur cotransciptionally and this significantly improves processing efficiency (reviewed in ref. 6). At the other end of the transcript, 3′-end cleavage and polyadenylation of mRNA can be promoted by splicing proteins (e.g., U2AF65 reviewed in ref. 7). It is clear that splicing (the processing of mRNA with snRNAs) connects to other mRNA-processes including RNA localisation, translational yield and mRNA decay.8 In another example, the Exon Junction Complex (EJC) is a set of proteins deposited on 5′ end of the exon during the second step of the splicing cycle, and remain bound to the spliced mRNA as it is exported to the cytoplasm.9 This complex interacts transiently with many factors that connect the mRNA to the downstream RNA processing network,10,11 as it is a major link between mRNA-splicing and mRNA export, as well as having a potential role in RNA degradation. The EJC appears to relay the previous location of introns,8 and thus detects incorrect splicing that introduces premature stop-codons. It has been shown that in mammals at least, spliceosomal proteins and especially those involved in exon-definition, remain associated with the pre-mRNA to be available for the splicing of the next introns, thus allowing for efficient splice site recognition for subsequent introns since splice site recognition only needs to be carried out once for a site.6 With splicing central to downstream RNA processing it is not surprising that many proteins are now seen as having roles in splicing as well as their own function (e.g., transcription or capping). However, it remains to be seen whether these proteins actually influence catalysis in the spliceosome, or are detected due to the close proximity of these RNA processing complexes.
Similarly, transcribed pre-tRNAs require processing before being able to function as amino acid transfer molecules for translation. Leader sequences at the 5′ and 3′ ends of the pre-tRNAs require cleaving, introns within the tRNA may need to be removed and in some cases a 3′ CCA tail needs to be added.12 In addition, certain nucleotides within the tRNA require modification by aminoacylation. The ribonucleoprotein RNase P is responsible for cleaving the 5′ leader sequence of pre-tRNAs in all cells, although the overall structure of this protein-RNA complex differs in eukaryotes, bacteria and archaea. In bacteria there is one small protein that plays diverse roles such as enhancing substrate binding, altering substrate recognition, stabilising RNA conformation, and aiding catalysis by discriminating between the substrate and product by binding to the 5′ leader sequence of the pre-tRNA.13,14 Eukaryotes have 9-10 proteins in the complex with a single RNA. Archaeal RNase P also has multiple proteins (five including the ribosomal protein L7Ae) which do show some homology to some of the eukaryotic RNase P proteins. The RNase P RNA from some representatives from each kingdom can be induced to perform weak catalysis without its accompanying proteins, but only with high salt and high cation conditions in vitro (summarised in ref. 15).
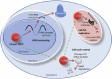
RNase P plays key networking roles in both the eukaryote's and prokaryote's RNA infrastructure, resulting in the cleavage of additional substrates and the repression of transcription (Fig. 2).16,17 In bacteria, as well as cleaving the 5′ leader sequence of tRNAs, it cleaves a similar leader sequence for tmRNA. tmRNA (transfer-messenger RNA) is a specialised tRNA molecule that together with the SmpB protein (small protein B) rescues stalled ribosomes in a process called trans-translation (reviewed in ref. 18).With a structure partly a tRNA molecule and partly an mRNA molecule,19 the tRNA part binds to the stalled ribosome, allows the translation to proceed along the mRNA part which encodes a distinctive degradation signal and a translation stop signal. When the mistranslated protein is released after the stop signal it is targeted for degradation. This process of trans-translation is conserved throughout bacteria and is also present in some mitochondria and chloroplasts.20,21 In prokaryotes other cleavage products by RNase P include some riboswitches and some viral RNAs as well as the 4.5S rRNA which is part of the Signal Recognition Particle involved in post-translational transport (reviewed in ref. 22).The eukaryotic counterpart of the 4.5S rRNA (7SL RNA) is also cleaved by RNase P (reviewed in ref. 13). In yeast the HRA1RNA and some C/D box snoRNAs are processed by RNase P although whether cleavage is the exact mechanism is yet to be completely determined. In humans MALAT1, another long ncRNA, is cleaved by RNase P. tRNAs in organelles within eukaryotes (in some species) either encode their own RNase P RNA (e.g., the yeast S. cerevisiae), use the nuclear counterpart (e.g., humans) or occasionally do without the RNA component altogether.23 Additionally, in the archaeans Nanoarchaeum equitans and Pyrobaculum aerophilum and the hyperthermophilic bacterium Aquifex aeolicus, there does not appear to be any RNase P-like RNA sequence in their genomes.24 In N. equitans the requirement for RNase P has been replaced by a strict placement of the promoter 26 nucleotides upstream of the mature tRNA sequence allowing transcription of leaderless tRNAs.25
There is a feedback affect of RNase P on its own polymerase RNA Pol III16 and the polymerase affecting rRNA transcription, RNA Pol I.17 Thus, RNase P in eukaryotes has a large effect on other aspects of RNA processing including splicing (U6 snRNA is transcribed by RNA Pol III), and RNA modification (by some yeast C/D box snoRNAs). It clearly plays a central role in the RNA infrastructure of both eukaryotes and prokaryotes and it is likely that other substrates and processing connections, especially in prokaryotes, are still to be uncovered.
RNase MRP is a ribonucleoprotein found only in eukaryotes, but closely related and sharing many of the same proteins with RNase P (for a review see ref. 13). It too has multiple roles (Fig. 3), processing the A3 site of rRNA in the nucleolus, a critical cell cycle control protein (Cyclin B2) in the cytoplasm, and the D-loop of mitochondrial DNA (MtDNA) in the mitochondria to generate RNA primers for Mt DNA replication. This is a good illustration of the spatial nature of RNA-Protein complexes that have different roles in different cellular compartments. RNase MRP is transcribed by RNA Pol III and thus is affected by the RNase P feedback on the polymerase.
Other aspects of rRNA processing in eukaryotes are linked to transcription and downstream rRNA maturation. Extensive modification of the pre-rRNAs includes methylation of 2′ hydroxyl groups of ribose (guided by C/D box snoRNAs [small nucleolar RNAs]) and pseudouridine formation from uracil (guided by H/ACA snoRNAs).26 In vertebrates, these snoRNAs are mostly found within introns, and are spliced out by snRNAs, illustrating the strong network of RNA biogenesis and splicing machineries. Yeast models (primarily in S. cerevisiae) indicate that RNA Pol I, elongation factors and rRNA sequence elements appear to optimize transcription elongation and co-ordinate interactions (including those with snoRNAs) with the pre-rRNA for correct rRNA processing and ribosome assembly.27 In addition, a protein complex of three transcription factors (the CURI complex comprising of Rap1, Fhl1 and Ifh1) links ribosomal protein production and pre-rRNA processing.28 Thus, rRNA processing also uses feedback from the later stages of processing to regulate transcription.
SPATIAL REGULATION OF EUKARYOTIC RNA PROCESSING
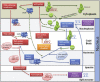
Spatial placement of both RNA and protein macromolecular components plays an important part in the regulation of RNA-processing. In eukaryotes, this is clearly demonstrated by how RNAs move through nuclear bodies (such as Cajal bodies, Gems and nucleoli) and for some of them, into cytoplasmic bodies such as P-bodies and RNA granules. As an example, Figure 4 illustrates the biogenesis of snRNAs and snoRNAs in humans. Typically there are different stages of RNA-processing taking place within different nuclear sub-compartments, but for the Sm-class-snRNAs, the processing moves to the cytoplasm, before the re-import of the snRNP-complexes back to the nucleus. In contrast, the Lsm-class snRNAs (U6 and U6atac snRNAs) in humans never leave the nucleus,29 although in yeast there may be some nuclear export and re-import of U6 snRNA.29 Cajal Bodies in particular appear to be important sub-nuclear compartments for RNPs since they are not only repositories for the biogenesis of RNPs. Mature snRNPs travel through Cajal Bodies, sometimes moving from one Cajal body to another suggesting that the Cajal Body is being used as a 'recycling center', enabling the re-assembly of the tri-snRNPs.30 In contrast, the assembly of C/D box snoRNPs appears to occur cotranscriptionally, but much of the intra-nuclear and intra-cellular trafficking of snoRNPs remain to be characterised.31
A feature of intra-cellular RNP trafficking is how some proteins assist these different RNPs in different manners. One such group of proteins linking snRNA and snoRNA biogenesis (Fig. 4) is the PHAX complex (consisting of PHAX, Cap Binding Protein (CBC), CRM1 and RanGTP) which in humans at least, transports snRNAs from the nucleus to the cytoplasm as well as transport of some snoRNAs (especially U3, U8, U13) around the nucleus to speckles, Cajal bodies and nucleoli.32 Although PHAX is a metazoan protein there has been a similar protein characterised in the protist Cryptosporidium parvum.32 Another important RNA-escorting macromolecule is the SMN protein complex, which is found in the nucleoplasm and nuclear bodies called Gems.33 The SMN complex scrutinizes cellular RNAs to ensure that Sm cores (of highly reactive RNA-binding Sm proteins) are only assembled on proper snRNAs,34 and the Gemin5 protein of this SMN complex can distinguish snRNAs from other cellular RNAs for snRNP biogenesis.34 The SMN complex also plays a role in other biogenesis pathways including those for hnRNPs and microRNPs.33 The above pathways for snRNP and snoRNP biogenesis have been largely characterised for mammalian and yeast systems, and although there is now some plant information,35 there is little known about how these RNPs complexes form in the many different groups of protists. As with plants we expect some different proteins to be involved and there will likely be different pathways.
After nuclear export some mRNAs are translated immediately, but many mRNAs are recruited to RNA granules in the cytoplasm until developmental or environmental cues signal their translation.36 Cytoplasmic RNA granules (reviewed in refs. 36,37)include Processing-bodies and Stress Granules as well as compartments found in germ cells (polar and germinal granules) and neurons (neuronal granules). Processing-bodies (P-bodies or GW bodies) are involved with post-transcriptional processes, including mRNA degradation, nonsense-mediated mRNA decay (NMD), translational repression and RNA-mediated gene silencing (reviewed in ref. 38). mRNA degradation is initiated by the deadenylation (shortening) of the 3′ polyA-tail followed by decapping.39 Stress Granules are a cytoplasmic RNA granule that typically forms during stress response (whereas P-bodies are present continuously).40 Stress Granules contain polyadenylated transcripts and are not degraded, making them available for rapid re-initiation after stress recovery,40 whereas mRNAs recruited to P-bodies are largely deadenylated.37 mRNAs within stress granules P-bodies and Stress Granules constantly exchange RNAs and proteins with the cytosol37 and mRNAs can move from one to the other. P-bodies have been investigated in yeasts, plants, trypanosomatids, insects and vertebrates (reviewed in ref. 37) and thus are likely to be important eukaryotic RNA-based cellular features. Evidence is suggesting that Stress Granules are the consequence, not the cause of the shut off of translation during stress, and the formation of critical macromolecules may be linked to the sequestering of key components (reviewed in ref. 37).
Although RNAs such as miRNAs, siRNAs and tRNAs typically act in the cytoplasm, there are some miRNAs that may re-enter the nucleus, possibly playing a role in modification or nuclear component assembly processes (summarised in refs. 41,42). tRNAs in particular show interesting nuclear-cytoplasmic dynamics. tRNA transcription and 5′ processing is typically in the nucleolus, however 3′ processing has been found mostly in the nuceloplasm; tRNA modification is usually in the cytoplasm but in some species, tRNA splicing takes place on the mitochondrial cytoplasmic surface (reviewed in ref. 42). Subsequently however, a retrograde pathway exists where the tRNAs are imported from the cytoplasm back into the nucleus43 but can then be re-exported to the cytoplasm in response to nutrient availability.44 Although major studies of tRNA cellular dynamics has been mainly in yeast, the retrograde process (moving cytoplasm to nucleus) at least appears conserved in vertebrates (summarised in ref. 42).
Spatiotemporal movement of RNAs or key components of RNA-based machineries, is not restricted to eukaryotes. The gram-negative bacteria Caulobacter crescentus is dimorphic in that it has a stalked form that adheres to surfaces (with a holdfast and stalk), and a swarmer form that is mobile with a flagellum. Often used as a model for bacterial cell cycle and cell differentiation studies, C. crescentus shows substructure localisation and temporal timing of trans-translation.45-47 tmRNA and its small protein SmpB are colocalised to a helix-like pattern in swarmer and predivisional cells but they are delocalised in stalked cells.46 However, the protein RNase R which interacts with the tmRNA is localised separately to another helix-like pattern that is out of phase with the tmRNA-SmpB pattern. Trans-translation requires that the individual tmRNA-SmpB molecules would have to disassociate from the helix-like structure in order to pass through the ribosome, and it is feasible that these structures facilitate the regulation of trans-translation.46 In a possible feedback mechanism, the tmRNA of C. crescentus is regulated in the cell cycle by temporally controlled transcription and translation.45 With trans-translation required for many functions across bacteria, including sporolation in Bacillus subtilis,48 symbiosis in Bradyrhizobium japonicum109, and pathogenecity in Salmonella enteria (summarised in ref. 47), the tight regulation of the RNA component is not unexpected. It is even possible that these helix-like structures seen in C. crescentus are analogous to the P-bodies we find in eukaryotes.
RNA REGULATION, CONNECTING COMPONENTS OF THE RNA-INFRASTRUCTURE
RNAi Networks
RNA regulation (including RNAi, riboswitches and RNA-editing), storage and degradation are linked to the processes discussed in the earlier sections. RNAi (RNA interference involving miRNAs, siRNAs and piRNAs are reviewed in refs. 49, 50). Although best known for roles in regulating mRNA levels, RNAi is also directly involved in many cellular processes including chromatin-mediated gene silencing and DNA re-arrangements. It is also not a matter of one target to one regulator. It has been shown51 that a single miRNA can directly or indirectly down-regulate the production of thousands of genes. Although RNAi as a mechanism appears general in eukaryotes, the timing and location of miRNA expression varies even between vertebrates due to changes in miRNA copy number, genomic context (either exclusively intergenic, or intronic and intergenic) or both.52 There can also be expression differences when there is conservation of the miRNA sequence.52
Other forms of RNA-based transcriptional regulation include regulation by RNase P which has a positive effect on Pol III promoter activity.16,53 RNase P associates with the chromatin of tRNA and 5S rRNA genes which contain the type-1 Pol III promoter sequences, but not with the U6 snRNA and 7SL-RNA that have type-3 Pol III promoter sequences. Transcription of these Pol III transcribed ncRNAs declines sharply in extracts depleted of active RNase P.16 RNase P may also have a role in the splicing-independent maturation of snoRNAs as recently demonstrated in yeast54 linking the production of tRNAs and rRNA.
A number of longer ncRNAs directly target transcription (reviewed in ref. 55) including SRA (a transcriptional co-activator for several steroid-hormone receptors), NRSE (Neuron-restrictive silencer element dsRNA), HSR1 (heat shock RNA-1) and 7SK RNA. This latter ncRNA is transcribed by Pol III and represses transcript elongation by Pol II (also reviewed in ref. 55). Another instance is a regulatory transcript from a minor promoter interfering with the expression of the main transcript.56 With the ongoing discovery57 of new ncRNAs in a wider range of eukaryotes we certainly expect the identification of other direct transcriptional regulators.
An interesting trend is the discovery that small regulatory RNAs can be derived from other ncRNAs. A number of studies have characterised miRNA-like RNAs derived from snoRNA-derived RNAs58 and RNAs derived from the Vault RNA.59 tRNA-derived RNAs are thought to be involved in translational repression.60,61 Studied in mammals, plants, fungi, and the protists Giardia and Tetrahymena, tRNAs are cleaved by members of the Ribonuclease A or T2 protein families in the anticodon loop forming 5′ and 3′ tRNA halves. Although how these different tRNA halves regulate translation inhibition is still very much under investigation, in mammals it has been shown that 5′ tRNA halves induces Stress Granule formation62 and that the original cleavage is enhanced by stress. In Trypanosomes, granules are formed that are distinct from Stress Granules.37,63 Other translational inhibition small RNAs include qiRNAs (QDE-2 associated RNAs) from the fungus Neurospora which inhibit protein translation during DNA damage response.64 With mass RNA sequencing still in its early days, these may represent only a fraction of the real amount of derived regulatory RNAs.
Transcription-initiation RNAsa are typically transcribed from a repeat motif called a 'spanion cluster'.65 These RNAs have a strong preference towards transcription initiation sites. Other small regulatory RNAs of note are the tiny RNAs of mammals, which are 17-18 bp in length and have a connection to splicing in that their 3′ ends map precisely to the splice donor site of internal exons.66 A subgroup of these splice-site RNAs are seen to be associated with highly expressed genes.66 How widespread these types of regulatory RNAs are remains to be investigated, but high throughput sequencing technology has enabled researchers to uncover these types and more.
RNAi is seen thus as a typical eukaryotic feature but there are some lineages that have lost their RNAi proteins but some still maintain some form of ncRNA-based regulation. The yeast S. cerevisiae does not have the 'standard' RNAi system since it lacks Dicer-like RNases, Argonaute or Piwi-like proteins, but it does have ncRNAs that act in the regulation of its genes.67 These ncRNAs including the 'cryptic unstable transripts' (CUTS), tend to stem from bidirectional transcription and may be passive by-products of transcriptional noise rather than any specific mechanism.67 In single celled protists, some species of Trypanosomes have lost their RNAi systems while others have retained them.68 Additionally, for some Trypanosomes the retention of associated viruses is necessary and it has been suggested that the loss of RNAi has facilitated viral retention. However genome plastidity is also a potential effect of this loss.68 Whether these RNAi-less Trypanosomes have evolved a different type of RNA-regulation system to compensate, is as yet unknown.
RNA Networks and Epigenetics
ncRNAs play a major role in epigenetics69-74 and include networks consisting of long ncRNAs (such as XIST and HOTAIR), and short ncRNAs69,74,75 such as miRNAs, siRNAs and piRNAs. miRNAs in particular have been shown to be important in RNA networks behind stem-cell self-renewal and differentiation (reviewed in ref. 76). In general, there are two types of stem-cell, tissue stem cells (which include somatic and germline cells which develop, maintain and repair tissues in developing and adult organisms), and embryonic stem cells (ES) which develop from an embryo to give rise to the foetus. In one example of a miRNA-epigenetic network, the expression of the miR-290-295 miRNA cluster (a group of miRNAs that share a 5′ proximal AAGUGC motif) increases during pre-implantation development and remains high in undifferentiated ES cells, but then decreases after ES cell differentiation.77 These miRNAs act as post-transcriptional regulators of retinoblastoma-like 2 (Rbl2) which in turn acts as a transcriptional repressor of DNA methyl transferases (DNMTs), Dnmt3a and Dnmt3b. DNMTs epigenetically silence OCT4, a key transcription factor of ES cell renewal and differentiation.77,78 Alternatively if Dicer is knocked out, miRNAs are depleted and the methylation of the Oct4 promoter is severely impaired during differentiation. Many other candidate targets of the AAGUGC seed-containing miRNAs have been identified as well as many indirectly regulated targets,77 but it remains to be seen how other aspects of self-renewal and differentiation are affected by the miR-290 cluster.
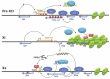
Networks involving multiple long ncRNAs (defined generally as having a length > 200 nt) are also known, with a classic example being the long ncRNA control of X-chromosome inactivation (reviewed in refs.74,79). In mammals the potential double dosage of gene expression from the X chromosome in XX females (when compared to XY males) is controlled by inactivating one of the X chromosomes. In mice there are two forms of X Chromosome inactivation (XCI)79 where XCI is imprinted in extra-embryonic tissues and the paternal X (Xp) is inactivated. Further along in development just before the embryo proper, the Xp is re-activated after which random XCI is initiated during early embryonic development. In humans XCI is randomly activated but it is not clear if the imprinted form is present.79 During random XCI in humans (Fig. 5), the long ncRNA Xist is repressed on the future active X chromosome Xa by another long ncRNA Tsix,80 and activated on the future inactive X chromosome by a third long ncRNA Jpx.81
This complex network of long ncRNA and methylation processes can be seen if we examine this system in more detail (Fig. 5). In mammals, in pre-XCI embryonic stem cells (ES) Tsix is transcribed at a higher level than Xist, and triggers H3-K4 dimethylation along both the Xist and Tsix genes. Xist becomes elevated when the major pluripotency factors Nanog, Oct3/4 and Sox2 dissociate from intron 1 within Xist initiating XCI. One of these proteins Oct4 is known to active Tsix and another RNA region Xite which is an activator of Tsix. Oct4 also acts as a repressor of Xist aiding in the control of the Xist:Tsix balance in XCI.82 During XCI different events occur upon the future active X chromosome (Xa) and the future inactive X chromosome (Xi).
On the future inactive X chromosome (Xi), Oct4 binding is lost so Tsix is downregulated and Xist is induced.82 A coating of Xist RNA forms a silent chromatin compartment where X-linked genes become 'localised' through binding to the Xist RNA.83 Xist RNA also recruits the chromatin repressive complex (CRC) to Xi. The inactive status is maintained by the polycomb repressive complex PCR2. In pre-XCI Tsix and another RNA, Repeat A (RepA), compete for PRC2 binding84 but upon XCI, RepA recruits PRC2 to Xist and PRC2 methylates Xist at H3K27 to upregulate Xist84. RepA also collaborates with the long ncRNA Jpx in an as yet unknown mechanism to transcriptionally activate Xist.81 On the future active X chromosome Xa, Oct4 is retained and maintains Tsix expression. Tsix, associated with methyltransferase Dnm3a, directs the methylation on the Xist promoter to repress Xist. With Xist repressed the Xa chromosome is active. Whether Jpx is still expressed or controlled by methylation is as yet unknown.
In addition, small ncRNAs may also be involved in X inactivation. Dicer-dependent XiRNAs can be produced from Xist and Tsix ncRNA duplexes80 and may direct methylation along the future Xi, and also direct methylation of CpG islands of the Xist promoter region in the Xa.85 Although XiRNAs are produced with Dicer, RNAi may not be directly involved in X chromosome inactivation, instead it could maintain the steady-state level of the Xist RNA.86 Recently other reports have contradicted the role of Dicer in XCI (summarised in ref. 87), and questions remain as where Xist:Tsix duplexes form, where Dicer could act, and the possible transport of XiRNAs between the cytoplasm and the nucleus. Although Xist and Tsix are long ncRNAs, they are both spliced and polyadenylated (reviewed in ref. 79), and it will be interesting to understand more in the future about how these mechanisms which have been demonstrated to have a regulatory affect, aid in XCI.
This type of Xist-linked random XCI is specific for eutherian mammals and is not seen in marsupials, flies or nematodes.88 Marsupial mammals lack Xist and only use imprinted XCI with the paternal X being inactivated.88 The presence of active and repressive histone modifications does suggest a related mechanism of dosage compensation and the presence of as yet unidentified ncRNAs is not ruled out.88 Insects possess a mechanism which instead of silencing one X copy in the female, instead promotes 2x transcriptional activity on the male X chromosome involving two long ncRNAs, rox1 and rox2.89 Rox1 and rox2 transcripts spread along the X chromosome recruiting the histone deacetylation protein complex, which then generates an open chromatin conformation to facilitate active transcription.90,91 Related mechanisms in other organisms is thus very likely.
RNA Regulation in Prokaryotes
Studies on RNA-based regulation tended in the past to concentrate on eukaryotes because the wealth of nonprotein-coding transcripts offered a large territory in which to find them. Prokaryotes have far less nonprotein coding territory, but in the last decade the importance of their RNA-based regulation has been found. There are some key networks of RNA regulation that have been studies mostly in bacteria but have some characterisation in archaea. This chapter will touch only briefly on these networks and will be expanded on in later chapters. Two areas of interest for prokaryotic RNA regulatory networks are how the CRISPR system interacts with cellular and viral RNAs to produce an effective viral defence mechanism, and secondly how environmental responses can be regulated using small RNA networks.
Clustered, regularly interspaced, short-palindromic repeats (CRISPRs) are an integral part of invader defence in bacteria and archaea (for reviews see refs. 92-94). Each CRISPR loci contains a short invader-derived spacer sequence (~40 nt) flanked by partial repeats (~30 nt), preceded by a leader sequence (150-550 nt). These units form clusters from one to over a hundred depending on species.93 Generally, upon invasion by viruses (also called in some species bacteriophages) or some plasmids, complexes of Cas proteins target and cleave short recognition motifs in the genome of the invader. These cleaved sequences are then incorporated into the leader sequence of the CRISPR locus. When the CRISPR locus is transcribed the CRISPR RNAs (crRNAs) then serve as templates to target invader sequences upon further infection (summarised in refs. 92,94). This prokaryotic immune mechanism allows for the acquisition of strain-specific immunity, a memory of past infections and has systems to ensure host integrity through self/nonself discrimination.94,95
Protein composition of the CRISPR system also varies with species. There are six core protein genes (cas1-cas6) having varied distribution throughout eukaryotes, with only cas1 nearly always being present92. These cas genes fall in clusters physically linked to the CRISPR loci. Another set of cmr genes are also clustered and some of them can be physically linked to the CRISPR loci.93 These proteins are involved in the maintenance of the CRISPR loci, the crRNA biogenesis and crRNA mediated resistance to invaders.92 The CRISPR/Cas system is found in most archaea with 70% of these carrying CRISPR/Cmr modules. In bacteria ~40% carry the CRISPR/Cas system and only 30% of these have CRISPR/Cmr modules. An important difference is that the CRISPR/Cas system specifically targets DNA,93,96 whereas the CRISPR/Cmr system targets RNA.93 Despite these two systems being mechanistically linked and sometimes physically linked it has been suggested that they have evolved interdependently.93
The closest parallel with the eukaryotes are the piRNAs that are processed from pre-piRNA cluster transcripts without the use of Dicer. These piRNAs increase in informational content by the insertion of transposon sequences. Unlike the CRISPR mechanism this increase is passive rather than active.93 There is no sequence homology between any of the CRISPR-associated proteins and proteins involved in RNAi in eukaryotes.92,93
CRISPR systems have an impact on plasmid stability and can affect antibiotic resistance. The CRISPR/Cas system has been demonstrated to target antibiotic resistance genes in Steptococcus thermophilus and cause plasmid loss.97 In another case, Enterococcus faecalis and Enterococcus faecium stains that had lost their CRISPR-cas systems, also lacked a plasmid mediated hospital-acquired antibiotic resistance98. It is possible in this case that modern drug therapy has shifted the evolution of these Enterococci with the loss of the CRISPR system, thus permitting the maintenance of an invading plasmid.98
Within bacteria there are other small RNAs responsible for negative and positive regulation of chromosomal genes and genes encoded on extra-chromosomal elements.99 Cis-antisense-RNAs are encoded in cis on the DNA strand opposite to their target share regions of complementarity (often 75 nt or more) to generally repress the target mRNA (reviewed in ref. 100). Many of these cis-RNAs reside in plasmids or other mobile elements, and are responsible for maintaining copy number, whereas others repress the translation of toxic proteins that remove cells that have lost the mobile element. Some chromosomally expressed cis-RNAs have been shown to repress the translation of proteins that are toxic at high levels (reviewed in ref. 100). Trans-encoded small RNAs share only limited complementarity with their targets and regulate expression by the pairing of the small RNA with their mRNA target. This group of small RNAs has been shown to interact in RNA networks that are critical for bacterial responses to stress and environmental cues, and regulate many outer membrane proteins.101
Most of these small RNAs repress translation (reviewed in refs. 99,102), but some can activate expression where pairing disrupts inhibitory secondary structure which would otherwise sequester the ribosome binding site (reviewed in ref. 100). The RNA chaperone protein Hfq binds preferentially to single-stranded RNAs interacting with both small RNAs and some target mRNAs by binding to a U-rich motif and an A-rich motif.103,104 This Sm-like protein is well conserved throughout bacteria,104 however, the precise mechanism by which it facilitates binding is not yet understood.103 The Hfq protein has been shown to localise close to the bacterial membrane and in Salmonella half of the small RNAs with known targets regulate the expression of outer membrane proteins (OMPs).104 From expression studies in Salmonella,105 about a fifth of all Salmonella genes are under the control of Hfq. The Hfq protein is not found in all bacteria (such as Bacillus subtilis and Staphlococcus aureus) and is not seen to be widespread in archaea, thus it remains to be seen if these organisms use another RNA chaperone protein to regulate large numbers of genes.99
The use of tRNAs in regulation is also not restricted to eukaryotes. The archaean, Nanoarchaeum equitans has a normal complement of tRNA genes (34 continuous and four with introns), but it also has six tRNAs genes that are coded in halves which become pieced together during processing (summarised in ref. 106). Other split tRNA genes have been found in Caldivirga maquilingensis where examples of tRNAs in three fragments have been discovered.106 It is possible that these split tRNAs could have resulted from genome re-arrangements at intron-containing tRNAs. In fact one split tRNA gene in N. equitans is located directly adjacent to a CRISPR element raising the idea that the insertion and excision of CRISPR elements could have played a role in the spreading of tRNA halves.106 How the splitting of these tRNAs fits into the regulation of tRNA processing, or are used in other cellular processes are not yet known.
CONCLUSION
Within the cell we see networks of RNAs involved in cleaving, modifiying or guiding the processing of other RNAs, and over time these networks have appeared to have exponentially expanded in complexity. Of course, the genes and their interactions were there all along and it is our knowledge that has grown. The modern RNA world is a tight-knit one where molecular machineries share proteins and sometimes RNAs. With the environment of the cells being so tightly packed (i.e., molecular crowding),107 this sharing permits connected machineries to respond together to cellular signals. Whether universal RNAs have remained because they are intrinsically better suited for this role remains an interesting evolutionary question.
It is clear that our view of the prokaryotic modern RNA world is changing as the emphasis on the biogenesis and processing of the core RNAs (mRNA, tRNA and rRNA) has shifted now to tackle RNA-based regulation. Although prokaryotic genomes do not carry anywhere near the same degree of noncoding transcripts we see in eukaryotes, they nevertheless carry a wealth of RNA genes with which we can already see networking. As more is uncovered other interesting studies will no doubt tackle the similarities and differences between prokaryotic and eukaryotic RNA networks. From here we can investigate whether these networks evolved separately or have stemmed from an 'RNP-world' ancestor.108 What is very clear is that in modern cellular networks, the role of RNA genes and how they connect in cellular networks makes them as important as their protein-coding cousins.
ACKNOWLEDGEMENTS
Many thanks go to Prof. David Penny, Prof. Mike Hendy and Prof. Pete Lockhart for stimulating discussions and support.
REFERENCES
- 1.
- Collins LJ, Penny D. RNA-infrastructure: dark matter of the eukaryotic cell? Trends in Genetics. 2009;25(3):120–128. [PubMed: 19171405]
- 2.
- Kornblihtt AR, de la Mata M, Fededa JP, et al. Multiple links between transcription and splicing. RNA. 2004;10(10):1489–1498. [PMC free article: PMC1370635] [PubMed: 15383674]
- 3.
- Hicks MJ, Yang CR, Kotlajich MV, et al. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4(6):e147. [PMC free article: PMC1450099] [PubMed: 16640457]
- 4.
- Gornemann J, Kotovic KM, Hujer K, et al. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19(1):53–63. [PubMed: 15989964]
- 5.
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA: 5′ss base pairing in yeast. Mol Cell. 2005;19(1):65–75. [PubMed: 15989965]
- 6.
- Crabb TL, Lam BJ, Hertel KJ. Retention of spliceosomal components along ligated exons ensures efficient removal of multiple introns. Rna. 2010;16(9):1786–1796. [PMC free article: PMC2924538] [PubMed: 20610656]
- 7.
- Millevoi S, Loulergue C, Dettwiler S, et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3' end processing machineries. The EMBO Journal. 2006;25(20):4854–4864. [PMC free article: PMC1618107] [PubMed: 17024186]
- 8.
- Chang YF, Imam JS, Wilkinson MF. Annu Rev Biochem. 2007. The nonsense-mediated decay RNA surveillance pathway. [PubMed: 17352659]
- 9.
- Nojima T, Hirose T, Kimura H, et al. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J Biol Chem. 2007 [PubMed: 17363367]
- 10.
- Shibuya T, Tange TO, Stroupe ME, et al. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA. 2006;12(3):360–374. [PMC free article: PMC1383576] [PubMed: 16495234]
- 11.
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Current Opinion in Cell Biology. 2004;16(3):279–284. [PubMed: 15145352]
- 12.
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 24(17):1832–1860. [PMC free article: PMC2932967] [PubMed: 20810645]
- 13.
- Esakova O, Krasilnikov AS. Of proteins and RNA: the RNase P/MRP family. Rna. 2010;16(9):1725–1747. [PMC free article: PMC2924533] [PubMed: 20627997]
- 14.
- Lai LB, Vioque A, Kirsebom LA, et al. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010;584(2):287–296. [PMC free article: PMC2799185] [PubMed: 19931535]
- 15.
- Pulukkunat DK, Gopalan V. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008;36(12):4172–4180. [PMC free article: PMC2475606] [PubMed: 18558617]
- 16.
- Reiner R, Ben-Asouli Y, Krilovetzky I, et al. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes and Development. 2006;20(12):1621–1635. [PMC free article: PMC1482482] [PubMed: 16778078]
- 17.
- Reiner R, Krasnov-Yoeli N, Dehtiar Y, et al. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS One. 2008;3(12):e4072. [PMC free article: PMC2605565] [PubMed: 19115013]
- 18.
- Hayes CS, Keiler KC. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Lett. 584(2):413–419. [PMC free article: PMC2794900] [PubMed: 19914241]
- 19.
- Wower J, Wower IK, Zwieb C. Making the jump: new insights into the mechanism of trans-translation. J Biol. 2008;7(5):17. [PMC free article: PMC2447533] [PubMed: 18598387]
- 20.
- Une M, Kurita D, Muto A, et al. Trans-translation by tmRNA and SmpB. Nucleic Acids Symp Ser (Oxf). 2009;53:305–306. [PubMed: 19749382]
- 21.
- Hayes CS, Keiler KC. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Lett. 2010;584(2):413–419. [PMC free article: PMC2794900] [PubMed: 19914241]
- 22.
- Marvin MC, Engelke DR. Broadening the mission of an RNA enzyme. J Cell Biochem. 2009;108(6):1244–1251. [PMC free article: PMC2800852] [PubMed: 19844921]
- 23.
- Holzmann J, Frank P, Loffler E, et al. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135(3):462–474. [PubMed: 18984158]
- 24.
- Li Y, Altman S. In search of RNase P RNA from microbial genomes. Rna. 2004;10(10):1533–1540. [PMC free article: PMC1370640] [PubMed: 15337843]
- 25.
- Randau L, Schroder I, Soll D. Life without RNase P. Nature. 2008;453(7191):120–123. [PubMed: 18451863]
- 26.
- Moss T, Langlois F, GagnonKugler T, et al. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cellular and Molecular Life Sciences. 2007;64(1):29–49. [PubMed: 17171232]
- 27.
- Schneider DA, Michel A, Sikes ML, et al. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell. 2007;26(2):217–229. [PMC free article: PMC1927085] [PubMed: 17466624]
- 28.
- Rudra D, Mallick J, Zhao Y, et al. Potential interface between ribosomal protein production and pre-rRNA processing. Mol Cell Biol. 2007;27(13):4815–4824. [PMC free article: PMC1951472] [PubMed: 17452446]
- 29.
- Hopper AK. Cellular dynamics of small RNAs. Crit Rev Biochem Mol Biol. 2006;41(1):3–19. [PubMed: 16455518]
- 30.
- Stanek D, Pridalova-Hnilicova J, Novotny I, et al. spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through cajal bodies. Mol Biol Cell. 2008;19(6):2534–2543. [PMC free article: PMC2397305] [PubMed: 18367544]
- 31.
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Genet. 2007;8(3):209–220. [PubMed: 17318225]
- 32.
- Mourao A, Varrot A, Mackereth CD, et al. Structure and RNA recognition by the snRNA and snoRNA transport factor PHAX. Rna. 16(6):1205–1216. [PMC free article: PMC2874172] [PubMed: 20430857]
- 33.
- Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Experimental Cell Research. 2004;296(1):51–56. [PubMed: 15120993]
- 34.
- Battle DJ, Lau CK, Wan L, et al. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell. 2006;23(2):273–279. [PubMed: 16857593]
- 35.
- Rodor J, Letelier I, Holuigue L, et al. Nucleolar RNPs: from genes to functional snoRNAs in plants. Biochem Soc Trans. 2010;38(2):672–676. [PubMed: 20298241]
- 36.
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172(6):803–808. [PMC free article: PMC2063724] [PubMed: 16520386]
- 37.
- Thomas MG, Loschi M, Desbats MA, et al. RNA granules: The good, the bad and the ugly. Cell Signal. 2010;23(2):324–334. [PMC free article: PMC3001194] [PubMed: 20813183]
- 38.
- Jakymiw A, Pauley KM, Li S, et al. The role of GW/P-bodies in RNA processing and silencing. J Cell Sci. 2007;120(Pt 8):1317–1323. [PubMed: 17401112]
- 39.
- Brengues M, Parker R. Mol Biol Cell. 2007. Accumulation of Polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-Bodies in Saccharomyces cerevisiae. [PMC free article: PMC1924816] [PubMed: 17475768]
- 40.
- Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169(6):871–884. [PMC free article: PMC2171635] [PubMed: 15967811]
- 41.
- Politz JC, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. Rna. 2009;15(9):1705–1715. [PMC free article: PMC2743059] [PubMed: 19628621]
- 42.
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes and Development. 2010;24(17):1832–1860. [PMC free article: PMC2932967] [PubMed: 20810645]
- 43.
- Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102(32):11290–11295. [PMC free article: PMC1183567] [PubMed: 16040803]
- 44.
- Whitney ML, Hurto RL, Shaheen HH, et al. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell. 2007;18(7):2678–2686. [PMC free article: PMC1924813] [PubMed: 17475781]
- 45.
- Keiler KC, Shapiro L. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J Bacteriol. 2003;185(2):573–580. [PMC free article: PMC145339] [PubMed: 12511504]
- 46.
- Russell JH, Keiler KC. Subcellular localization of a bacterial regulatory RNA. Proc Natl Acad Sci U S A. 2009;106(38):16405–16409. [PMC free article: PMC2752561] [PubMed: 19805312]
- 47.
- Cheng L, Keiler KC. Correct timing of dnaA transcription and initiation of DNA replication requires trans translation. J Bacteriol. 2009;191(13):4268–4275. [PMC free article: PMC2698507] [PubMed: 19429626]
- 48.
- Abe T, Sakaki K, Fujihara A, et al. tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Mol Microbiol. 2008;69(6):1491–1498. [PubMed: 18673456]
- 49.
- Munroe SH, Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cellular and Molecular Life Sciences. 2006;63(18):2102–2118. [PubMed: 16847578]
- 50.
- Hartig JV, Tomari Y, Forstemann K. piRNAs—the ancient hunters of genome invaders. Genes and Development. 2007;21(14):1707–1713. [PubMed: 17639076]
- 51.
- Selbach M, Schwanhausser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. [PubMed: 18668040]
- 52.
- Ason B, Darnell DK, Wittbrodt B, et al. Differences in vertebrate microRNA expression. Proc Natl Acad Sci U S A. 2006;103(39):14385–14389. [PMC free article: PMC1599972] [PubMed: 16983084]
- 53.
- Jarrous N, Reiner R. Human RNase P: a tRNA-processing enzyme and transcription factor. Nucleic Acids Res. 2007;35(11):3519–3524. [PMC free article: PMC1920233] [PubMed: 17483522]
- 54.
- Coughlin DJ, Pleiss JA, Walker SC, et al. Genome-wide search for yeast RNase P substrates reveals role in maturation of intron-encoded box C/D small nucleolar RNAs. Proc Natl Acad Sci U S A. 2008;105(34):12218–12223. [PMC free article: PMC2527892] [PubMed: 18713869]
- 55.
- Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7(8):612–616. [PubMed: 16723972]
- 56.
- Martianov I, Ramadass A, Serra Barros A, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670. [PubMed: 17237763]
- 57.
- Washietl S, Pedersen JS, Korbel JO, et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2007;17(6):852–864. [PMC free article: PMC1891344] [PubMed: 17568003]
- 58.
- Taft RJ, Glazov EA, Lassmann T, et al. Small RNAs derived from snoRNAs. Rna. 2009;15(7):1233–1240. [PMC free article: PMC2704076] [PubMed: 19474147]
- 59.
- Persson H, Kvist A, Vallon-Christersson J, et al. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11(10):1268–1271. [PubMed: 19749744]
- 60.
- Cole C, Sobala A, Lu C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. Rna. 2009;15(12):2147–2160. [PMC free article: PMC2779667] [PubMed: 19850906]
- 61.
- Pederson T. Regulatory RNAs derived from transfer RNA? Rna. 2010;16(10):1865–1869. [PMC free article: PMC2941095] [PubMed: 20719919]
- 62.
- Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959–10968. [PMC free article: PMC2856301] [PubMed: 20129916]
- 63.
- Garcia-Silva MR, Frugier M, Tosar JP, et al. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol. 2010;171(2):64–73. [PubMed: 20156490]
- 64.
- Lee HC, Chang SS, Choudhary S, et al. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459(7244):274–277. [PMC free article: PMC2859615] [PubMed: 19444217]
- 65.
- Cserzo M, Turu G, Varnai P, et al. Relating underrepresented genomic DNA patterns and tiRNAs: the rule behind the observation and beyond. Biol Direct. 2010;5:56. [PMC free article: PMC3583238] [PubMed: 20860791]
- 66.
- Taft RJ, Simons C, Nahkuri S, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol. 2010;17(8):1030–1034. [PubMed: 20622877]
- 67.
- Harrison BR, Yazgan O, Krebs JE. Life without RNAi: noncoding RNAs and their functions in Saccharomyces cerevisiae. Biochem Cell Biol. 2009;87(5):767–779. [PubMed: 19898526]
- 68.
- Lye LF, Owens K, Shi H, et al. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6(10):e1001161. [PMC free article: PMC2965760] [PubMed: 21060810]
- 69.
- Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol Cell. 2008;100(3):149–166. [PubMed: 18271756]
- 70.
- Whitehead J, Pandey GK, Kanduri C. Biochim Biophys Acta. 2008. Regulation of the mammalian epigenome by long noncoding RNAs. [PubMed: 19015002]
- 71.
- Kurokawa R, Rosenfeld MG, Glass CK. Transcriptional regulation through noncoding RNAs and epigenetic modifications. RNA Biol. 2009;6(3) [PubMed: 19411842]
- 72.
- Verdel A, Vavasseur A, Le Gorrec M, et al. Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol. 2009;53(2-3):245–257. [PubMed: 19412884]
- 73.
- Costanzo M, Baryshnikova A, Bellay J, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. [PMC free article: PMC5600254] [PubMed: 20093466]
- 74.
- Collins LJ, Chen XS, Schonfeld B. The Epigenetics of Non-coding RNA. In: Tollefsbol T, ed. Handbook of Epigenetics. Oxford: Academic Press. 2010:49–61.
- 75.
- Collins LJ, Chen XS. Ancestral RNA: The RNA biology of the eukaryotic ancestor. RNA Biology. 2009. Invited review, In press. [PubMed: 19713749]
- 76.
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10(2):116–125. [PMC free article: PMC4118578] [PubMed: 19165214]
- 77.
- Sinkkonen L, Hugenschmidt T, Berninger P, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15(3):259–267. [PubMed: 18311153]
- 78.
- Benetti R, Gonzalo S, Jaco I, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15(3):268–279. [PMC free article: PMC2990406] [PubMed: 18311151]
- 79.
- Barakat TS, Jonkers I, Monkhorst K, et al. X-changing information on X inactivation. Exp Cell Res. 2010;316(5):679–687. [PubMed: 20083102]
- 80.
- Navarro P, Pichard S, Ciaudo C, et al. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19(12):1474–1484. [PMC free article: PMC1151664] [PubMed: 15964997]
- 81.
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. [PMC free article: PMC2994261] [PubMed: 21029862]
- 82.
- Donohoe ME, Silva SS, Pinter SF, et al. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460(7251):128–132. [PMC free article: PMC3057664] [PubMed: 19536159]
- 83.
- Chaumeil J, Le Baccon P, Wutz A, et al. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20(16):2223–2237. [PMC free article: PMC1553206] [PubMed: 16912274]
- 84.
- Zhao J, Sun BK, Erwin JA, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. [PMC free article: PMC2748911] [PubMed: 18974356]
- 85.
- Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320(5881):1336–1341. [PMC free article: PMC2584363] [PubMed: 18535243]
- 86.
- Kanellopoulou C, Muljo SA, Dimitrov SD, et al. X chromosome inactivation in the absence of Dicer. Proc Natl Acad Sci U S A. 2009;106(4):1122–1127. [PMC free article: PMC2633553] [PubMed: 19164542]
- 87.
- Kota SK. RNAi in X inactivation: contrasting findings on the role of interference. Bioessays. 2009;31(12):1280–1283. [PubMed: 19921656]
- 88.
- Rens W, Wallduck MS, Lovell FL, et al. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci U S A. 2010;107(41):17657–17662. [PMC free article: PMC2955130] [PubMed: 20861449]
- 89.
- Franke A, Baker BS. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 1999;4(1):117–122. [PubMed: 10445033]
- 90.
- Brown JW, Simpson CG, Thow G, et al. Splicing signals and factors in plant intron removal. Biochem Soc Trans. 2002;30(2):146–149. [PubMed: 12023842]
- 91.
- Oh H, Park Y, Kuroda MI. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 2003;17(11):1334–1339. [PMC free article: PMC196065] [PubMed: 12782651]
- 92.
- Hale CR, Zhao P, Olson S, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–956. [PMC free article: PMC2951265] [PubMed: 19945378]
- 93.
- Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2010 [PubMed: 20863886]
- 94.
- Vale PF, Little TJ. CRISPR-mediated phage resistance and the ghost of coevolution past. Proc Biol Sci. 2010;277(1691):2097–2103. [PMC free article: PMC2880148] [PubMed: 20236977]
- 95.
- Marraffini LA, Sontheimer EJ. Self versus nonself discrimination during CRISPR RNA-directed immunity. Nature. 2010;463(7280):568–571. [PMC free article: PMC2813891] [PubMed: 20072129]
- 96.
- Marraffini LA, Sontheimer EJ. Invasive DNA, chopped and in the CRISPR. Structure. 2009;17(6):786–788. [PMC free article: PMC2711432] [PubMed: 19523896]
- 97.
- Garneau JE, Dupuis ME, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. [PubMed: 21048762]
- 98.
- Palmer KL, Gilmore MS. multidrug-resistant enterococci lack CRISPR-cas. MBio. 2010;1(4) [PMC free article: PMC2975353] [PubMed: 21060735]
- 99.
- Gottesman S, Storz G. Cold Spring Harb Perspect Biol. 2010. bacterial small RNA Regulators: versatile roles and rapidly evolving variations. [PMC free article: PMC3225950] [PubMed: 20980440]
- 100.
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. [PMC free article: PMC3132550] [PubMed: 19239884]
- 101.
- Vogel J, Papenfort K. Small noncoding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006;9(6):605–611. [PubMed: 17055775]
- 102.
- Gottesman S. Small RNAs shed some light. Cell. 2004;118(1):1–2. [PubMed: 15242637]
- 103.
- Soper T, Mandin P, Majdalani N, et al. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107(21):9602–9607. [PMC free article: PMC2906882] [PubMed: 20457943]
- 104.
- Diestra E, Cayrol B, Arluison V, et al. Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS One. 2009;4(12):e8301. [PMC free article: PMC2789413] [PubMed: 20011543]
- 105.
- Sittka A, Lucchini S, Papenfort K, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global posttranscriptional regulator, Hfq. PLoS Genet. 2008;4(8):e1000163. [PMC free article: PMC2515195] [PubMed: 18725932]
- 106.
- Heinemann IU, Soll D, Randau L. Transfer RNA processing in archaea: unusual pathways and enzymes. FEBS Lett. 2010;584(2):303–309. [PMC free article: PMC2796832] [PubMed: 19878676]
- 107.
- Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26(10):597–604. [PubMed: 11590012]
- 108.
- Collins LJ, Kurland CG, Biggs P, et al. The modern RNP world of eukaryotes. J Hered. 2009;100(5):597–604. [PubMed: 19643816]
- 109.
- Ebeling S, Kundig C, Hennecke H. Discovery of a rhizobial RNA that is essential for symbiotic root nodule development. J. Bacteriol. 1991;173(20):6373–6382. [PMC free article: PMC208969] [PubMed: 1717438]
Footnotes
- a
- THE RNA INFRASTRUCTURE: An Introduction to ncRNA Networks - Madame Curie Bioscie...THE RNA INFRASTRUCTURE: An Introduction to ncRNA Networks - Madame Curie Bioscience Database
- The Tn4371 ICE Family of Bacterial Mobile Genetic Elements - Madame Curie Biosci...The Tn4371 ICE Family of Bacterial Mobile Genetic Elements - Madame Curie Bioscience Database
- Idiopathic Parkinson's Disease: Staging an α-Synucleinopathy with a Predictable ...Idiopathic Parkinson's Disease: Staging an α-Synucleinopathy with a Predictable Pathoanatomy - Madame Curie Bioscience Database
- The Role of Heat Shock Proteins during Neurodegeneration in Alzheimer's, Parkins...The Role of Heat Shock Proteins during Neurodegeneration in Alzheimer's, Parkinson's and Huntington's Disease - Madame Curie Bioscience Database
- Abl Family Kinases in Mammalian Development - Madame Curie Bioscience DatabaseAbl Family Kinases in Mammalian Development - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...